Abstract
ATP-dependent proteases, like FtsH (HflB), recognize specific protein substrates. One of these is the λ CII protein, which plays a key role in the phage lysis-lysogeny decision. Here we provide evidence that the conserved C-terminal end of CII acts as a necessary and sufficient cis-acting target for rapid proteolysis. Deletions of this conserved tag, or a mutation that confers two aspartic residues at its C terminus do not affect the structure or activity of CII. However, the mutations abrogate CII degradation by FtsH. We have established an in vitro assay for the λ CIII protein and demonstrated that CIII directly inhibits proteolysis by FtsH to protect CII and CII mutants from degradation. Phage λ carrying mutations in the C terminus of CII show increased frequency of lysogenization, which indicates that this segment of CII may itself be sensitive to regulation that affects the lysis-lysogeny development. In addition, the region coding for the C-terminal end of CII overlaps with a gene that encodes a small antisense RNA called OOP. We show that deletion of the end of the cII gene can prevent OOP RNA, supplied in trans, interfering with CII activity. These findings provide an example of a gene that carries a region that modulates stability at the level of mRNA and protein.
Proteolysis by ATP-dependent proteases is an important mechanism for the rapid control of gene activity, the removal of unfolded inactive proteins, and the elimination of incomplete polypeptides. In bacteria, proteolysis acts on key regulatory transcription factors affecting the heat shock (σ32), stationary phase (σs), and the SOS DNA repair system (LexA) responses, capsular polysaccharide biosynthesis, and the control of the lysis-lysogeny decision of phage λ (1). FtsH is a membrane-bound ATP-dependent protease in which the ATPase domain and the protease domain are linked in one polypeptide (2). FtsH orthologs are found within mitochondria and chloroplasts in higher organisms (3, 4). The number of native proteins known to be substrates for FtsH is rather small and includes the heat shock sigma factor σ32, SecY, YccA, subunit a of the membrane-embedded F0 part of the H+-ATPase, as well as phage λ CII, CIII, and Xis proteins (see ref. 5). However, the signal(s) by which these native substrates are recognized by FtsH is not known. FtsH, as well as Tsp, ClpAP, and ClpXP recognize the SsrA peptide tag that is added to the C terminus of incomplete proteins by trans-translation (6).
The lysogenic response established after infection of Escherichia coli by the temperate bacteriophage λ requires the initial synthesis of the CI repressor from the pE promoter and the integration protein Int, from the pI promoter. In addition, establishment of viable lysogenic cells requires the expression of the paQ promoter that inhibits lytic gene expression. The phage CII protein, which is 97 amino acid residues long, plays a key initiating role in these processes by activating the pE, pI, and paQ promoters (7). CII itself is regulated at many levels: transcription, translation initiation, mRNA stability, and proteolysis. Studies strongly indicate that increased CII stability favors the lysogenic response (8). It was shown that FtsH rapidly degrades CII protein (9, 10); and that the CIII protein favors the lysogenic decision by stabilizing CII (11, 12).
Materials and Methods
Plasmids and Proteins.
Plasmids carrying derivatives of cII were constructed by inserting a PCR product of the cII gene into plasmid pET-15b (Novagen) between the NdeI and BamHI restriction sites, CII protein has been expressed and purified as described (13). For in vivo assays of CII activity, pGB2 plasmid derivatives were constructed (8). These plasmids, in which the CII is expressed from the tac promoter, were introduced into strain A5039 E. coli lac IQ lacZ− cells. For CIII expression the cIII gene was PCR-amplified and cloned between the BamHI and HindIII sites of plasmid pQE30 (Qiagen, Valencia, CA), generating plasmid pOK8 in which CIII is made as a fusion to NH2-MRGSHHHHHHGS, CIII is expressed and purified under denaturing conditions by using the Qiagen protocol. Refolding was done on Ni-nitrilotriacetic acid matrix with PBS, CIII was eluted with PBS containing 500 mM imidazole, and the imidazole was removed by G25 spin column. For the construction of CI-CII protein fusions, the N terminus of cI (codons 1–94) was PCR-amplified from a plasmid carrying the cI gene (obtained from C. Herman, University of California, San Francisco) as part of a 3′ oligo coding for the last nine amino acids of CII. This construct was then cloned between the BamHI and HindIII sites of pQE30. The CI-CII fusions, His-6-CI-CII89–97 and His-6-CI-CII89–95DD were expressed in A9863 (W3110 clpP:cat ftsH1) and purified under native conditions on HiTrap-chelating HP column (Amersham Pharmacia). Strain A9863 was constructed by P1 transduction by introducing the clpP:cat mutation from strain SG22098 (S. Gottesman, National Institutes of Health) into W3110 and ftsH1:tet from strain AR754 (2). Plasmid pJV937-7 carries the OOP gene under the control of PLlacO-1 promoter (14) and the lac IQ gene, was kindly provided by J. Vogel. GST-FtsH fusion protein was expressed and purified as described (10).
Proteolysis Experiments.
In vitro assays for FtsH proteolysis were performed under standard conditions as described (10) and the proteins were resolved on a NuPAGE 4–12% Bis-Tris precast gel (Invitrogen) by using Mes buffer and visualized by Coomassie stain or immunostaining. Similar conditions were used for the inhibition of FtsH by CIII. Digestion of CII with trypsin (Sigma) (5 ng/μl) was performed at 4°C in a buffer containing: 50 mM Tris acetate, pH 8/5 mM Mg acetate/12.5 μM Zn acetate/80 mM NaCl/1.4 mM β-mercaptoethanol. The proteins were precipitated by 10% trichloroacetic acid and the protein bands were resolved on a NuPAGE 4–12% Bis-Tris precast gel.
CD Spectra.
The CD spectra of the CII proteins were recorded in PBS at 25°C by using a Jobin et Yvon CD6 spectropolarimeter as described (15). Thermal melts of CII WT and CII1–82 were performed by using identical conditions that were monitored by changes in CD ellipticity at 222 nm with a 2-min equilibration time and a 15-s averaging time at each temperature. The normalized ellipticity was calculated as (θ − θ75)/(θ25 − θ75), where θ is the ellipticity at 222 nm at each temperature and θ25 and θ75 are the ellipticities at 222 nm at 25 and 75°C, respectively.
Construction of λ Mutant by Recombineering.
For the introduction of the cII mutations into phage λ, strain DY374 (W3110 λ cI857Δcro-bio nadA:Tn10 gal490) (16) was grown at 30°C and concentrated by centrifugation in TM (10 mM Tris, pH 7.4/10 mM MgSO4) and infected for 15 min on ice with λ+. Infected cells were diluted in LB medium and induced for recombinant Red functions at 42°C for 15 min, rapidly cooled in ice, and electroporated with 70-nt-long oligonucleotides allowing for a 35-nt-long homology from both sides of the mutations. The cells were immediately diluted into 5 ml of warm LB, shaken vigorously for 90 min at 39°C, and plaques carrying the mutation, at a level of ≈4%, were identified by PCR or hybridization (the DD mutant) and confirmed by DNA sequencing. To test for the importance of the sequence coding for amino acid residues 77–81, we first generated the deletion mutant λcII1–77 that forms clear plaques. In further crosses with λcII1–77 and a set of oligonucleotides to add additional codons, we searched for recombinants able to form turbid plaque, only the oligo that generated the deletion λcII1–81 was found to yield turbid plaque-forming recombinants.
Results
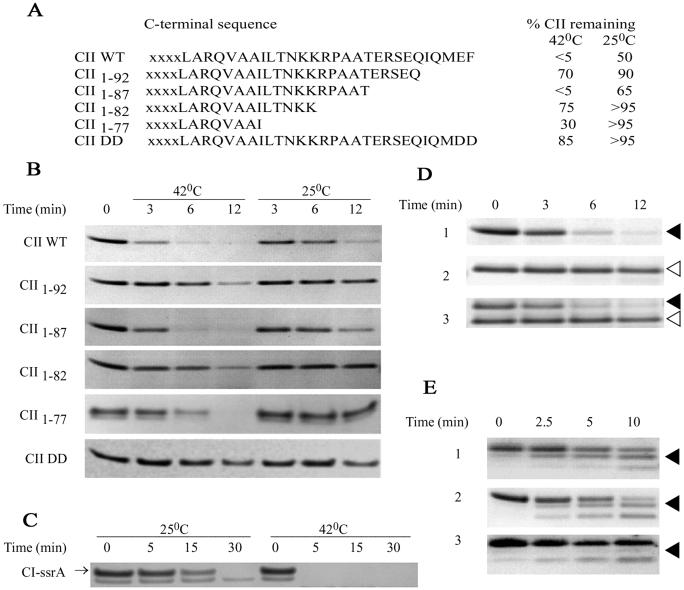
To discover the signal for rapid FtsH-dependent degradation of CII, several CII mutations were constructed, and protein stability was determined in vitro (Fig. 1 A and B). A set of C-terminal truncations, CII1–92 CII1–82, and CII1–77, greatly stabilize CII against degradation by FtsH, reducing the rate of proteolysis by ≈5-fold. A larger deletion of the C terminus CII1–70 has also been shown to be proteolytically stable in vivo and in vitro (M. Gonzales, Y. Shotland, and A.B.O., unpublished data). The CII1–87 mutant protein, lacking the last 10 residues of CII, is rapidly degraded. In contrast, we found that a deletion of five residues from the N terminus of CII did not stabilize the protein (not shown). The CIIcan1 mutant, a Val-to-Ala change at residue 2 was originally selected as a phage mutant forming turbid plaques in the absence of CIII (17). We found that this mutation had little effect in stabilizing CII in vitro (not shown). In other studies, SsrA-tagged proteins carrying two Asp residues at the C terminus (DD mutants) become more resistant to proteolysis then normal SsrA-tagged protein (6). We found that the mutant protein in which the Glu-Phe at the C terminus of CII was replaced by the DD mutation is highly resistant to proteolysis by FtsH (Fig. 1 A and B). These findings demonstrate that the composition of the C terminus is important in FtsH-mediated proteolysis of CII.
Fig 1.
Proteolysis of carboxyl-terminal mutations. (A) The sequence of the CII mutations and the percent of CII remaining after 6 min of degradation as determined by quantitative analysis of Coomassie staining of SDS/PAGE. (B) A representative proteolysis experiment visualizing CII by Coomassie staining after separation on SDS/PAGE. Lane 1 shows the initial amount of CII and lanes 2–4 and lanes 5–7 show the amount of CII remaining after 3, 6, and 12 min of incubation under standard conditions, at 42 and 25°C, respectively. (C) His-6-CI-ssrA (180 pmol) were incubated with 13.6 pmol of FtsH, in 80 μl, and 20-μl samples were taken in the indicated time at 25 (lanes 1–4) and 42°C (lanes 5–8) and visualized as in B. (D) CII WT (reaction 1, filled arrowhead) or mutant protein CII1–82 (reaction 2, open arrowhead) were incubated with FtsH under standard conditions at 42°C as described in C. In reaction 3, CII WT and CII1–82 were incubated together during the proteolysis reaction. The proteins were visualized as in B. (E) Purified WT CII (90 pmol) (reactions 1 and 3) and CII-DD (90 pmol) (reaction 2) were digested with trypsin for the indicated time. CII was visualized with CII antibodies raised against the last 20 residues of CII (reaction 3), or by Coomassie staining (reactions 1 and 2). The arrowhead denotes the position of the truncated CII.
It was previously found that low temperature favors the lysogenic response and greatly reduces the lytic response of phage λ. This phenotype was correlated with greatly increased CII protein stability in vivo (8). This increased stability at low temperature may result from CII structural changes, reduced FtsH activity, or the presence of additional host factors that may affect CII proteolysis. We tested the temperature-dependent degradation of CII in vitro. WT CII and the CII mutants are degraded more rapidly at 42 than at 25°C (Fig. 1 A and B). Likewise, the CI protein tagged with the SsrA peptide, CI-SsrA, is much more resistant at 25°C (Fig. 1C). These results suggest that FtsH has a reduced activity at low temperature.
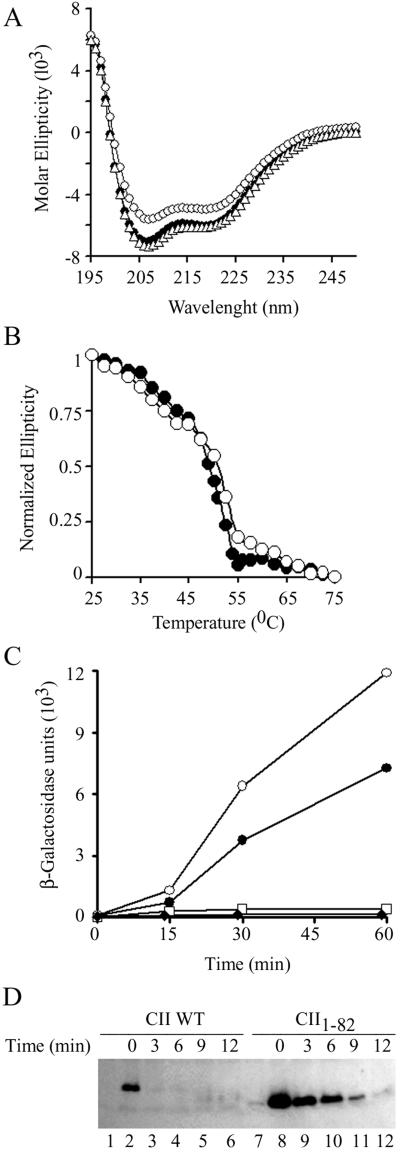
Misfolded protein aggregates are known to resist proteolysis (18). To check whether the increased stability of the mutant proteins is due to protein misfolding, we performed far-UV CD analysis of purified CII WT, CII-DD and CII1–82. The complete spectrum of the three proteins was found to be very similar (Fig. 2A). However, the spectrum of CII1–82 indicates a lower α-helical content suggesting that the deletion eliminates a short α-helical region. Furthermore, denaturation curves of WT and CII1–82 as a function of temperature show that the two proteins possess the same denaturation profile (Fig. 2B). We conclude that the mutations did not affect CII protein structure suggesting that the increased resistance to proteolysis is due to the intrinsic property of the C terminus and not to an indirect effect on CII protein structure.
Fig 2.
Structural and functional characterization of C-terminal mutant proteins. (A) Circular dichroism UV spectra of CII WT, 10 μM (•); CII-DD, 20 μM (▵), and CII1–82, 10 μM (○). (B) Denaturation of the CII WT (•) and CII1–82 (○) proteins as a function of temperature monitored by changes in circular dichroism ellipticity at 222 nm. (C) E. coli A5039 strain (lac IQ lacZ−) carrying two compatible plasmids were grown at 37°C on LB supplemented with 50 μg/ml ampicillin and 50 μg/ml spectinomycin. When the cultures reached an OD600 of 0.2, they were transferred to 42°C, the CII, expressed from the tac promoter, was induced with 1 mM isopropyl β-D-thiogalactoside, and samples were assayed for β-galactosidase activity at the indicated time points. This strain carries the plasmid pHG92 that expresses the lacZ gene under the control of the CII-regulated pE promoter. It also carries either the compatible vector plasmid pGB2 (⧫), pSKCII (WT cII cloned in pGB2) (•), pSKCII1–82 (○), or pSK CII1–77 (□). (D) To cells from the same experiment 200 μg/ml chloramphenicol was added at 60 min after induction and samples were taken at the indicated times after chloramphenicol addition (pSKCII, lanes 2–6, and pSKCII1–82, lanes 8–12) and placed on ice. The cells were then centrifuged, lysed, and loaded on an SDS-polyacrylamide gel and CII was visualized by antibodies raised against full-length CII. Lanes 1 and 7 show the level of CII in cells before the addition of isopropyl β-D-thiogalactoside.
CII was found in equilibrium between the monomeric and tetrameric forms (19). To determine whether the C terminus acts only in cis, we incubated CII WT together with CII1–82 and followed their proteolysis by FtsH. The results presented in Fig. 1D show that the presence of both proteins in the same reaction mixture did not affect the individual rate of degradation of either protein. The WT CII was rapidly degraded whereas CII1–82 was degraded very slowly. This result was not affected by the relative ratio of the two proteins (data not shown). Thus, the WT CII cannot stimulate the proteolysis of the CII1–82 deletion mutant suggesting that the C terminus act only in cis. These results further suggest that CII interact with the catalytic site of FtsH as a monomer.
It is possible that the C terminus of CII is partially exposed enabling it to act as a target for recognition and proteolysis by FtsH. To test whether the CII C terminus is exposed, purified CII and CII-DD were partially hydrolyzed by trypsin and intermediates were separated on SDS/PAGE (Fig. 1E). We found that two major degradation products accumulated. The larger degradation product of ≈8 kDa indicates that ≈20 residues of CII were cleaved away (Fig. 1E, lines 1 and 2) probably at the Lys-Lys-Arg cluster (see Fig. 1A). Antibodies raised against the last 20 amino acids of CII recognize only the full-length protein but do not recognize the 8-kDa degradation product, showing that trypsin treatment removed the C terminus of CII (Fig. 1E, line 3). These results suggest that the C-terminal end of CII may be exposed and available to interact not only with trypsin but also with FtsH.
We tested the ability of CII and the deletion mutants to activate transcription from the pE promoter. Two plasmids were introduced into a bacterial strain deleted for the LacZ gene. One plasmid carries the CII gene under the control of the Lac repressor. The second plasmid carries the LacZ reporter gene downstream to the CII-dependent pE promoter. Induction of CII WT leads to an ≈30-fold increase in β-galactosidase activity within 60 min (Fig. 2C). Expression of CII1–82 led to a 50-fold increase in β-galactosidase activity. In contrast, the CII1–77 was defective and unable to stimulate β-galactosidase expression. We conclude that the terminal 15 amino acids of CII are not essential for transcriptional activation of pE. Direct measurements of CII levels in vivo show higher levels and increased stability of the CII1–82 mutant over WT CII (Fig. 2D). The 3-fold increase in level of accumulated CII may account for the increase in pE promoter activity (Fig. 2C).
The plasmids expressing CII and its derivatives were tested for their ability to complement for λ phage defective in CII. Cells carrying the vector or the plasmid that express CII1–77 did not complement for CII. WT CII and CII1–82 expression from the plasmid inhibited λ plaque formation including that of λ cI mutants (with or without isopropyl β-D-thiogalactoside induction). This finding suggests that CII inhibits lytic phage functions, perhaps through an effect on phage DNA replication and/or Q gene expression.
Other genetic studies also suggest that the C terminus of CII is not required for its lysogenic activity (20). By using an assay for the detection of λ CII mutations in transgenic mice, on the basis of the recovery of functionally defective CII mutants from an integrated λ genome, more than 100 single amino acid changes in CII were analyzed. All mutations were found to cluster within the first 77 codons of CII (S. Garges and S. Adhya, personal communication) (21, 22).
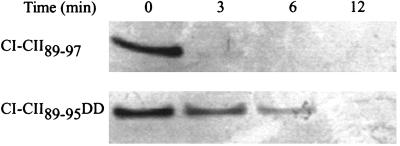
To test whether the C-terminal end of CII is necessary and sufficient for proteolysis by FtsH, a fusion protein was made in which the last nine residues of CII are fused to the N-terminal domain of CI. This protein named CI-CII89–97 was degraded in vitro by FtsH (Fig. 3), at about the same rate as CI-SsrA (not shown), whereas CI-CII89–95DD was more stable (Fig. 3). However, both the CI-CII89–97 and CI-CII89–95DD are less stable than CII and CII-DD, respectively (see Fig. 1B) suggesting that the protease target region is more accessible in the CI-CII fusions. These findings show that the C-terminal end of CII is sufficient for proteolysis and indicates that FtsH recognizes the full-length CII and the nine-residue tag present in CI-CII89–97. In preliminary experiments we found that we could not isolate the CI-CII89–97 protein from WT cells or in a strain carrying a temperature-sensitive FtsH but only from a double-mutant strain carrying both the ftsH1ts and a clpP:Kn double mutant suggesting that this hybrid protein may be degraded by more than one protease. We recently found that CII can be degraded by ClpAP and ClpYQ in vitro (O.K, S. K. Singh, S.K., M. R. Maurizi, and A.B.O., unpublished results).
Fig 3.
The C terminus of CII is a sufficient signal for degradation by FtsH. CI-CII protein fusions (160 pmol) were incubated in the presence of 13.6 pmol of FtsH, in 80 μl, at 42°C for the indicated times, separated on SDS/PAGE, and visualized by Coomassie staining.
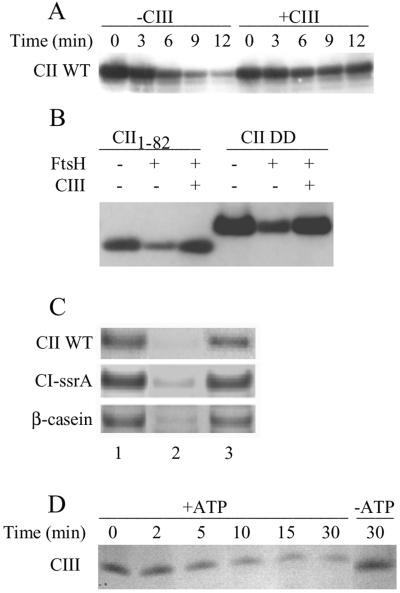
To test whether the CII C terminus is required for the action of CIII in preventing CII proteolysis, we set up an in vitro system to detect the antiproteolytic activity of CIII. Purified CIII greatly increased the stability of WT CII, CII1–82 and CII-DD against FtsH (Fig. 4 A and B). These results lead us to conclude that the action of CIII, in protecting CII from FtsH, does not require the CII C terminus. In addition, FtsH degradation of CI-SsrA and of β-casein was also inhibited by CIII (Fig. 4C). Thus, CIII seems to be a direct and general inhibitor of FtsH that does not require additional phages or host factors for its action. CIII was found to be protective only when present in excess relative to FtsH (data not shown), which presumably is because FtsH can also degrade CIII in an ATP-dependent manner (Fig. 4D).
Fig 4.
CIII prevents degradation by FtsH. (A) Proteolysis of 50 pmol of CII by 12.5 pmol of FtsH under standard conditions in the absence or presence of 250 pmol of His-6-CIII in 100 μl. Samples were taken at the indicated times, and CII was separated on SDS/PAGE and visualized by CII antibodies raised against full-length CII. (B) Inhibition of CII1–82 and CII-DD degradation by His-6-CIII. Reactions were incubated for 30 min at 42°C assayed as in A. (C) Inhibition by CIII (70 pmol) of degradation of CII (45 pmol), CI-SsrA (45 pmol), or β-casein (25 pmol) by FtsH (3.4 pmol). Lane 1, initial input; lane 2, degradation for 5 min without CIII; lane 3, degradation for 5 min in the presence of His-6-CIII. CII was separated on SDS/PAGE and the substrate was visualized by Coomassie staining. (D) Degradation of 462 pmol of His-6-CIII by 23.8 pmol of FtsH. Samples were taken in the indicated times in the presence (lanes 2–6) or absence (lane 7) of 5 mM ATP. CIII was separated on SDS/PAGE and the substrate was visualized by Coomassie staining.
We introduced the mutation CII-DD and the deletions CII1–77, CII1–81, and CII1–82 into the λ genome by oligonucleotide recombination (16). The phage mutants λ cII1–81, λ cII1–82, and λ cIIDD mutants were found to form turbid plaques on WT host indistinguishable from WT phage, whereas λ cII1–77 formed clear plaques. Genetic analysis indicates that λ cII1–78, λ cII1–79, and λ cII1–80 form clear plaques like λ cII1–77 (see Materials and Methods) suggesting that only the first 80 amino acids are essential for CII activity.
Quantitative assays of lysogenization frequencies showed that the highest yield of lysogens was obtained at 37°C when infecting stationary cells. Lysogenization frequencies were greatly reduced on infection of exponentially growing cells. Infection by λ cII1–82 yielded a similar number of lysogens to that obtained with WT phage. We consistently found that λ cIIDD yielded a lower number of lysogens at 37°C when infecting exponentially growing cells. However, the rate of lysogeny by WT phage was greatly reduced (16-fold) at 42°C, whereas the decrease in the number of lysogens after infection by λ cIIDD and λ cII1–82 mutants was much smaller (8.3 and 5.5, respectively; Table 1). These results are in line with our finding that FtsH can efficiently degrade CII at 42°C (Fig. 1). At 25°C, no significant differences in lysogeny frequency were found between the various phages; at this temperature, FtsH proteolytic activity is greatly reduced. These results support the hypothesis that the C terminus of CII sensitizes CII to proteolysis and in this way participates in determining the level of lysogeny.
Table 1.
Increased lysogenic response by λ cll mutants
| Infecting phage
|
% Lysogeny | Relative lysogenization | ||||
|---|---|---|---|---|---|---|
| Stationary phase | Exponential phase | 25/37°C | 42/37°C | OOP+/OOP− (low moi) | OOP+/OOP− (high moi) | |
| λ c+ | 41 | 0.55 | 0.12 | 0.06 | 0.11 | 0.24 |
| λ cII-DD | 60 | 0.15 | 0.14 | 0.12 | 1.1 | 0.13 |
| λ cII1–82 | 42 | 0.59 | 0.15 | 0.18 | 2.3 | 0.5 |
Bacterial cultures (W3110; W3110/pJV937-7) were grown, in LB medium and in LB-ampicilin medium in the presence of isopropyl β-d-thiogalactoside, respectively, and phage was adsorbed at room temperature for 15 min, at a multiplicity of 0.01 except for the high-multiplicity experiment [multiplicity of infection (moi) of 3]. The infected cells were diluted into LB medium and incubated at the indicated temperature for 30 min before spreading on LB plates containing 30 μg of kanamycin per ml, and the number of kanamycin-resistant colonies was counted. Lysogens were scored as KmR colonies and the numbers are given as a percentage of the input-infecting phage (in high moi, lysogens were scored as KmR colonies as a percentage of the total cells infected).
Percent lysogeny was determined for cells grown at 37°C throughout the experiment.
Relative lysogenization was calculated as the ratio between the percent lysogens at the indicated temperature to the percent lysogens at 37°C or the ratio between the percent lysogens on infection of strain W3110/pJV937-7 (+OOP) to the percent lysogens obtained on infection of strain W3110 (−OOP).
The infecting phage for all experiments carried the kan cassette (36).
For the experiment at 25°C, cells were grown exponentially at 30°C before infection. For the experiment at 42°C, cells were grown exponentially at 42°C before infection.
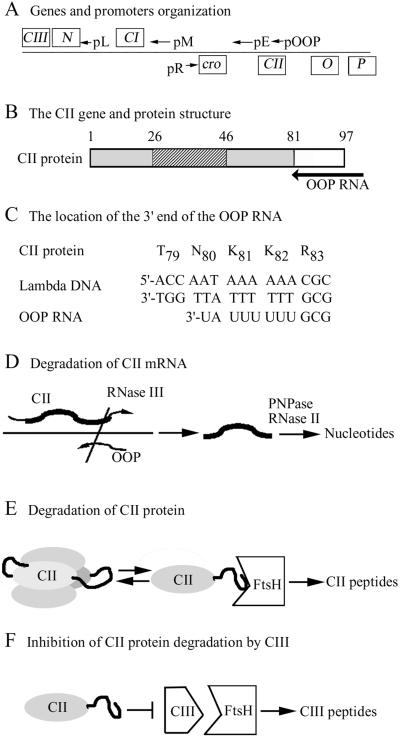
The C-terminal region of the CII gene overlaps with the cis-encoding sequence for OOP, a short 77-base-long antisense RNA, initiating downstream of CII and ending with a Rho-independent terminator within CII. This RNA covers the last 17 codons of CII (Fig. 5 A–C) (23) and coincides with the region encoding the CII protease-sensitive target. OOP acts as an antisense RNA to CII and reduces CII expression by increasing the rate of CII mRNA hydrolysis, which is initiated by cleavage of the double-stranded RNA by RNase III (24). On cells, in which OOP RNA is expressed, in trans from a plasmid, λ WT and λ cIIDD form clear plaques, whereas λ cII1–81 and λ cII1–82 form turbid plaques. Quantitative assays showed that OOP RNA reduces the lysogenic frequency ≈10-fold by λ WT phage infection at low multiplicity, but had no effect on λ cIIDD or λ cII1–82 mutant phages (Table 1). At high multiplicity of infection the differential effects of the OOP RNA was not so great. These results demonstrate that the C-terminal region of the cII gene is essential for the response to OOP RNA.
Fig 5.
A schematic model for posttranscriptional control of CII. (A) A Map of the immunity region of λ phage showing the early promoters and the direction of transcription (arrows) and the relevant genes (boxed). (B) The CII protein domains: striped, DNA-binding domain; gray, regions essential for CII activity (1–81); open box, the C-terminal domain. The position of the region coding for OOP RNA is shown by an arrow. The coordinates of the domain are indicated. (C) The position of the CII-coding sequence around the 3′ end of the OOP antisense RNA. (D) CII mRNA degradation induced by OOP RNA. The coding region of CII mRNA is shown in bold; the OOP antisense RNA is presented by an arrow. Cleavage of the CII/OOP double-stranded RNA by RNase III initiate the hydrolysis of CII mRNA by RNase II and polynucleotide phosphorylase (PNPase). (E) Proteolytic degradation of CII by FtsH. The C-terminal end of CII is shown as a thick line. (F) CIII inhibit CII degradation by interacting with FtsH. CIII is a substrate for FtsH.
Discussion
We have developed an in vitro system that allowed us to extend our investigation of the molecular basis of substrate selectivity by FtsH. We have found that although both σ32 and CII are degraded at a similarly rapid rate in vivo, degradation of σ32 in a purified system was at least 10-fold slower than that of CII (10). Furthermore, proteolysis of σ32, but not of CII, requires the presence of the DnaK-DnaJ-GrpE chaperone machine (10, 25, 26). The region of σ32 that is recognized by FtsH is not known (27, 28). In contrast, no chaperone activity was found to be required for CII degradation by FtsH. Our results show that the region in CII recognized by FtsH is restricted to a short C-terminal tail. It acts as a necessary and sufficient cis-acting target for rapid proteolysis. The sequence of the last five amino acids of CII, which is highly conserved among other CII-like proteins of lambdoid phages, is unique to the phage and is not found in any E. coli protein. As recently shown by NMR studies, the C-terminal part of CII is flexible (29), which is supported by its sensitivity to trypsin. Our results define the native integral target for a cytoplasmic protein recognized by FtsH. Deletion of up to 15 residues of the C terminus does not affect the structure or activity of CII in the stimulation of transcription initiation We found that CII shorter than 81 residues was nonfunctional. Very limited sequence similarity exists between the C terminus of CII and the SsrA peptide tag, both of which are recognized by FtsH. The SsrA tag provides a set of overlapping determinants for the interaction of the tag with proteases ClpX, ClpA, and with SspB (30).
Active CII is found as a tetramer in equilibrium with monomers, dimers, and trimers. It was suggested that in the activation of transcription, CII binds to DNA as a tetramer (19). We interpret our finding of a cis-acting C-terminal protease target to suggest that CII interact with the catalytic site of FtsH as a monomer. In support of this hypothesis, it was shown that CII mutants that are defective in tetramer formation, are less stable in vivo (20). Furthermore, in vitro proteolysis of chemically cross-linked CII showed that CII trimers and tetramers were resistant to proteolysis (31). Thus, it is possible that in the tetramer form the C-terminal peptide is inaccessible to FtsH.
We have established an in vitro assay for CIII and demonstrated that it inhibits FtsH proteolytic activity. Our initial experiments suggest that it may act as a competitive inhibitor. It was previously shown that the central region of CIII, residues 14–37, is sufficient for its protective action in vivo (32). We observed that a peptide coding for residues 14–37 of CIII acts both as a substrate and an inhibitor of FtsH in vitro depending on its relative concentration and time of incubation (data not shown).
Control by antisense RNA is an efficient and specific means of regulating gene expression. For its function, antisense RNA assumes a specific structure and requires base pairing with target RNA. Trans-action by antisense RNA is found to regulate a large number of genes of E. coli (33–35). In contrast, several genes that are found on extra chromosomal elements, such as transposons, phages, and plasmids, were found to be regulated by a cis-acting antisense RNA. Our genetic studies indicate that the last 16 codons of CII that are overlapped by the OOP coding region are not essential for CII protein activity. Thus, the 3′ end of the cII gene has evolved to encode information for regulating both mRNA and protein stability (Fig. 5 D and E).
Acknowledgments
We thank C. Herman and M. Gonzales for unpublished results; C. Herman for the plasmid expressing CI-SsrA; J. Vogel for the plasmid expressing OOP RNA; M. Gonzales for CII antibodies; and S. Gottesman for bacterial strains. We thank S. Gottesman, M. Maurizi, Y. Shlomai, and S. Altuvia for stimulating discussion. This research was supported by Israel Science Foundation Grant 489/01-1.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gottesman S. (1996) Annu. Rev. Genet. 30, 465-506. [DOI] [PubMed] [Google Scholar]
- 2.Tomoyasu T., Gamer, J., Bukau, B., Kanemori, M., Mori, H., Rutman, A. J., Oppenheim, A. B., Yura, T., Yamanaka, K., Niki, H., et al. (1995) EMBO J. 14, 2551-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindahl M., Spetea, C., Hundal, T., Oppenheim, A. B., Adam, Z. & Andersson, B. (2000) Plant Cell 12, 419-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casari G., De Fusco, M., Ciarmatori, S., Zeviani, M., Mora, M., Fernandez, P., De Michele, G., Filla, A., Cocozza, S., Marconi, R., et al. (1998) Cell 93, 973-983. [DOI] [PubMed] [Google Scholar]
- 5.Karata K., Inagawa, T., Wilkinson, A. J., Tatsuta, T. & Ogura, T. (1999) J. Biol. Chem. 274, 26225-26232. [DOI] [PubMed] [Google Scholar]
- 6.Gottesman S., Roche, E., Zhou, Y. & Sauer, R. T. (1998) Genes Dev. 12, 1338-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman D. I. (1992) Curr. Opin. Genet. Dev. 2, 727-738. [DOI] [PubMed] [Google Scholar]
- 8.Obuchowski M., Shotland, Y., Koby, S., Giladi, H., Gabig, M., Wegrzyn, G. & Oppenheim, A. B. (1997) J. Bacteriol. 179, 5987-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kihara A., Akiyama, Y. & Ito, K. (1997) Proc. Natl. Acad. Sci. USA 94, 5544-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shotland Y., Koby, S., Teff, D., Mansur, N., Oren, D. A., Tatematsu, K., Tomoyasu, T., Kessel, M., Bukau, B., Ogura, T. & Oppenheim, A. B. (1997) Mol. Microbiol. 24, 1303-1310. [DOI] [PubMed] [Google Scholar]
- 11.Hoyt M. A., Knight, D. M., Das, A., Miller, H. I. & Echols, H. (1982) Cell 31, 565-573. [DOI] [PubMed] [Google Scholar]
- 12.Akiyama Y., Kihara, A. & Ito, K. (1996) FEBS Lett. 399, 26-28. [DOI] [PubMed] [Google Scholar]
- 13.Shotland Y., Shifrin, A., Ziv, T., Teff, D., Koby, S., Kobiler, O. & Oppenheim, A. B. (2000) J. Bacteriol. 182, 3111-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lutz R. & Bujard, H. (1997) Nucleic Acids Res. 25, 1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shotland Y., Teff, D., Koby, S., Kobiler, O. & Oppenheim, A. B. (2000) J. Mol. Biol. 299, 953-964. [DOI] [PubMed] [Google Scholar]
- 16.Ellis H. M., Yu, D., DiTizio, T. & Court, D. L. (2001) Proc. Natl. Acad. Sci. USA 98, 6742-6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones M. O. & Herskowitz, I. (1978) Virology 88, 199-212. [DOI] [PubMed] [Google Scholar]
- 18.Wickner S., Maurizi, M. R. & Gottesman, S. (1999) Science 286, 1888-1893. [DOI] [PubMed] [Google Scholar]
- 19.Ho Y., Lewis, M. & Rosenberg, M. (1982) J. Biol. Chem. 257, 9128-9134. [PubMed] [Google Scholar]
- 20.Ho Y. S., Mahoney, M. E., Wulff, D. L. & Rosenberg, M. (1988) Genes Dev. 2, 184-195. [DOI] [PubMed] [Google Scholar]
- 21.Jakubczak J. L., Merlino, G., French, J. E., Muller, W. J., Paul, B., Adhya, S. & Garges, S. (1996) Proc. Natl. Acad. Sci. USA 93, 9073-9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harbach P. R., Zimmer, D. M., Filipunas, A. L., Mattes, W. B. & Aaron, C. S. (1999) Environ. Mol. Mutagen. 33, 132-143. [DOI] [PubMed] [Google Scholar]
- 23.Hendrix R. W., Roberts, J. W., Franklin, S. W. & Weisberg, R. A., (1983) Lambda II (Cold Spring Harbor Lab. Press, Plainview, NY).
- 24.Krinke L. & Wulff, D. L. (1990) Genes Dev. 4, 2223-2233. [DOI] [PubMed] [Google Scholar]
- 25.Tatsuta T., Tomoyasu, T., Bukau, B., Kitagawa, M., Mori, H., Karata, K. & Ogura, T. (1998) Mol. Microbiol. 30, 583-593. [DOI] [PubMed] [Google Scholar]
- 26.Tilly K., McKittrick, N., Zylicz, M. & Georgopoulos, C. (1983) Cell 34, 641-646. [DOI] [PubMed] [Google Scholar]
- 27.Bertani D., Oppenheim, A. B. & Narberhaus, F. (2001) FEBS Lett. 493, 17-20. [DOI] [PubMed] [Google Scholar]
- 28.Tomoyasu T., Arsene, F., Ogura, T. & Bukau, B. (2001) J. Bacteriol. 183, 5911-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Datta A. B., Chakrabarti, P., Subramanya, H. S. & Parrack, P. (2001) Biochem. Biophys. Res. Commun. 288, 997-1000. [DOI] [PubMed] [Google Scholar]
- 30.Flynn J. M., Levchenko, I., Seidel, M., Wickner, S. H., Sauer, R. T. & Baker, T. A. (2001) Proc. Natl. Acad. Sci. USA 98, 10584-10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shotland Y., (1999) Ph.D. thesis (Hebrew Univ., Jerusalem).
- 32.Kornitzer D., Altuvia, S. & Oppenheim, A. B. (1991) Proc. Natl. Acad. Sci. USA 88, 5217-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wassarman K. M., Repoila, F., Rosenow, C., Storz, G. & Gottesman, S. (2001) Genes Dev. 15, 1637-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Argaman L., Hershberg, R., Vogel, J., Bejerano, G., Wagner, E. G., Margalit, H. & Altuvia, S. (2001) Curr. Biol. 11, 941-950. [DOI] [PubMed] [Google Scholar]
- 35.Lease R. A. & Belfort, M. (2000) Proc. Natl. Acad. Sci. USA 97, 9919-9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dennis J. J. & Zylstra, G. J. (1998) Appl. Environ. Microbiol. 64, 2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]