Abstract
Usher syndrome type 1 (USH1) patients suffer from sensorineuronal deafness, vestibular dysfunction, and visual impairment. Several genetic loci have been linked to USH1, and four of the relevant genes have been identified. They encode the unconventional myosin VIIa, the PDZ-domain protein harmonin, and the putative adhesion receptors cadherin 23 (CDH23) and protocadherin 15 (PCDH15). We show here that CDH23 and harmonin form a protein complex. Two PDZ domains in harmonin interact with two complementary binding surfaces in the CDH23 cytoplasmic domain. One of the binding surfaces is disrupted by sequences encoded by an alternatively spliced CDH23 exon that is expressed in the ear, but not the retina. In the ear, CDH23 and harmonin are expressed in the stereocilia of hair cells, and in the retina within the photoreceptor cell layer. Because CDH23-deficient mice have splayed stereocilia, our data suggest that CDH23 and harmonin are part of a transmembrane complex that connects stereocilia into a bundle. Defects in the formation of this complex are predicted to disrupt stereocilia bundles and cause deafness in USH1 patients.
Approximately 1 in 800 children is born with hearing impairment, and large parts of the aging population are afflicted by age-related hearing loss. Single gene defects are responsible for ≈50% of the hearing defects in newborns, and genetic predisposition is an important determinant in some forms of age-related hearing loss. Approximately 50 deafness genes have been identified, but the mechanisms by which these genes exert their functions are largely unclear (1–3).
Many forms of deafness involve defects in hair cells, the mechanosensors for sound waves in the cochlea of the inner ear. Each hair cell has an apical bundle of 30–300 stereocilia that contain mechanotransduction channels. The channels change their open probability during stereocilia deflection. Individual stereocilia of a hair cell do not act alone. They are connected through linkage systems such as tip links, side links, ankle links, and top connectors into a bundle that moves as a whole during sound stimulation (Fig. 1a) (1, 2). Antibodies that recognize extracellular linkages in stereocilia have been raised (4, 5), but the molecules that form the linkages are not known. The tip link may serve as a gating spring that regulates opening and closing of the transduction channel (6, 7). Side and ankle links may mediate adhesion between stereocilia to connect them and to coordinate their movement (8, 9).
Fig 1.
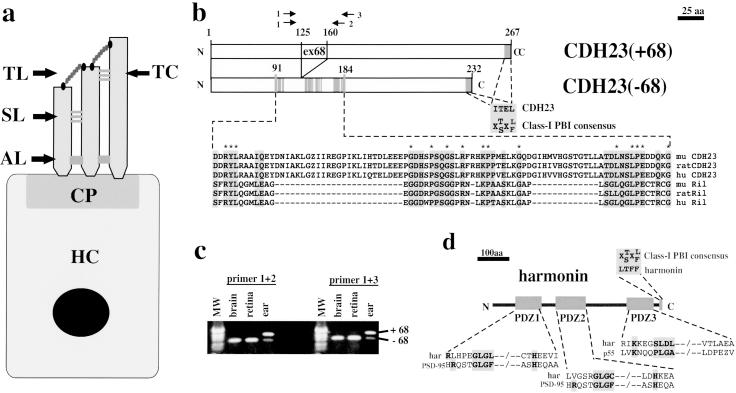
Hair cell diagram, protein–protein interaction domains in CDH23 and harmonin, and alternative splicing of CDH23. (a) A diagram of a hair cell (HC) is shown. The stereocilia are anchored at the cuticular plate (CP) to the hair cell body. They are connected to each other through extracellular linkages such as the tip link (TL), top connectors (TC), side links (SL), and ankle links (AL). (b) The cytoplasmic domain of CDH23 contains two putative PBIs. The C terminus of CDH23 fits the consensus sequence for class-I PBIs (30). An internal domain in the CDH23(−68) cytoplasmic domain shows homology to a PBI in Ril (35). In the CDH23(+68) isoform, 35 aa that are encoded by the alternatively spliced exon 68 are inserted into the Ril homology region. Gray boxes outline amino acids that are identical/conserved between CDH23 and Ril. Identical amino acids are indicated by an asterisk (*). (c) RT-PCR was conducted with RNA from adult mice with two sets of CDH23-specific primers (see b). The identity of the fragments was confirmed by DNA sequencing. The CDH23 isoform containing exon 68 was expressed in the inner ear but not in the brain or retina. MW, molecular weight marker. (d) Harmonin contains three PDZ domains (19, 20) and a class-I PBI. PDZ1 and PDZ2 have similarity to PDZ domains in PSD-95 (43, 44). PDZ3 shows similarity to a PDZ domain in p55 (38).
By positional cloning and through the analysis of genetically modified mice several deafness genes have been identified that affect the integrity of the stereocilia bundle. Promising candidate molecules to connect stereocilia are CDH23 and PCDH15, members of the cadherin superfamily of cell adhesion molecules (10). Mutations in the CDH23 and PCDH15 gene cause USH1D and USH1F, respectively, as well as other forms of deafness (11–14). Stereocilia are splayed in Cdh23- and Pcdh15-deficient waltzer and Ames waltzer mice, suggesting that both molecules have specialized adhesive functions important to maintain the integrity of the stereocilia bundle (14–17). PCDH15 is expressed in hair cells (14), but the expression pattern of CDH23 has not been reported. Likewise, downstream effectors of these cadherins are not known. The PCDH15 cytoplasmic domain contains proline-rich regions that may mediate protein–protein interactions (14). No protein interaction surfaces or catalytic domains have been described in the CDH23 cytoplasmic domain. Candidate molecules to mediate PCDH15 and CDH23 functions are myosin VIIa, and the PDZ domain protein harmonin. Both proteins are expressed in hair cells, and mutations in their genes lead to USH1B and USH1C, respectively (18–20). Furthermore, myosin VIIa-deficient shaker-1 mice have splayed stereocilia (21). Myosin VIIa is expressed at adherence junctions in several tissues, where it binds by means of vezatin and catenins to classical cadherins. In hair cells, myosin VIIa and vezatin colocalize at ankle links (22). Because classical cadherins and catenins are not expressed in stereocilia, other receptors such as CDH23 and PCDH15 may recruit the vezatin/moysin VIIa complex in hair cells.
As a first step toward understanding the function of CDH23, we have determined its expression pattern in the inner ear and retina and searched for proteins that interact with its cytoplasmic domain. We show that CDH23 is expressed in hair cell stereocilia and the photoreceptor layer in the retina, and that it binds to harmonin. Complex formation between CDH23 and harmonin is regulated by an alternatively spliced exon in the CDH23 gene that is specifically expressed in the ear. Because stereocilia are splayed in Cdh23-deficient mice (17), our data suggest that CDH23 and harmonin cooperate to assemble a transmembrane complex that connects stereocilia.
Materials and Methods
Immunohistochemistry.
Rabbit antisera were raised against GST-CDH23 expressed in and purified from Escherichia coli (23). Inner ear RNA was reverse transcribed and the CDH23 cytoplasmic domain amplified by PCR (5′-GGGGGATCCAACT GGTACTACAGGACCA-3′; 5′-CGCGAATTCTCACAGC TCCGTGATTTCCAG AG-3′). The fragments were cloned into pGEX-2TK (Amersham Pharmacia). The antisera were purified against a fusion protein between the CDH23 cytodomain and the maltose-binding protein expressed from pMAL-c2 (New England Biolabs) (23). Harmonin antisera were raised against a mixture of two peptides (NH2-MDRKVAREFRHKVDFC-COOH, and NH2-CRSRKLKEVR LDRLHP-COOH). Immunization was performed at Eurogentec (Brussels). Antisera were purified against the peptides used as an immunogen (24). Other tools were as follows: anti-IL-2 antibodies (Santa Cruz Biotechnology; Upstate Biotechnology, Lake Placid, NY); secondary antibodies coupled to horseradish peroxidase (Amersham Pharmacia); rhodamine conjugated anti-rabbit antibodies (Jackson ImmunoResearch); FITC-phalloidin (Sigma). Immunohistochemistry was performed as described (25).
Expression Analysis of CDH23 Splice Variants.
RNA was prepared by using Trizol (Life Technologies, Grand Island, NY) and RT-PCR was performed with the following primers (1: 5′-GACAACATCGCCAAGCTG-3′; 2: 5′-ATGAACCATGTG GATTCCATC-3′; 3: 5′-GCAAGCTGTTGAGATCAGTGG-3′).
Yeast Two-Hybrid Assays.
CDH23 constructs were expressed as fusions with the Gal4 DNA-binding domain from pGBKT7 (CLONTECH). Harmonin constructs were expressed from pGBKT7, or as fusions to the Gal4 activation domain by using pGADT7 (CLONTECH). DNA fragments were amplified by PCR and cloned into NdeI/BamHI sites (CDH23) or SmaI/BamHI sites (harmonin). Protein–protein interactions were measured as yeast growth under selective pressure (26).
GST Pull-Down Experiments.
Harmonin and PDZ domains were expressed from pGADT7 by in vitro translation with the TNT Coupled Reticulocyte Lysate System (Promega). GST-CDH23 fusions were expressed in E. coli. GST pull-down experiments were performed as described (23). For quantification, harmonin bands were cut out from SDS-polyacrylamide gels and radioactivity measured by scintillation counting. Input amounts of GST-CDH23 were determined by densitometrical scanning of Coomassie-stained SDS-polyacrylamide gels. The amount of radioactivity was normalized against the amount of GST-CDH23 fusion protein.
Immunoprecipitations.
The cDNA encoding a harmonin splice variant (amino acid 1–548) was cloned into the C1-GFP vector (CLONTECH). Harmonin was amplified by PCR (5′-TAAC CCGGGGATGGACCGGAAGGTGGCCC-3′; 5′-CGCGGATCCTTAAAAGAAGGTTAGCTCGTCATC-3′). The CDH23 cytoplasmic domains were amplified by PCR (5′-GGGAATTCAAGCTTAACTGGTACTACAGGAC-3′; 5′-GGGCCGCTCGAGTCACAGCTCCGTGATTTCCAG-3′) and inserted into pCMVIL2R (27). Cells were transfected with the calcium-phosphate method (28) or by using Fugene (Roche Diagnostics). Forty-eight hours after transfection, extracts were prepared [50 mM Hepes, pH 7.5/150 mM NaCl/1.5 mM MgCl2/1 mM EDTA/10% (vol/vol) glycerol/1% Triton X-100/100 mM NaF/1 mM phenylmethylsulfonyl fluoride/10 μg/ml aprotinin] and used for Western blots or immunoprecipitations (29). Enhanced chemiluminescence reagent was used for detection (Amersham Pharmacia). For immunoprecipitation from tissue, the brain of a C57BL/6 mouse was homogenized in RIPA buffer (50 mM Tris, pH 7.5/150 mM NaCl/1% Triton X-100/0.5% deoxycholate/0.1% SDS/1 mM phenylmethylsulfonyl fluoride/10 μg/ml aprotinin), and diluted 1:1 with 50 mM Tris/150 mM NaCl, pH 7.5. Proteins were detected by Western blot. Quantification of band intensity was performed by using nih image software.
Results
The CDH23 Cytoplasmic Domain: PDZ-Binding Domains and Alternative Splicing.
To identify downstream effectors of CDH23, we analyzed the sequence of its cytoplasmic domain. We identified two putative binding sites for PDZ domains (referred to as PDZ-binding interfaces, PBIs). PBIs are frequently found at the C terminus of cell surface receptors and have been categorized on the basis of their sequence and binding specificity as class-I, -II, -III, and -IV PBIs (30–32). A putative class-I PBI is present at the C terminus of CDH23 (Fig. 1b). Less frequently, proteins contain internal PBIs, but no consensus sequence had emerged (33, 34). The CDH23 cytoplasmic domain contains a stretch of amino acids with significant homology to a protein domain in the adaptor protein Ril, which functions as an internal PBI (ref. 35; Fig. 1b). This observation suggests that amino acids 91–184 of CDH23 are part of a PBI.
Two CDH23 mRNAs that differ by an insert of 105 bp have been described. The 105 bp are encoded by an alternatively spliced exon (exon 68) (12, 36), and insert 35 aa into the putative internal PBI of CDH23 (Fig. 1b). mRNA containing exon 68 is expressed preferentially in inner ear sensory epithelia. Other tissues such as heart, kidney, spleen, brain, and retina contain CDH23 mRNA lacking exon 68 (Fig. 1c and data not shown), which suggests that exon 68-encoded sequences regulate CDH23 function in the inner ear.
Harmonin: PDZ Domains and Interactions with CDH23 in Yeast.
To identify proteins that bind to the CDH23 cytoplasmic domain, we searched for PDZ domains in known deafness genes. Three putative PDZ domains are present in harmonin (19, 20), and we identified a putative class-I PBI at its C terminus (Fig. 1d). We refer to the PDZ domains, from the N to the C terminus, as PDZ1, PDZ2, and PDZ3. PDZ domains harbor characteristic amino acid sequences, such as the R/K-XXX-GLGF motif that forms part of the protein interaction surface. Differences in surrounding amino acids provide interaction specificity for target proteins (30, 37). PDZ1 and PDZ2 of harmonin contain variations of the R/K-XXX-GLGF motif, with only the GLG sequence being conserved. PDZ3 does not contain the GLG motif, but a distant variant with homology to a PDZ domain in p55 (Fig. 1d; ref. 38), which suggests that each PDZ domain of harmonin engages in interactions with different PBIs. Candidate target PBIs include the two PBIs of CDH23, and the C-terminal PBI of harmonin.
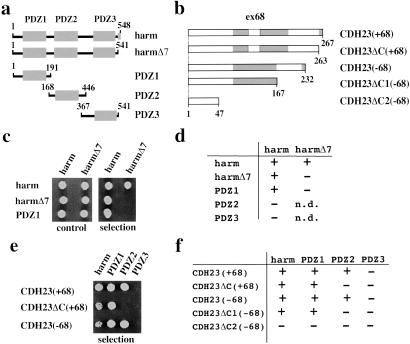
We tested whether harmonin interacts with itself and with CDH23 by yeast two-hybrid analysis (Fig. 2). Harmonin interacted with harmonin, but interactions were disrupted when the C-terminal PBI was deleted in both harmonin molecules used to measure interactions (harmΔ7). Harmonin fragments containing PDZ1, but not PDZ2 and PDZ3, interacted with full-length harmonin, suggesting that PDZ1 interacted with the C-terminal PBI in harmonin (Fig. 2 c and d). The CDH23(−68) and CDH23(+68) cytoplasmic domains also interacted with harmonin, but not with enigma, a protein that contains a PDZ domain homologous to PDZ1 and PDZ2 of harmonin (39) (Fig. 2 e and f; and data not shown). Truncations that deleted the putative C-terminal PBI of CDH23 [CDH23ΔC(+68) and CDH23ΔC1(−68)] interacted with harmonin, while deletion of both putative PBIs [CDH23ΔC2(−68)] abolished interaction (Fig. 2 e and f). We next identified domains in harmonin that mediate interactions with CDH23. Harmonin fragments containing PDZ1 or PDZ2 interacted with CDH23(−68) and CDH23(+68). Interactions with PDZ2 were disrupted when the C-terminal PBI of CDH23 was deleted. Interactions with PDZ1 were abolished by deletion of both PBIs. PDZ3 did not interact with CDH23 (Fig. 2 e and f).
Fig 2.
In yeast, harmonin interacts with harmonin and with CDH23. (a and b) Diagram of the harmonin and CDH23 fragments used in the two-hybrid assays. (c) Representative example of a yeast growth assay to measure harmonin–harmonin interactions. (Left) Growth in nonselective medium (control); (Right) growth in selective medium lacking histidine (selection). (d) Summary of the results establishing harmonin–harmonin interactions. (e) Representative example of a yeast growth assay to determine CDH23–harmonin interactions. (f) Summary of the results establishing CDH23–harmonin interaction. +, growth; −, no growth under selective pressure; n.d., not determined.
We conclude that PDZ1 of harmonin can bind to the C-terminal PBI of harmonin, and to the internal PBI of CDH23. PDZ2 of harmonin can bind to the C-terminal PBI of CDH23. Surprisingly, interactions of PDZ1 with the internal PBI of CDH23 are not affected by exon-68-encoded sequences. Because different expression levels of the fusion proteins in yeast could mask differences in interaction strength, we analyzed the effect of exon 68 in biochemical assays.
Regulation of CDH23–Harmonin Interactions by Alternative Splicing.
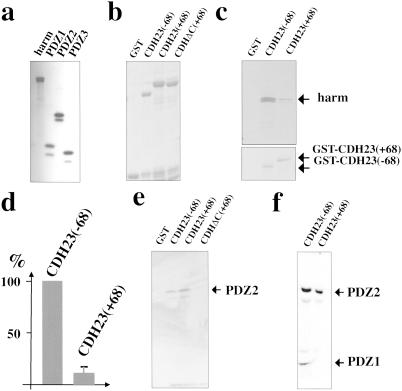
To analyze CDH23–harmonin interactions biochemically, we performed GST-pull-down and coimmunoprecipitation experiments. For pull-down experiments, harmonin and fragments containing individual PDZ domains were radiolabeled by in vitro transcription/translation (Fig. 3a), and analyzed for complex formation with GST-CDH23 fusion proteins (Fig. 3b). To allow for a quantitative comparison of complex formation, similar amounts of the GST-CDH23(−68) and GST-CDH23(+68) proteins were used (Fig. 3c Lower). Harmonin interacted with GST-CDH23(−68) and GST-CDH23(+68), but ≈10-fold greater amounts of harmonin were recovered with GST-CDH23(−68) (Fig. 3 c and d), which suggests that amino acids encoded by exon 68 affect the interaction of harmonin with CDH23.
Fig 3.
Exon 68-encoded amino acids affect CDH23–harmonin interactions in GST pull-down experiments. (a) Harmonin and its PDZ domains were expressed by in vitro transcription/translation in the presence of [35S]Met, resolved by SDS/PAGE, and visualized by autoradiography. (b) Purified GST-CDH23 fusion proteins were resolved on SDS-polyacrylamide gels and visualized by Coomassie staining. (c) Representative GST pull-down experiment. The input amount of GST-fusion proteins is shown (Lower). (d) The amount of harmonin that formed a complex with GST-CDH23 isoforms was quantified by scintillation counting and normalized against the input amount of GST-CDH23. The amount of harmonin interacting with GST-CDH23(−68) was set at 100%. Three experiments were performed, and the mean ± SD was determined. (e and f) Individual PDZ domains were used in GST pull-down experiments. The input proteins are shown in a. (e) PDZ2 was used for the GST pull-down experiments. (f) PDZ1 and PDZ2 were independently generated by in vitro translation and mixed.
To define the mechanism by which alternative splicing affects CDH23–harmonin complex formation, we analyzed fragments containing individual PDZ domains of harmonin for interaction with CDH23. In agreement with the two-hybrid results, PDZ2-containing fragments interacted with CDH23(−68) and CDH23(+68), but not with a truncated CDH23(+68) lacking the C-terminal PBI [CDH23ΔC(+68); Fig. 3e]. Deletion of the C-terminal PBI did not affect binding of PDZ1 to CDH23 (data not shown). We next mixed PDZ1 and PDZ2 fragments that were generated independently by in vitro transcription/translation, which allowed us to compare directly the amount of PDZ1 and PDZ2 recovered with GST-CDH23(−68) and GST-CDH23(+68). PDZ2 interacted equally well with both CDH23 isoforms, whereas PDZ1 interacted preferentially with the CDH23(−68) isoform (Fig. 3f). PDZ3 did not bind to either CDH23 isoform (data not shown).
We conclude that binding of CDH23 to harmonin is mediated by binding of the internal and C-terminal PBIs in CDH23 to PDZ1 and PDZ2 of harmonin, respectively. Sequences encoded by exon 68 perturb interactions between the internal PBI of CDH23 with PDZ1, but do not affect interactions of the C-terminal PBI of CDH23 with PDZ2 of harmonin.
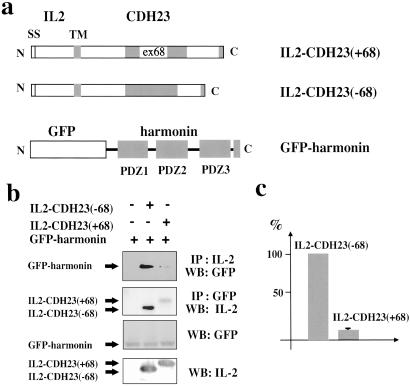
We next analyzed whether CDH23 and harmonin interact in the physiologically more relevant cellular context, and whether complex formation under these conditions is affected by alternative splicing. To mimic the membrane localization of CDH23, we generated a chimera containing the signal sequence, extracellular domain, and transmembrane domain of the IL-2 receptor fused to the CDH23 cytoplasmic domain. We also generated a fusion between the GFP and harmonin (Fig. 4a). The constructs were expressed in COS and 293 cells, and complex formation was analyzed in coimmunoprecipitation experiments by using antibodies to IL-2 and GFP (Fig. 4 b and c). Both proteins could be coimmunoprecipitated as a complex. Substantially less harmonin coprecipitated with the CDH23(+68) than with the CDH23(−68) isoform (Fig. 4 b and c), demonstrating that complex formation under physiological conditions is modulated by sequences encoded by exon 68.
Fig 4.
Coimmunoprecipitation experiments from transfected cells. (a) Diagram of the fusion proteins (SS, signal sequence; TM, transmembrane domain). (b) Extracts from cells transfected to express the constructs shown in a were used for immunoprecipitation (IP) experiments with antibodies specific for IL-2 or GFP. Proteins were separated by SDS/PAGE and visualized by Western blotting (WB) with GFP or IL-2 antibodies. As a control, proteins were separated by SDS/PAGE without immunoprecipitation and visualized by Western blotting. (c) The amount of harmonin that coprecipitated with IL-2 antibodies was densitometrically quantified. Three independent experiments were performed, and harmonin levels recovered with CDH23(−68) were set at 100%. The mean ± SD is indicated.
CDH23–Harmonin Interaction in Vivo, and CDH23 Expression in Hair Cells and the Retina.
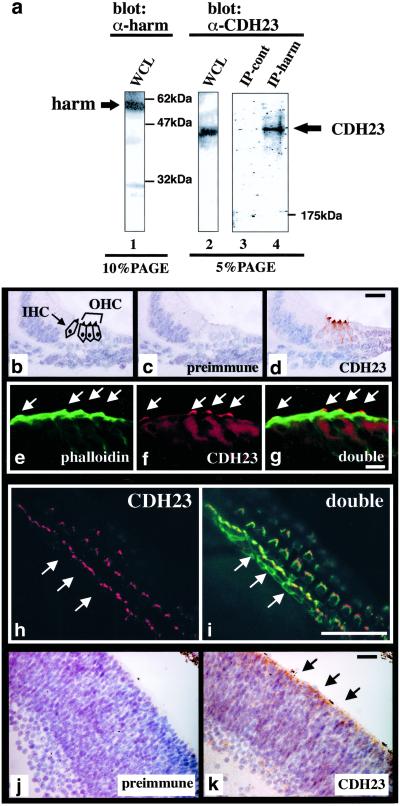
To analyze whether CDH23 and harmonin form a protein complex in vivo, we performed coimmunoprecipitation experiments with antisera raised against CDH23 and harmonin by using protein extracts from mouse tissue. The affinity-purified antisera recognized their respective antigen with high specificity in Western blots and immunoprecipitations performed with extracts from mouse tissues, or from cells transfected to express CDH23 or harmonin (Fig. 5a, lanes 1 and 2; data not shown). To analyze complex formation, extracts were prepared from P6 mouse brains, and immunoprecipitations were performed with the harmonin-specific antiserum. CDH23 was specifically detectable in the harmonin immunoprecipitates, but not in control precipitates (Fig. 5a, lanes 3 and 4), demonstrating that CDH23 and harmonin form a complex in vivo.
Fig 5.
CDH23–harmonin complex formation in vivo, and CDH23 expression in hair cells and the retina. (a) Whole-cell extracts (WCL) from P6 mouse brains were resolved on SDS-polyacrylamide gels directly (lanes 1 and 2), after immunoprecipitation with harmonin-specific antiserum (lane 4), or after a mock immunoprecipitation without antiserum (lane 3). Proteins were detected by Western blot with harmonin (lane 1) or CDH23-specific antiserum (lanes 2–4). (b–i) Analysis of CDH23 expression in the apical part of the cochlea at P0. (b) The section shows the position of the inner and outer hair cells (IHC, OHC). (c) No signal was detected with preimmune serum. (d) CDH23 is expressed in hair cell stereocilia. (e–g) Immunofluorescence analysis: (e) F-actin, labeled with FITC-phalloidin, is present in the cuticular plate and stereocilia (arrows); (f) CDH23 is expressed in stereocilia only; (g) double exposure. (h) Immunofluorescence analysis of a P6 whole mount cochlea stained with CDH23-specific antiserum, followed by rhodamine-conjugated secondary antibody. CDH23 is expressed in stereocilia of inner and outer hair cells. The arrows point to the one row of inner hair cells. (i) Whole mounts were labeled with FITC-phalloidin to reveal F-actin. The double exposure shows localization of CDH23 and F-actin. (j and k) Sections of P0 retinas were stained with preimmune serum or with CDH23-specific serum. CDH23 expression is strongest in the photoreceptor layer (arrows). [Bars = 15 μm (b–d), 5 μm (e–g), 30 μm (h and i), and 20 μm (j and k).]
Previous studies have shown that stereocilia are splayed in CDH23-deficient waltzer mice (17), and that harmonin is expressed in hair cells and their stereocilia (19). Expression of CDH23 in the inner ear has not been reported. We therefore stained cochlear sections and whole mounts with a CDH23-specific antiserum (Fig. 5 b–i). CDH23 was specifically expressed in the stereocilia of hair cells (Fig. 5 d, f, and h), where it is expected to exert its function as a complex with harmonin. Because harmonin is more widely expressed in hair cells (19), it may interact with additional proteins in other locations in hair cells.
Because Usher 1 patients suffer from visual impairment, we also stained retinas with CDH23 antibodies (Fig. 5 j and k). A low signal was detectable throughout the retina, but the signal was particularly strong in the photoreceptor layer, suggesting that CDH23 regulates photoreceptors development or function.
Discussion
Our data provide evidence that the two Usher syndrome proteins CDH23 and harmonin act in a common molecular pathway. The two proteins bind to each other, are coexpressed in hair cell stereocilia, and mutations in the genes encoding CDH23 and harmonin cause similar symptoms in USH1 patients (11, 12, 19, 20). The observation that stereocilia are splayed in CDH23-deficient waltzer mice (17) suggests that CDH23 and harmonin are components of the tip-, ankle-, side-links, or top connectors that link stereocilia into a bundle. Top connectors have not been observed in rodents, and the tip and ankle links, but not side links are sensitive to treatment with Ca2+ chelators (5, 40, 41). Because Ca2+ chelators abolish cadherin function, it is likely that CDH23 is a component of the tip- or ankle-link, or both. Future three-dimensional reconstructions using quantitative immunoelectron microscopy should help to clarify this issue.
Previously no sequence motifs have been described in the CDH23 cytoplasmic domain that may mediate protein–protein interactions. We now describe two PBIs in the CDH23 cytoplasmic domain: an internal domain with homology to a PBI in the adaptor protein Ril (35), and a PBI at the C terminus fitting the consensus for class I PBIs (30). The Ril homology region in CDH23 mediates interactions with the PDZ1 of harmonin, whereas its C-terminal PBI interacts with PDZ2. PDZ1 of harmonin also interacts with the C terminus of harmonin itself. Because the structure of PDZ domains is optimized to interact with free carboxylate groups (30, 31), we expected that the C terminus of CDH23 and harmonin would interact with PDZ1 of harmonin. We were surprised to identify an internal PBI within the CDH23 cytoplasmic domain. Internal PBIs have been found only in a few proteins including Ril and the neuronal nitric oxide synthase (33, 35). The internal PBI of neuronal nitric oxide synthase adopts a conformation with an exposed hairpin turn that mimics the overall structure of C-terminal PBIs (33), suggesting that the internal PBI in CDH23 has a similar structure. The homology between Ril and CDH23 is an example of sequence conservation between internal PBIs, raising the possibility that additional similar PBIs remain to be discovered.
Formation of the CDH23–harmonin complex is regulated by sequences encoded in the alternatively spliced exon 68 that is expressed in inner ear, but not the retina. Our data suggest that PDZ1 and PDZ2 of harmonin bind to CDH23(−68) isoforms in the retina. In the ear, interactions of PDZ1 with CDH23 are perturbed by the insertion of 35 amino acids encoded by exon 68. PDZ1 is therefore free to engage with other proteins, for example, with the C-terminal PBI in harmonin, which suggests that the overall composition and function of the CDH23–harmonin complex differs in ear and retina. For example, CDH23(+68) may be clustered more efficiently than CDH23(−68) by harmonin–harmonin interactions (Fig. 6). PDZ1, as well as other protein interaction motifs in harmonin, such as PDZ3, may recruit additional proteins. This finding is consistent with the function of other PDZ domain proteins that serve as assembly platforms for signaling complexes, for example, at epithelial cell junctions (42). Alternatively spliced isoforms have been described not only for CDH23 but also for harmonin (19), suggesting that different CDH23–harmonin-containing protein complexes are assembled in different cell types. Understanding the differences in complex composition may shed light on the molecular pathogenesis leading to sensorineuronal deafness and retinitis pigmentosa in USH1 patients.
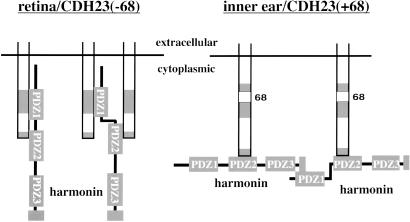
Fig 6.
Model for CDH23–harmonin complex formation and its regulation by alternative splicing. On the basis of our data, we propose the following model. PDZ1 and PDZ2 of harmonin interact with the internal and C-terminal PBI of CDH23(−68), respectively, which could lead to binding of harmonin to one CDH23 molecule, or it could cross-link two receptors (Left). Insertion of the sequence encoded by exon 68 perturbs interactions between PDZ1 of harmonin and the internal PBI of CDH23(+68). This perturbation poises the complex for alternative interactions (Right). Notably, the PDZ1 domain of harmonin may be available to interact with other proteins when harmonin is bound to CDH23(+68), but not to CDH23(−68). Binding partners for PDZ1 include harmonin, leading to the formation of harmonin–harmonin oligomers. Because CDH23(+68) is expressed in the ear but not the retina, splicing may control the architecture and composition of CDH23–harmonin complexes in a tissue-specific manner.
Acknowledgments
We thank M. Senften, C. Wirbelouer, and A. Kralli for advice and comments on the manuscript. The Novartis Research Foundation supported this work. J.S. is supported by the Boehringer Ingelheim Fond and A.R. is supported by a C. J. Martin Fellowship (no. 148939) from the National Health and Medical Research Council (Australia).
Abbreviations
PBI, PDZ-binding interface
References
- 1.Müller U. & Evans, A. L. (2001) Trends Cell Biol. 11, 334-342. [DOI] [PubMed] [Google Scholar]
- 2.Steel K. P. & Kros, C. J. (2001) Nat. Genet. 27, 143-149. [DOI] [PubMed] [Google Scholar]
- 3.Hone S. W. & Smith, R. J. (2001) Semin. Neonatol. 6, 531-541. [DOI] [PubMed] [Google Scholar]
- 4.Richardson G. P., Bartolami, S. & Russell, I. J. (1990) J. Cell Biol. 110, 1055-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodyear R. & Richardson, G. (1999) J. Neurosci. 19, 3761-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corey D. P. & Hudspeth, A. J. (1983) J. Neurosci. 3, 962-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pickles J. O., Comis, S. D. & Osborne, M. P. (1984) Hear. Res. 15, 103-112. [DOI] [PubMed] [Google Scholar]
- 8.Hudspeth A. J. (1992) Soc. Gen. Physiol. Ser. 47, 357-370. [PubMed] [Google Scholar]
- 9.Gillespie P. G. & Walker, R. G. (2001) Nature 413, 194-202. [DOI] [PubMed] [Google Scholar]
- 10.Yagi T. & Takeichi, M. (2000) Genes Dev. 14, 1169-1180. [PubMed] [Google Scholar]
- 11.Bolz H., von Brederlow, B., Ramirez, A., Bryda, E. C., Kutsche, K., Nothwang, H. G., Seeliger, M., Salcedo-Cabera, M. C., Vila, M. C., Molina, O. P., et al. (2001) Nat. Genet. 27, 108-112. [DOI] [PubMed] [Google Scholar]
- 12.Bork J. M., Peters, L. M., Riazuddin, S., Bernstein, S. L., Ahmed, Z. M., Ness, S. L., Polomeno, R., Ramesh, A., Schloss, M., Srisailpathy, C. R., et al. (2001) Am. J. Hum. Genet. 68, 26-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed Z. M., Riazuddin, S., Bernstein, S. L., Ahmed, Z., Khan, S., Griffith, A. J., Morell, R. J., Friedman, T. B. & Wilcox, E. R. (2001) Am. J. Hum. Genet. 69, 25-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alagramam K. N., Yuan, H., Kuehn, M. H., Murcia, C. L., Wayne, S., Srisailpathy, C. R., Lowry, R. B., Knaus, R., Van Laer, L., Bernier, F. P., et al. (2001) Hum. Mol. Genet. 10, 1709-1718. [DOI] [PubMed] [Google Scholar]
- 15.Alagramam K. N., Zahorsky-Reeves, J., Wright, C. G., Pawlowski, K. S., Erway, L. C., Stubbs, L. & Woychik, R. P. (2000) Hear. Res. 148, 181-191. [DOI] [PubMed] [Google Scholar]
- 16.Raphael Y., Kobayashi, K. N., Dootz, G. A., Beyer, L. A., Dolan, D. F. & Burmeister, M. (2001) Hear. Res. 151, 237-249. [DOI] [PubMed] [Google Scholar]
- 17.Di Palma F., Holme, R. H., Bryda, E. C., Belyantseva, I. A., Pellegrino, R., Kachar, B., Steel, K. P. & Noben-Trauth, K. (2001) Nat. Genet. 27, 103-107. [DOI] [PubMed] [Google Scholar]
- 18.Weil D., Blanchard, S., Kaplan, J., Guilford, P., Gibson, F., Walsh, J., Mburu, P., Varela, A., Levilliers, J., Weston, M. D., et al. (1995) Nature 374, 60-61. [DOI] [PubMed] [Google Scholar]
- 19.Verpy E., Leibovici, M., Zwaenepoel, I., Liu, X. Z., Gal, A., Salem, N., Mansour, A., Blanchard, S., Kobayashi, I., Keats, B. J., et al. (2000) Nat. Genet. 26, 51-55. [DOI] [PubMed] [Google Scholar]
- 20.Bitner-Glindzicz M., Lindley, K. J., Rutland, P., Blaydon, D., Smith, V. V., Milla, P. J., Hussain, K., Furth-Lavi, J., Cosgrove, K. E., Shepherd, R. M., et al. (2000) Nat. Genet. 26, 56-60. [DOI] [PubMed] [Google Scholar]
- 21.Self T., Mahony, M., Fleming, J., Walsh, J., Brown, S. D. & Steel, K. P. (1998) Development (Cambridge, U.K.) 125, 557-566. [DOI] [PubMed] [Google Scholar]
- 22.Kussel-Andermann P., El-Amraoui, A., Safieddine, S., Nouaille, S., Perfettini, I., Lecuit, M., Cossart, P., Wolfrum, U. & Petit, C. (2000) EMBO J. 19, 6020-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J. & Russell, D. W., (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 24.Harlow E. & Lane, D., (1999) Using Antibodies: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 25.Littlewood Evans A. & Muller, U. (2000) Nat. Genet. 24, 424-428. [DOI] [PubMed] [Google Scholar]
- 26.Gietz R. D. & Woods, R. A. (2002) Methods Mol. Biol. 185, 471-486. [DOI] [PubMed] [Google Scholar]
- 27.Akiyama S. K., Yamada, S. S., Yamada, K. M. & LaFlamme, S. E. (1994) J. Biol. Chem. 269, 15961-15964. [PubMed] [Google Scholar]
- 28.Muller U. & Doerfler, W. (1987) J. Virol. 61, 3710-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller U., Bossy, B., Venstrom, K. & Reichardt, L. F. (1995) Mol. Biol. Cell 6, 433-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheng M. & Sala, C. (2001) Annu. Rev. Neurosci. 24, 1-29. [DOI] [PubMed] [Google Scholar]
- 31.Harris B. Z. & Lim, W. A. (2001) J. Cell Sci. 114, 3219-3231. [DOI] [PubMed] [Google Scholar]
- 32.Vaccaro P. & Dente, L. (2002) FEBS Lett. 512, 345-349. [DOI] [PubMed] [Google Scholar]
- 33.Hillier B. J., Christopherson, K. S., Prehoda, K. E., Bredt, D. S. & Lim, W. A. (1999) Science 284, 812-815. [PubMed] [Google Scholar]
- 34.Harris B. Z., Hillier, B. J. & Lim, W. A. (2001) Biochemistry 40, 5921-5930. [DOI] [PubMed] [Google Scholar]
- 35.Cuppen E., Gerrits, H., Pepers, B., Wieringa, B. & Hendriks, W. (1998) Mol. Biol. Cell 9, 671-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Palma F., Pellegrino, R. & Noben-Trauth, K. (2001) Gene 281, 31-41. [DOI] [PubMed] [Google Scholar]
- 37.Songyang Z., Fanning, A. S., Fu, C., Xu, J., Marfatia, S. M., Chishti, A. H., Crompton, A., Chan, A. C., Anderson, J. M. & Cantley, L. C. (1997) Science 275, 73-77. [DOI] [PubMed] [Google Scholar]
- 38.Marfatia S. M., Morais-Cabral, J. H., Kim, A. C., Byron, O. & Chishti, A. H. (1997) J. Biol. Chem. 272, 24191-24197. [DOI] [PubMed] [Google Scholar]
- 39.Guy P. M., Kenny, D. A. & Gill, G. N. (1999) Mol. Biol. Cell 10, 1973-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Y., Yamoah, E. N. & Gillespie, P. G. (1996) Proc. Natl. Acad. Sci. USA 93, 15469-15474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Assad J. A., Shepherd, G. M. & Corey, D. P. (1991) Neuron 7, 985-994. [DOI] [PubMed] [Google Scholar]
- 42.Nagafuchi A. (2001) Curr. Opin. Cell Biol. 13, 600-603. [DOI] [PubMed] [Google Scholar]
- 43.Kim E., Niethammer, M., Rothschild, A., Jan, Y. N. & Sheng, M. (1995) Nature 378, 85-88. [DOI] [PubMed] [Google Scholar]
- 44.Kornau H. C., Schenker, L. T., Kennedy, M. B. & Seeburg, P. H. (1995) Science 269, 1737-1740. [DOI] [PubMed] [Google Scholar]