Abstract
Mitotic sister-chromatid cohesion (SCC) is known to depend in part on conserved proteins called adherins, which although necessary for SCC are not themselves localized between sister chromatids. We have examined mitotic DNA-repair and meiotic chromosome behavior in the Coprinus cinereus adherin mutant rad9-1. Genetic pathway analysis established that Rad9 functions in an Mre11-dependent pathway of DNA repair. Using fluorescence in situ hybridization, we found that the rad9-1 mutant is defective in the establishment of meiotic homolog pairing at both interstitial and subtelomeric sites but in the maintenance of pairing at only interstitial loci. To determine the role of Rad9 in meiotic SCC, we hybridized nuclear spreads simultaneously with a homolog-specific probe and a probe that recognizes both members of a homologous pair. We found that Rad9 is required for wild-type levels of meiotic SCC, and that nuclei showing loss of cohesion were twice as likely also to fail at homolog pairing. To ask whether the contribution of Rad9 to homolog pairing is solely in the establishment of SCC, we examined a rad9-1;msh5-22 double mutant, in which premeiotic DNA replication is inhibited. The msh5-22 mutation partially suppressed the deleterious effects of the rad9-1 mutation on homolog pairing; however, pairing in the double mutant still was significantly lower than in the msh5-22 single mutant control. Because the role of Rad9 in homolog pairing is not obviated by the absence of a sister chromatid, we conclude that adherins have one or more early meiotic functions distinct from the establishment of cohesion.
Sister-chromatid cohesion (SCC) is established during DNA replication (1, 2) and is required for the proper segregation of chromosomes (reviewed in refs. 3 and 4). In both mitosis and meiosis, SCC allows attached metaphase chromosomes to counteract the poleward forces imposed on them by the spindle until the onset of anaphase (2), and recent studies have shown that cohesion also plays an important role in spindle–kinetochore interactions (5) and the establishment of a bioriented spindle (6).
In meiosis, the tight association of sister chromatids by cohesion is central to the formation of axial elements, which later give rise to the lateral elements of the synaptonemal complex (SC; ref. 7). Therefore, although the inability to form SC structures is not always indicative of a cohesion defect, mutants that are defective in SCC fail to form axial elements and SC structures (8). After the dissolution of the SC, cohesion at sites along chromosomal arms distal to chiasmata are thought to block these crossover points from migrating off the chromosomal ends (4, 9). Arm cohesion is released just before anaphase I, chiasmata are resolved, and homologous chromosomes, containing pairs of sister chromatids still attached at the centromeres, are pulled toward opposite spindle poles (4, 9). Immediately before anaphase II, cohesion at the centromeres is released, and sister chromatids then segregate in opposite directions (4, 9).
Mutations in some genes implicated in SCC exhibit sensitivity to DNA-damaging agents (8, 10–15). Models for the role of chromatid cohesion in DNA repair center around its role in recombination (reviewed in refs. 8 and 16). In these models, cohesion ensures the availability of a closely associated, probably undamaged sister chromatid, which can serve as a template for repair during the G2 phase of the cell cycle. Components of some DNA-repair complexes such as Rad50 in the Mre11 complex are structurally related to components of the cohesin complex (16, 17). Therefore, there may be similarities between DNA structures involved in SCC and those required for DNA repair (8). Mutations in any of the genes encoding the Mre11–Rad50–Xrs2 complex members in Saccharomyces cerevisiae cause a hyperrecombination phenotype (18–22). One interpretation of these observations is that lesions that normally would be repaired by homologous recombination with the sister chromatid are repaired instead by using a homologous (nonsister) chromatid as a recombination partner. Experiments in Schizosaccharomyces pombe strains support this interpretation, because the observed increase in recombination between homologous chromosomes in rad50Δ strains is accompanied by a decrease in recombination between sister chromatids (12). These results suggest that one of the roles of the Mre11–Rad50 complex may be in mediating specialized interactions between sister chromatids, which facilitate repair. Thus, the cohesin complex functions as the primary level of sister-chromatid interactions, and these may be built on or modulated by the Mre11 complex. Mre11 complex-dependent sister-chromatid interactions also may facilitate nonhomologous end-joining repair of double-strand breaks (DSBs) by protecting DNA ends from exonucleolytic degradation after DSB formation (23).
We study meiosis and DNA repair in the basidiomycete Coprinus cinereus. The meiotic process is synchronous in this organism (24) and tightly associated with the development of the fruit body, or mushroom. Previously, we demonstrated that the rad9-1 mutant demonstrates the phenotypes of hypersensitivity to ionizing radiation and defects in chromosome condensation and synapsis (25–27). More recently, the S. cerevisiae (28, 29) and S. pombe (30) homologs of C. cinereus Rad9 have been demonstrated to function in mitotic SCC. Scc2, the Rad9 homolog from budding yeast, has been shown to participate in the loading of the cohesin complex on chromatin after DNA replication (28). The scc2-4 mutant does not demonstrate defects in the chromatin loading of the condensin complex (28), and Ciosk et al. (28) have proposed that the principal role of Scc2 during vegetative growth in S. cerevisiae is in the establishment of SCC. Although scc2-4 is defective in DNA DSB repair (14), this phenotype can be attributed to a primary defect in SCC. However, observations from two other organisms suggest that Rad9 homologs may have other functions possibly absent in S. cerevisiae. The S. pombe homolog, Mis4, was demonstrated to be required during M phase (30), a function inconsistent with a sole requirement for this protein in setting up cohesin during S phase, and Mis4 has been shown recently to be required for kinetochore–microtubule interactions (5). Also, Nipped-B, the Rad9 ortholog in Drosophila melanogaster, has been demonstrated to oppose the insulating activities of a gypsy transposon inserted between an enhancer and a promoter of two different genes (31), indicating a structural role for that protein in active chromatin.
These observations compelled us to examine further the role of Rad9 in both meiosis and DNA repair to determine whether its function in these processes in C. cinereus can be attributed completely to a role in the establishment of SCC. In this study, we demonstrate that Rad9 functions in an Mre11-dependent DNA-repair pathway, supporting evidence for the interdependence of SCC with the functions of the Mre11–Rad50 complex. Furthermore, we demonstrate that Rad9 is required for wild-type levels of homolog pairing and SCC during meiosis I in C. cinereus. However, we also show that the contribution of Rad9 to homolog pairing is not explained completely by its requirement for meiotic SCC. Thus, the contributions of Rad9 to meiotic chromosome behavior are not restricted to a role in sister-chromatid associations.
Materials and Methods
Strains and Culture Conditions.
Culture conditions, matings, and fruitings were as described (26). To generate rad9-1 and rad9-1;msh5-22 strains, the rad9-1;4-23 strain was crossed to the msh5-22 strain (strain PJP337, previously called spo22-1; ref. 32). The resulting dikaryon was fruited, and spore progeny were isolated. Individual progeny were mated to the original rad9-1;4-23 (33) and msh5-22 strains and screened for both hypersensitivity to γ radiation and a defect in meiosis to indicate the presence of the rad9-1 and msh5-22 mutations, respectively. The msh5-22 dikaryon used in this study comes from a cross between an msh5-22 single mutant isolated in the rad9-1;4-23 × msh5-22 cross and the original PJP337 strain. The wild-type, rad9-1, mre11-1, and rad9-1;mre11-1 strains used in the γ-radiation survival curves were isolated as progeny from a cross between the rad9-1;5-20 strain (33) and the mre11-1;5-6 strain (34).
γ-Radiation Survival Curves.
Methods for 137Cs survival curves were similar to those described by Valentine et al. (35) and Ramesh and Zolan (36). Oidia were harvested, plated in triplicate, irradiated (or held for an unirradiated control), and assayed for colony-forming ability. The wild-type and rad9-1 strains were analyzed twice, and the mre11-1 and rad9-1;mre11-1 double mutant were analyzed three times; data represent either six or nine total plates per experimental point. Data were analyzed as described by Rice (37).
Fluorescence in Situ Hybridization (FISH).
FISH experiments were carried out as described by Li et al. (38). All probes used for FISH except the B42-locus cosmid (a generous gift from Lorna Casselton; ref. 39), have been described (38). Each rad9-1 experiment was performed in parallel with a similar experiment by using wild-type chromosome spreads. Each probe and every reagent were tested by using wild-type strains at the K + 6 time point. Statistical differences for pairing data in comparisons of strains and time points were evaluated by using χ2 tests.
Determination of Nuclear DNA Content.
Basidial nuclei were examined at 6 h after karyogamy (K + 6; as defined in ref. 25). Single gill sections were fixed and stained with propidium iodide as described (35). Fluorescent digital images of stained nuclei were captured by using a Nikon Eclipse E600 microscope, and METAMORPH imaging-system software (Universal Imaging, Media, PA) was used to determine relative DNA content. The relative intensity for a single nucleus was determined by subtracting the background value from the integrated gray value of each nucleus.
Results
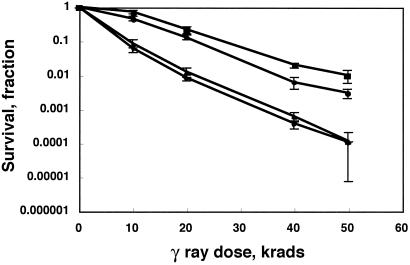
Previously, we demonstrated that the rad9-1 mutant shows the same dual phenotypes, hypersensitivity to ionizing radiation, and meiotic dysfunction as the mre11-1 mutant (27, 34, 35). In addition, indirect evidence from pathway analysis indicated that Rad9 likely functions in the same DNA-repair pathway as Mre11 (26, 35). To address the connection between Rad9 and Mre11 directly, we measured the extent of survival of wild-type, rad9-1, mre11-1, and rad9-1;mre11-1 strains after exposure to increasing doses of γ radiation (Fig. 1). Both the mre11-1 and rad9-1 strains showed greater sensitivity to ionizing radiation than the wild-type strain, with the mre11-1 single mutant strain demonstrating greater sensitivity than the rad9-1 single mutant. This observation is consistent with our previous observations (26, 27, 35), which demonstrated that the rad9-1 strain shows a moderate but reproducible sensitivity to ionizing radiation. The rad9-1;mre11-1 double-mutant strain exhibits an average sensitivity that overlaps that of the mre11-1 single mutant (Fig. 1). Therefore, Rad9 functions in an Mre11-dependent pathway of γ-radiation survival.
Fig 1.
Pathway analysis for radiation sensitivity. Oidia were plated, irradiated, and incubated at 37°C. Survival at each time point was calculated relative to the unirradiated control for that strain. The error bars show 95% confidence intervals for the mean survival values reported. Squares, wild-type; circles, rad9-1; diamonds, mre11-1; and triangles, rad9-1, mre11-1 double mutant.
Our prior studies of rad9-1 revealed its defects in meiotic chromosome synapsis (25). However, this observation did not necessarily indicate that this mutant is also deficient in homolog pairing. Because synapsis does depend on homology (40–42), it was also possible that homologous associations could occur in rad9-1 without ensuring synapsis. To determine whether the rad9-1 mutant is defective in homolog pairing, we performed FISH analysis on surface-spread nuclei using probes for chromosomes 8 and 13 at time points of 1 and 6 h after karyogamy (K + 1 and K + 6, respectively). At K + 1, most nuclei in both wild-type and rad9-1 strains contain chromosomes that appear relatively diffuse when using our spreading and staining methods (25, 38). At K + 6, >90% of wild-type meiotic nuclei show full condensation and synapsis, whereas all rad9-1 nuclei exhibit short stretches of SC against a diffuse background (25).
A comprehensive FISH analysis of homology pairing in a wild-type strain congenic with rad9-1 was reported by Li et al. (38). In studies performed in parallel with this published work, we found that the rad9-1 strain exhibits an obvious pairing defect early in meiosis. Individually, homologous pairing at interstitial sites sampled for chromosome 8 (probe 1) or chromosome 13 (probe 5) at K + 1 was ≈65% (Table 1). This amount of pairing is significantly less (P < 0.005) than the ≈95% levels observed for these probes in the simultaneously executed wild-type control (all cited data for the wild-type strain have been reported by Li et al. in ref. 38). Only 48% of rad9-1 nuclei exhibited pairing of both of these chromosome 8 and 13 sites (Table 1) compared with 91% at K + 1 in the wild-type control. The early pairing defect of rad9-1 cells also was observed at interstitial (probe 3) and subtelomeric (probe 6) sites of chromosome 13 (Table 2). For each of these sites, pairing occurred in an average of 60% of the nuclei at K + 1, significantly less than the wild-type values for those same sites; similarly, the 42% of nuclei showing pairing for both probes was significantly less than the 79% observed in the wild-type strain (P < 0.005 for all comparisons).
Table 1.
Homolog pairing in the rad9-1 mutant
| Time point
|
Chr. 8 probe
|
Chr. 13 probe
|
N
|
Percent pairing | |||
|---|---|---|---|---|---|---|---|
| Chr. 8 | Chr. 13 | Chr. 8 and chr. 13 | Chr. 8 and/or chr. 13 | ||||
| K + 1 | 1 | 5 | 70 | 57 | 70 | 47 | 80 |
| 42 | 79 | 62 | 50 | 90 | |||
| 112 | 65 | 67 | 48 | 84 | |||
| K + 6 | 1 | 5 | 65 | 45 | 54 | 26 | 72 |
| 1 | 5 | 58 | 37 | 55 | 31 | 62 | |
| 123 | 41 | 54 | 28 | 67 | |||
Data are arrayed by time point and probes. Within a category, each nonbold line of data represents one experiment, which refers to chromosome (Chr.) spreads from one mushroom, and one hybridization. Up to four slides were included per experiment. Boldface lines represent the total nuclei (N) examined for that category and averages (weighted by the number of nuclei for each experiment) for other categories.
K + 1 and K + 6 refer to 1 and 6 h after karyogamy (25), respectively.
Probes were as described by Li et al. (38).
Table 2.
Homolog pairing at different sites on the same chromosome
| Time point
|
Probe A
|
Probe B
|
N
|
Percent pairing | |||
|---|---|---|---|---|---|---|---|
| Probe A | Probe B | Probe A and probe B | Probe A and/or probe B | ||||
| K + 6 | 1 | 2 | 36 | 56 | 50 | 36 | 69 |
| 13 | 46 | 62 | 31 | 77 | |||
| 23 | 39 | 30 | 22 | 48 | |||
| 23 | 65 | 70 | 48 | 87 | |||
| 95 | 53 | 52 | 35 | 69 | |||
| K + 1 | 3 | 6 | 86 | 60 | 60 | 42 | 81 |
| K + 6 | 3 | 6 | 15 | 53 | 53 | 33 | 73 |
| 105 | 27 | 57 | 18 | 65 | |||
| 120 | 30 | 57 | 20 | 66 | |||
| K + 6 | 5 | 6 | 28 | 36 | 46 | 21 | 61 |
Data are arrayed by time point and probes. Other table aspects are as described for Table 1. If all data for a category are boldface, then only one experiment was performed.
K + 1 and K + 6 refer to 1 and 6 h after karyogamy (25), respectively.
Probes were as described by Li et al. (38). Probes 1 and 2 were for chromosome 8; probes 3, 5, and 6 were for chromosome 13. N, number of nuclei examined.
The homolog-pairing defect of the rad9-1 strain was observed also with all probes at K + 6. Interstitial probes for both chromosome 8 (probe 1) and chromosome 13 (probe 5) were paired in 41% and 54%, respectively (Table 1), of nuclei examined. When used in the same hybridizations, chromosome 8 interstitial probes 1 and 2 individually exhibited an average of 53% pairing in rad9-1 nuclei (Table 2). Chromosome 13 interstitial probe 3 and subtelomeric probe 6 showed 30% and 57% homolog pairing, respectively (Table 2). All K + 6 values for rad9-1 were significantly less (P < 0.005) than the respective levels observed in wild-type nuclei. Thus, homolog pairing in the rad9-1 mutant was significantly less than in a congenic wild-type strain at both K + 1 and K + 6 at all tested loci.
To address whether pairing of two different sites on the same chromosome occurs independently, we compared the expected frequency of simultaneous pairing of two different probes targeting the same chromosome with the observed average frequency of nuclei in which both probes actually demonstrated simultaneous pairing (Table 2). Expected values (not shown) were calculated by multiplying the product of the average pairing frequencies of the two individual probes by the total number of nuclei examined. The observed values at K + 6 for probe-combinations 1 and 2 (chromosome 8), 3 and 6 (chromosome 13), and 5 and 6 (chromosome 13) exhibited no statistical differences (P > 0.1) from expected values. Expected and observed values were also indistinguishable for probe-combination 3 and 6 in K + 1 nuclei. Therefore, the pairing of one locus does not affect the likelihood that another site on the same chromosome will also pair in a given nucleus.
Although probes for all loci revealed large defects in pairing in rad9-1, it is also notable that the average pairing for interstitial probes listed in Tables 1 and 2 actually decreased between K + 1 and K + 6. To further address the issue of whether pairing decreases during prophase I, we combined data from Tables 1 and 2 with data from additional experiments to determine the average pairing percentages for individual probes at K + 1 and K + 6 (Table 3). Pooled data for chromosome 8 (probe 1) demonstrate that homolog pairing at this interstitial site exhibited a significant decrease (P < 0.005) between early (K + 1) and late (K + 6) prophase I. Similarly, pooled data for chromosome 13 interstitial probes 3 and 5 also reveal a significant decrease (P < 0.005 for both) between early and late homolog-pairing percentages. Strikingly, only the chromosome 13 subtelomeric site (probe 6) showed average pairing percentages that were indistinguishable (P > 0.1) between K + 1 and K + 6.
Table 3.
Homolog pairing at different meiotic time points
| Probe | Chromosome | Time point | Number of experiments | N | Average percent pairing |
|---|---|---|---|---|---|
| 1 | 8 | K + 1 | 2 | 112 | 65 |
| K + 6 | 6 | 218 | 46 | ||
| 3 | 13 | K + 1 | 1 | 86 | 60 |
| K + 6 | 2 | 120 | 30 | ||
| 5 | 13 | K + 1 | 2 | 112 | 67 |
| K + 6 | 7 | 231 | 48 | ||
| 6 | 13 | K + 1 | 1 | 86 | 60 |
| K + 6 | 3 | 193 | 62 |
Probes were as described by Li et al. (38).
K + 1 and K + 6 refer to 1 and 6 h after karyogamy (25), respectively.
Number of experiments from which data for the relevant probe were pooled. In some cases, additional experiments not shown in Tables 1 or 2 were used.
Total number of nuclei (N) in pooled data.
Averages were weighted by the number of nuclei for each component experiment.
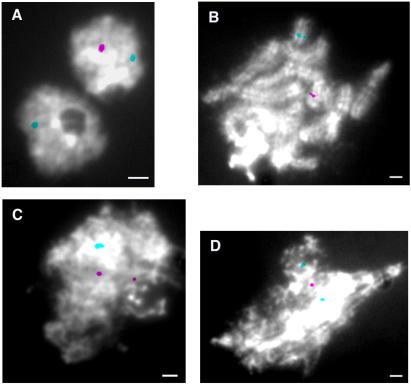
To test whether the rad9-1 mutant exhibits an impairment in meiotic SCC, which might underlie its known meiotic defects, we performed FISH on meiotic chromosome spreads by simultaneously hybridizing with two different probes. The first probe was specific for the B42 mating-type locus from the Java-6 (43) parental strain. Mating-type loci exhibit substantial sequence variation in different strains of C. cinereus (39, 44, 45). Use of the B42 locus in FISH to prekaryogamy nuclei confirmed that this probe hybridized to only one of the two parental nuclei, as expected (Fig. 2A). Thus, the B42-locus probe is homolog-specific and can be used to evaluate the level of association between sister chromatids at that chromosomal position. The second probe (probe 3; ref. 38) hybridizes to an interstitial position on chromosome 13 in both parental strains and therefore can be used to indicate whether in the same nuclei the chromosome 13 homologs are paired (Fig. 2). Four experiments, indicating four separate hybridizations, were performed by using cap tissue isolated from wild-type or rad9-1 strains at K + 6 (Table 4). The wild-type strain rarely showed sister-chromatid separation at the B locus; the B42 probe hybridized to two spots in an average of 8% of wild-type nuclei. However, for the rad9-1 mutant, 31% of the nuclei had two spots with the B-locus probe. Thus, the rad9-1 mutant exhibited a 4-fold increase in the percentage of nuclei with separation of sister chromatids as compared with the wild-type strain, which is a significant change (P < 0.005). These data indicate that the Rad9 protein is required to achieve or maintain wild-type levels of meiotic SCC, at least at the B locus analyzed here.
Fig 2.
Homolog pairing and SCC in the rad9-1 mutant. Chromosome spreads were hybridized with a probe specific for the B42 mating-type locus (red) and a probe that detects both chromosome 13 homologs (green). (A) Prefusion wild-type nuclei. (B) Wild-type pachytene chromosome spread. (C and D) Spreads of rad9-1 nuclei from the time point (K + 6) at which wild-type strains are in pachytene. (Scale bars, 1 μm.)
Table 4.
SCC is defective in rad9-1, and homolog pairing correlates with cohesion
| Strain
|
N
|
Percent pairing of chr. 13
|
Percent nuclei with loss of SCC
|
Percent pairing of chr. 13 | |
|---|---|---|---|---|---|
| SCC present | SCC absent | ||||
| Wild type | 60 | 100 | 5 | — | — |
| 55 | 100 | 15 | — | — | |
| 24 | 100 | 4 | — | — | |
| 22 | 100 | 0 | — | — | |
| 161 | 100 | 8 | — | — | |
| rad9-1 | 28 | 68 | 21 | 82 | 17 |
| 51 | 35 | 53 | 46 | 26 | |
| 33 | 45 | 33 | 50 | 36 | |
| 48 | 50 | 13 | 55 | 17 | |
| 160 | 48 | 31 | 57 | 26 | |
Because we used one homolog-specific probe (to the B42 locus, on chromosome 10; M. Celerin and M.E.Z., unpublished data) and one probe that hybridized to both copies of chromosome 13, our experiments also allowed us to address whether rad9-1 nuclei demonstrating SCC defects were more likely to show a defect in homolog pairing than those nuclei that did not exhibit a cohesion defect. Table 4 shows the percentages of rad9-1 nuclei exhibiting successful homolog pairing in nuclei with either one or two B-locus spots. In the class of nuclei with one B-locus spot, an average of 57% also exhibited pairing for the chromosome 13 probe. In contrast, the percentage of nuclei with two B-locus spots that also successfully paired the chromosome 13 locus was only 26%; this is a significant difference (P < 0.005). Therefore, despite the fact that the regions used to assay SCC and homolog pairing reside on different chromosomes, loss of cohesion correlates with and may underlie a loss of pairing. These results raise the possibility that an observed loss of homolog pairing may reflect a widespread loss of SCC within a nucleus.
We wanted to address whether the homologous chromosome pairing defect of the rad9-1 mutant reflects only the role of the Rad9 protein as an effector of SCC. Therefore, we took advantage of a C. cinereus mutant, msh5-22, which is defective in premeiotic DNA replication; prophase I genomic DNA content levels are approximately half those of wild-type strains (refs. 32 and 46 and M. Celerin, W.J.C., S.T.M., C.W.J., E.A.S., P. J. Pukkila, and M.E.Z., unpublished data). Despite the replication defect, msh5-22 strains exhibit complete synapsis (32, 46, 47) and pairing levels statistically indistinguishable from those in wild-type strains (Table 5).
Table 5.
The msh5-22 mutation partially restores pairing in a rad9-1 mutant
| Strain
|
N
|
Percent pairing | |||
|---|---|---|---|---|---|
| Chr. 8 probe | Chr. 13 probe | Chr. 8 and chr. 13 | Chr. 8 and/or chr. 13 | ||
| rad9-1 | 46 | 72 | 65 | 52 | 85 |
| 54 | 52 | 56 | 28 | 80 | |
| 100 | 61 | 60 | 39 | 82 | |
| msh5-22 | 68 | 91 | 93 | 85 | 100 |
| rad9-1;msh5-22 | 70 | 83 | 71 | 60 | 94 |
| 30 | 73 | 70 | 53 | 90 | |
| 21 | 76 | 76 | 67 | 86 | |
| 121 | 79 | 72 | 60 | 92 | |
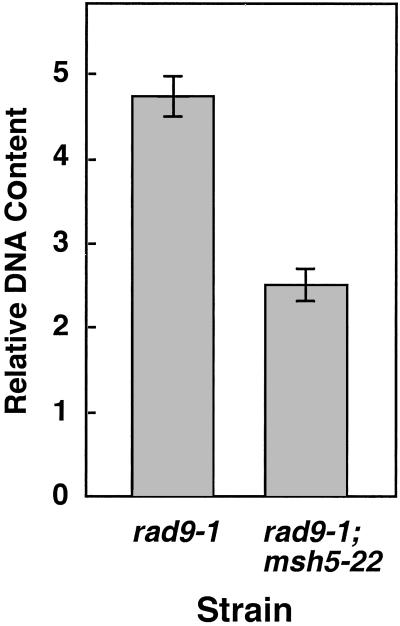
We constructed a rad9-1;msh5-22 double-mutant strain and, as expected, found that its prophase I DNA content is approximately one-half that of the rad9-1 single mutant (Fig. 3). Thus, even in a rad9-1 background, the msh5-22 mutation causes a severe defect in premeiotic DNA replication. If the role of Rad9 in homolog pairing were limited to its contribution in establishing appropriate sister-chromatid interactions, then the homolog-pairing defect of the rad9-1 mutant should be reduced greatly in an msh5-22 background. Thus, the msh5-22 mutation serves as a “test system” in which the effects of the rad9-1 mutation on homolog pairing can be assayed independently of defects in sister-chromatid interactions.
Fig 3.
Analysis of DNA replication. Basidial nuclei were examined at midpachytene (K + 6; ref. 25). Data were collected and normalized to the 1C (i.e., haploid unreplicated) value obtained at the same time from a sample of tissue at K + 12 after both meiotic divisions (not shown). Forty nuclei were measured per strain. The error bars show 95% confidence intervals.
Interstitial probes for chromosomes 8 and 13 were used to measure homolog pairing in a rad9-1;msh5-22 strain and a sibling rad9-1 strain (Table 5). Nuclei were examined at K + 6. In two experiments, the rad9-1 single mutant showed pairing in an average of 61% and 60% of nuclei for the chromosome 8 and chromosome 13 probes, respectively. Both chromosomes were paired in 39% of the nuclei. The rad9-1;msh5-22 strain showed higher levels of pairing than the rad9-1 single mutant for both probes. The double mutant had an average of 79% and 72% of nuclei containing paired chromosome 8 and chromosome 13 sites, respectively. Simultaneous pairing of both probes was observed in 60% of the nuclei; this was significantly greater (0.01 > P > 0.001) than in the rad9-1 single mutant. Therefore, we conclude that the msh5-22 mutation has a suppressive effect on the meiotic homolog-pairing defect caused by the rad9-1 mutation. However, the levels of pairing observed in the double mutant are not restored to the levels observed in the msh5-22 strain; they remain significantly lower (60% versus 85%; P < 0.001; Table 5). Thus, the absence of sister chromatids does not correct the homolog-pairing defect of the rad9-1 mutant completely.
Discussion
We have demonstrated by γ-radiation survival curves that Rad9 functions in at least one Mre11-dependent pathway of DNA repair (Fig. 1). Similarly, previous studies (35) showed that rad9 is in the same pathway as the C. cinereus rad12 gene, which we now know encodes an ortholog of Rad50 (ref. 47; S. N. Acharya, E. A. Friedle, J. T. Grubb, J. A. Vincent, A. P. Schroeder, and M.E.Z., unpublished data). In addition, a recent study (14) revealed a role for the Rad9 ortholog Scc2 in mitotic G2 phase DSB repair. Thus, three studies have positioned the adherin Rad9 within DSB-repair pathways.
The rad9-1 single mutant demonstrates less sensitivity to ionizing radiation than the mre11-1 mutant. This observation may simply mean that the rad9-1 mutation is a relatively weak allele for this phenotype. Preliminary, Western blot analysis using antibodies against an amino-terminal peptide fragment of Rad9 revealed a protein of approximately the predicted size (239 kDa; ref. 25) in wild-type strains. The same antibodies recognize a protein of ≈190 kDa in meiotic extracts from the rad9-1 mutant, the size predicted from the sequence of the mutant allele (E.A.S., S.T.M., W.J.C., A. Amiri, and M.E.Z., unpublished data). Therefore, rad9-1 is probably not a null allele. The extent of the function of Rad9 in DNA repair might not be fully apparent from the moderate hypersensitivity of the rad9-1 mutant to ionizing radiation, and alleles more severe for this phenotype may not be viable. Alternatively, only one or a few aspects of Mre11-dependent DNA repair may require Rad9.
Our pathway-analysis data (Fig. 1; ref. 35) differ from the observations of Hartsuiker et al. (12), who reported that the fission yeast cohesin Rad21 functions in the same methyl methanesulfonate-induced repair pathway as Rad50 but in a different γ-radiation survival pathway. There are two likely explanations for the discrepancy between their data and ours. First, Rad21 and its S. cerevisiae ortholog Scc1 are mitotic cohesins, the major proteins responsible for SCC (2), whereas Rad9 and its characterized fungal orthologs Scc2 (29) and Mis4 (30) are classified as adherins, which are necessary for the loading of cohesins onto chromosomes but are not part of the cohesion complex per se (2, 28, 30, 48). Because Rad9 is in the Mre11/Rad50 pathway for the repair of γ radiation-induced damage and Rad21 is apparently in a different pathway (12), cohesins must have Mre11 complex-independent roles in DSB repair, whereas adherins do not. Alternatively, the physiology of DNA repair between C. cinereus and S. pombe may differ such that γ-induced DNA damage is processed differentially in the two organisms. It is already known that both Mis4 and the S. pombe ortholog of mre11, rad32, are required for wild-type levels of resistance to UV light (30, 49), whereas the C. cinereus rad9 and mre11 genes are not (26, 34).
We have shown that the rad9-1 mutant is impaired severely in both early and midprophase I homolog pairing (Tables 1). Because the rad9-1 mutant exhibits homolog-pairing defects at the time when homologous chromosomes are initiating physical interactions with each other, we conclude that Rad9 functions in the establishment of homolog pairing. The decrease in pairing observed at interstitial sites between K + 1 and K + 6 supports the idea that Rad9 also functions in the maintenance of homolog pairing through prophase I. Weiner and Kleckner (50) have proposed that homolog pairing in budding yeast occurs in two stages that are separated by the disruptive effect of premeiotic DNA replication. The first stage represents the formation of unstable interactions between homologous chromosomes. This stage then is followed by a second phase, in which interactions are stabilized by interhomolog recombination. In zygotic meiosis, such as that of C. cinereus, both stages would occur after replication, during prophase I. Although the rad9-1 mutation negatively affects pairing at all tested sites and times, one explanation for the greater contribution of Rad9 to later homolog pairing (K + 6) is that it contributes to the recombination-based stabilization of homolog interactions. Consistent with this idea is the observation that the C. cinereus mre11-1 mutant (34) exhibits pairing levels similar to those observed in the rad9-1 mutant. Our results are compatible with the idea that the pairing role of Rad9 depends on or is necessary for the function of Mre11 in meiotic DSB formation and recombination initiation (51, 52). However, the C. cinereus spo11-1 mutant (53) pairs significantly better than rad9-1 at K + 6 (74% versus 49% and 80% versus 30% for probes 2 and 3, respectively; P < 0.005 for both comparisons). In addition, spo11-1 actually exhibits an increase in the percentage of nuclei with paired homologs between K + 3 and K + 6 (53), in contrast to the decrease observed in rad9-1. Therefore, Rad9 (and probably Mre11) cannot require Spo11-dependent recombination for its entire contribution to homolog pairing.
The only site that did not undergo a decrease in pairing levels through time was the subtelomeric site on chromosome 13. If the limited synapsis in the rad9-1 mutant is primarily homologous, then SC formation might occur principally at sites that have not lost homolog pairing. In this case, telomeres would be more likely to synapse, because they have maintained the pairing levels established in early prophase I, likely because of their specialized role in homolog pairing (reviewed in refs. 54–57). However, we have observed instances of triple synapsis and pairing partner switches in the rad9-1 mutant (25), an observation that argues against purely homologous synapsis.
When mitotic cells harboring conditional mutations in the SCC2 (29) and mis4 (30) genes are shifted to nonpermissive temperatures, they exhibit precocious (preanaphase) sister-chromatid separation. We have shown that a mutation in the C. cinereus ortholog of these genes, rad9, causes defects in meiotic SCC. We interpret this observation to mean that Rad9, and probably Scc2 and Mis4 as well, function in meiotic SCC. However, if the role of Rad9 in homolog pairing were limited to its contribution to sister-chromatid interactions via cohesion, then the function of Rad9 in homolog pairing would have become dispensable in an msh5-22 background, in which sister chromatids are not made. We observed (Table 5) that the level of pairing in the msh5-22;rad9-1 double mutant is significantly greater than the level observed in the rad9-1 single mutant. Therefore, the homolog-pairing defect is explained partially by the role of this protein in establishing proper sister-chromatid interactions when sister chromatids are produced. It is formally possible that the replication defect of msh5-22 results in other changes in meiotic chromatin that suppress the pairing defect of rad9-1. However, the distinguishing characteristic of msh5-22 is an absence of sister chromatids during meiosis (32), and no other early meiotic defects have been reported for this mutant.
The pairing levels in the msh5-22;rad9-1 double mutant do not reach the levels seen in the msh5-22 strain (Table 5). Thus, the meiotic homolog-pairing defect of rad9-1 is not explained completely by poor sister-chromatid interactions, and it is evident that the Rad9 protein has an additional role(s) in homolog pairing that is distinct from the establishment of SCC (28). It must be noted, however, that this assertion does not necessarily mean that the additional roles of Rad9 in homolog pairing are mediated independently of cohesin, because this complex still may be loaded in the unreplicated msh5-22 mutant. Such loading would suggest that the cohesin components, similar to Rad9, may have roles beyond sister-chromatid association events, which are important for homologous chromosomal interactions and possibly DNA repair.
Acknowledgments
We thank Lorna Casselton for the C. cinereus B mating-type probe used in this study; Erin Gerecke for help with the analysis of FISH experiments; Sonia Acharya, Martina Celerin, and Bob Metzenberg for helpful discussion and comments on the manuscript; and Pat Foster and Martina Celerin for advice about statistics. We also thank Claire Walczak for the use of her microscopy system. This work was supported by National Institutes of Health Grant GM43930 (to M.E.Z.). W.J.C. was supported in part by National Institutes of Health Training Grant 2T32GM07757 (to the Department of Biology, Indiana University), a McCormick Science fellowship (to the College of Arts and Sciences, Indiana University), a J. Stewart and Dagmar K. Riley fellowship (College of Arts and Sciences, Indiana University), and a Bayard Ogg fellowship (to the Department of Biology, Indiana University). S.T.M. was supported in part by National Science Foundation Minority Postdoctoral Research Fellowship DBI-9628899. C.W.J. was supported in part by an LS McClung Undergraduate Summer Research award and an Honors College Thesis award, and C.W.J. and E.A.S. were supported by Undergraduate Research and Creative Activity Partnership awards (Indiana University).
Abbreviations
SCC, sister-chromatid cohesion
SC, synaptonemal complex
DSB, double-strand break
FISH, fluorescence in situ hybridization
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Uhlmann F. & Nasmyth, K. (1998) Curr. Biol. 8, 1095-1101. [DOI] [PubMed] [Google Scholar]
- 2.Nasmyth K. (2001) Annu. Rev. Genet. 35, 673-745. [DOI] [PubMed] [Google Scholar]
- 3.Miyazaki W. Y. & Orr-Weaver, T. L. (1994) Annu. Rev. Genet. 28, 167-187. [DOI] [PubMed] [Google Scholar]
- 4.Moore D. P. & Orr-Weaver, T. L. (1998) Curr. Top. Dev. Biol. 37, 263-299. [DOI] [PubMed] [Google Scholar]
- 5.Toyoda Y., Furuya, K., Goshima, G., Nagao, K., Takahashi, K. & Yanagida, M. (2002) Curr. Biol. 12, 347-358. [DOI] [PubMed] [Google Scholar]
- 6.Goshima G. & Yanagida, M. (2000) Cell 100, 619-633. [DOI] [PubMed] [Google Scholar]
- 7.Klein F., Mahr, P., Galova, M., Buonomo, S. B., Michaelis, C., Nairz, K. & Nasmyth, K. (1999) Cell 98, 91-103. [DOI] [PubMed] [Google Scholar]
- 8.van Heemst D. & Heyting, C. (2000) Chromosoma 109, 10-26. [DOI] [PubMed] [Google Scholar]
- 9.Lee J. Y. & Orr-Weaver, T. L. (2001) Annu. Rev. Cell Dev. Biol. 17, 753-777. [DOI] [PubMed] [Google Scholar]
- 10.Birkenbihl R. P. & Subramani, S. (1992) Nucleic Acids Res. 20, 6605-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guacci V., Koshland, D. & Strunnikov, A. (1997) Cell 91, 47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartsuiker E., Vaessen, E., Carr, A. M. & Kohli, J. (2001) EMBO J. 20, 6660-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huynh A. D., Leblon, G. & Zickler, D. (1986) Curr. Genet. 10, 545-555. [DOI] [PubMed] [Google Scholar]
- 14.Sjogren C. & Nasmyth, K. (2001) Curr. Biol. 11, 991-995. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto A., Guacci, V. & Koshland, D. (1996) J. Cell Biol. 133, 99-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strunnikov A. V. & Jessberger, R. (1999) Eur. J. Biochem. 263, 6-13. [DOI] [PubMed] [Google Scholar]
- 17.Jessberger R., Frei, C. & Gasser, S. M. (1998) Curr. Opin. Genet. Dev. 8, 254-259. [DOI] [PubMed] [Google Scholar]
- 18.Bressan D. A., Olivares, H. A., Nelms, B. E. & Petrini, J. H. (1998) Genetics 150, 591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanov E. L., Korolev, V. G. & Fabre, F. (1992) Genetics 132, 651-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malone R. E., Ward, T., Lin, S. & Waring, J. (1990) Curr. Genet. 18, 111-116. [DOI] [PubMed] [Google Scholar]
- 21.Moreau S., Ferguson, J. R. & Symington, L. S. (1999) Mol. Cell. Biol. 19, 556-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bressan D. A., Baxter, B. K. & Petrini, J. H. (1999) Mol. Cell. Biol. 19, 7681-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore J. K. & Haber, J. E. (1996) Mol. Cell. Biol. 16, 2164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raju N. B. & Lu, B. C. (1970) Can. J. Bot. 48, 2183-2186. [Google Scholar]
- 25.Seitz L. C., Tang, K., Cummings, W. J. & Zolan, M. E. (1996) Genetics 142, 1105-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zolan M. E., Tremel, C. J. & Pukkila, P. J. (1988) Genetics 120, 379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zolan M. E., Crittenden, J. R., Heyler, N. K. & Seitz, L. C. (1992) Nucleic Acids Res. 20, 3993-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciosk R., Shirayama, M., Shevchenko, A., Tanaka, T., Toth, A. & Nasmyth, K. (2000) Mol. Cell 5, 243-254. [DOI] [PubMed] [Google Scholar]
- 29.Michaelis C., Ciosk, R. & Nasmyth, K. (1997) Cell 91, 35-45. [DOI] [PubMed] [Google Scholar]
- 30.Furuya K., Takahashi, K. & Yanagida, M. (1998) Genes Dev. 12, 3408-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rollins R. A., Morcillo, P. & Dorsett, D. (1999) Genetics 152, 577-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pukkila P. J., Shannon, K. B. & Skrzynia, C. (1995) Can. J. Bot. 73, Suppl. 1, S215-S220. [Google Scholar]
- 33.Zolan M. E., Heyler, N. K. & Stassen, N. Y. (1994) Genetics 137, 87-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerecke E. E. & Zolan, M. E. (2000) Genetics 154, 1125-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valentine G., Wallace, Y. J., Turner, F. R. & Zolan, M. E. (1995) Mol. Gen. Genet. 247, 169-179. [DOI] [PubMed] [Google Scholar]
- 36.Ramesh M. A. & Zolan, M. E. (1995) Chromosoma 104, 189-202. [DOI] [PubMed] [Google Scholar]
- 37.Rice J. A., (1995) Mathematical Statistics and Data Analysis (Wadsworth, Belmont, CA).
- 38.Li L., Gerecke, E. E. & Zolan, M. E. (1999) Chromosoma 108, 384-392. [DOI] [PubMed] [Google Scholar]
- 39.Halsall J. R., Milner, M. J. & Casselton, L. A. (2000) Genetics 154, 1115-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loidl J., Nairz, K. & Klein, F. (1991) Chromosoma 100, 221-228. [DOI] [PubMed] [Google Scholar]
- 41.Maguire M. P. & Riess, R. W. (1994) Genetics 137, 281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McClintock B. (1933) in Zeitschrift fur Zellforschung und Mikroskopische Anatomie, eds. Goldschmidt, R. & von Mollendroff, W. (Springer, Berlin), pp. 191–237.
- 43.Binninger D. M., Skrzynia, C., Pukkila, P. J. & Casselton, L. A. (1987) EMBO J. 6, 835-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casselton L. A. & Olesnicky, N. S. (1998) Microbiol. Mol. Biol. Rev. 62, 55-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Shea S. F., Chaure, P. T., Halsall, J. R., Olesnicky, N. S., Leibbrandt, A., Connerton, I. F. & Casselton, L. A. (1998) Genetics 148, 1081-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanda T., Arakawa, H., Yasuda, Y. & Takemaru, T. (1990) Exp. Mycol. 14, 218-226. [Google Scholar]
- 47.Merino S. T., Cummings, W. J., Acharya, S. N. & Zolan, M. E. (2000) Proc. Natl. Acad. Sci. USA 97, 10477-10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirano T. (2002) Genes Dev. 16, 399-414. [DOI] [PubMed] [Google Scholar]
- 49.Tavassoli M., Shayeghi, M., Nasim, A. & Watts, F. Z. (1995) Nucleic Acids Res. 23, 383-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiner B. M. & Kleckner, N. (1994) Cell 77, 977-991. [DOI] [PubMed] [Google Scholar]
- 51.Nairz K. & Klein, F. (1997) Genes Dev. 11, 2272-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Usui T., Ohta, T., Oshiumi, H., Tomizawa, J., Ogawa, H. & Ogawa, T. (1998) Cell 95, 705-716. [DOI] [PubMed] [Google Scholar]
- 53.Celerin M., Merino, S. T., Stone, J. E., Menzie, A. M. & Zolan, M. E. (2000) EMBO J. 19, 2739-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dernburg A. F., Sedat, J. W., Cande, W. Z. & Bass, H. W. (1995) in Telomeres, eds. Blackburn, E. H. & Greider, C. W. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 295–338.
- 55.Scherthan H. (1997) Chromosomes Today 12, 217-248. [Google Scholar]
- 56.Walker M. Y. & Hawley, R. S. (2000) Chromosoma 109, 3-9. [DOI] [PubMed] [Google Scholar]
- 57.Zickler D. & Kleckner, N. (1998) Annu. Rev. Genet. 32, 619-697. [DOI] [PubMed] [Google Scholar]