Abstract
Tumor reversion is the process by which some cancer cells lose their malignant phenotype. This study was aimed at defining some of the molecular and phenotypic properties of this process. Biological models of tumor reversion were isolated from human leukemia and breast cancer cell lines by using the H-1 parvovirus as a selective agent. Differential gene expression analysis was performed between the parental malignant cells and their revertants or alternatively between these parental cells and their SIAH-1 transfectant counterparts. These SIAH-1 transfectants have a suppressed malignant phenotype and were used as a control for a viral-free system. Two hundred sixty-three genes were found to be either activated or inhibited during the reversion process, as confirmed by Northern blot analysis or quantitative PCR. Of these, 32% were differentially expressed in all systems, irrespective of whether parvovirus-selected, SIAH-1 overexpressing, or p53 mutant or wild-type cell lines were used, suggesting the existence of a universal mechanism underlying tumor reversion. Translationally Controlled Tumor Protein (tpt1/TCTP) has the strongest differential expression, down-regulated in the reversion of U937- and SIAH-1-overexpressing cells. Inhibition of TCTP expression by anti-sense cDNA or small interfering RNA molecules results in suppression of the malignant phenotype and in cellular reorganization, similar to the effect of SIAH-1. Hence, tumor reversion can be defined at the molecular level, not just as the reversal of malignant transformation, but as a biological process in its own right involving a cellular reprogramming mechanism, overriding genetic changes in cancer, by triggering an alternative pathway leading to suppression of tumorigenicity.
Most cancer research is primarily focused on understanding how normal cells become malignant and on what the genomic alterations that underlie tumor formation are. By examining the problem of “Expression genetics in cancer: Shifting the focus from DNA to RNA” (1), research was already striving to understand the consequences of gene expression variations by making a comparison between tumor cells and “well matched normal counterparts.” However, these normal counterparts never acquired the know-how to quit cancer.
As an alternative, we suggest that the study of tumor reversion can ultimately enable us to understand how a tumor cell acquires the molecular knowledge of how to quit malignancy. We thus suggest the use of biological models of reversion, as will be described in this study, to proceed with an adequate comparison, namely between parental tumor cells and derived daughter cells that display a reverted malignant phenotype. This approach is to prevent us from missing the crucial events involved in the process of quitting the malignant phenotype.
Tumor reversion has so far been widely studied for “single oncogene” transformed cells, because they can easily be detected due to their specific “ flat” morphology (2, 3). A more comprehensive study of reversion can be achieved by using, as starting material, cells reverted from human tumors. This possibility, so far, has been almost entirely ignored because of the unavailability of proper biological models. Our initial hypothesis (4, 5) that the H-1 parvovirus selects for revertant cells was based on experiments demonstrating that it preferentially kills tumor cells while sparing their normal counterparts (6, 7).
The differential gene expression screening between the parental tumor cells and the H-1 parvovirus selected revertants was complemented by the differential gene expression analysis between the human leukemia U937 and breast cancer MCF7 cells and their counterparts, stably transfected with the human SIAH-1 gene (8, 9), the homologue of the Drosophila seven in absentia (sina) (10). SIAH-1 is a p53-inducible gene, active in the process of cell death and tumor suppression by a mechanism consisting of ubiquitination and proteasomal degradation of specific target proteins (5, 11–18). We compared the pattern generated by differential gene expression analysis between the tumor cells and revertants to the one between the tumor cells versus SIAH-1 stable transfectants to understand whether there was a common core of effector genes in tumor reversion in a parvovirus-free environment, and whether this core was at least partially achieved by reexpression of a single gene, like SIAH-1. This study takes into account that the gene profiling in SIAH-1 reexpressing cells will be the consequence and reflects the reorganization due to the elimination of target proteins by the proteasomal machinery, accounting for both the direct interaction with SIAH-1 targets and also the indirect consequences that such interaction could generate on gene expression.
We found the Translationally Controlled Tumor Protein (tpt1/TCTP) to be the most strikingly down-regulated in tumor reversion. tpt1/TCTP is highly conserved and abundantly expressed across a wide range of eukaryotes (19–22), and its function has been associated with cell growth. Indeed, the expression at the protein level is up-regulated by growth stimuli and repressed when growth arrest is induced. TCTPs were found among the 20 most abundantly produced proteins during exponential growth in Saccharomyces cerevisiae (100,000 copies per cell compared with 60,000 for actin). It was shown that tpt1/TCTP form a structural superfamily with the MSS4/DSS4 proteins that bind to the GDP/GTP-free form of Rab proteins (22).
From the data presented, it appears that in the process of reversion, the imbalance in gene expression produced by the cancer cell would be at least partially corrected by (i) inhibition of tpt1/TCTP, and (ii) activation of SIAH-1. Ultimately, tumor reversion may take advantage of an existing alternative pathway that is dormant in the tumor cells but, once activated, could lead to suppression of tumorigenicity.
Materials and Methods
Revertant Cells.
The tumor cells K562, U937, T47D, MDA-MB231, BT20, and 184B5 were obtained from the American Type Culture Collection. Isolation of revertants started 12–18 weeks after routine culture. Different concentrations of H-1 parvovirus were used to infect the tumor cells (multiplicity of infection 10–1,000 plaque-forming units per cell). The plates were observed and the medium replaced once per week. The adherent tumor cell lines were isolated with cloning cylinders (Sigma) by using collagenase/dispase (Roche Diagnostics). Isolated revertants and parental tumor cell lines were tested for their ability to form colonies in semisolid medium (agar-noble, Difco). For in vivo tumorigenicity, female scid/scid mice were injected with 10 × 106 cells per site. Statistical analysis on the growth curves was performed as described (23). H-1 parvovirus DNA was amplified by using as primers: 5′-CTAGCAACTCTGCTGAAGGAACTC-3′and 5′-TAGTGATGCTGTTGCTGTATCTGATG-3′, giving rise to a PCR product of 254 base pairs.
Differential Gene Expression Analysis.
Differential cDNA display was performed as described (24). megasort and Massively Parallel Signature Sequencing (mpss) were performed at Lynx Therapeutics (Hayward, CA) as described (25, 26). Quantitative PCR (Applied Biosystems 7900) were performed following the manufacturer's instructions. Northern and Western blot analyses were performed as described (23). Western blot, with the series of tumors from patients and normal controls, was purchased from Geno Technology (St. Louis). The following probes, tpt1/TCTP and GAPDH, and antibodies were used: antihistamine-releasing factor (Medical and Biological Laboratories, Nagoya, Japan), anti-parp clone C2.10 (Enzyme Systems Products, Livermore, CA), anti-actin (Santa Cruz Biotechnology), and anti-β tubulin (ICN). Flow cytometry was performed as described (23).
Transfectants and tpt1/TCTP Small Interfering RNA Molecules (siRNA) Knock Down.
U937 (8) and MCF7 (9) stably transfected with SIAH-1 were described before. Transfection of U937 cells with anti-sense TCTP was performed by using Lipofectin (Invitrogen). The cDNA corresponding to the coding region of tpt1 was cloned in an inverted way in pBK-RSV (Stratagene), and the transfection was followed by selection with 1.5 mg/ml G418. tpt1 was knocked down with siRNA (27). RNA duplex with 3′ dTdT overhang directed against tpt1 mRNA 5′-AAGGTACCGAAAGCACAGTAA-3′(siRNA1), or 5′-AACCATCACCTGCAGGAAACA-3′ (siRNA2) were synthesized (Dharmacon Research, Lafayette, CO). Mouse trt/TCTP siRNA duplex 5′-AACCATCACTTACAAGAAACC-3′ was used as control. Cells were transfected with 1 nM siRNA by using oligofectamine (Invitrogen). Cells were further incubated for 3 days, detached, counted, and mixed 1:1 with matrigel (Becton Dickinson), resulting in a final concentration of 2 × 105 cells per ml, matrigel concentration 6.25 mg/ml. Cells were stained with anti-E-cadherin antibodies (Transduction Laboratories, Lexington, KY), and nuclei were propidium iodide-stained and analyzed by confocal microscopy.
Results
Models of Tumor Reversion.
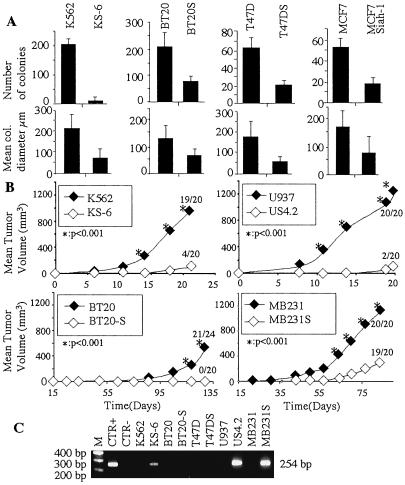
Five models of tumor reversion were obtained by using as selective agent the H-1 parvovirus. The rescued clones are thus resistant to the cytopathic effect of the virus. The whole procedure, including the assessment of tumorigenicity in scid/scid mice in selecting a biological model using this methodology, takes between 10 and 12 mo, depending on the cell type. The final clones retain many of the properties of the parental transformed cells, such as capacity to undergo multiple passages, abnormal chromosome number, and chromosomal translocations (data not shown). For human leukemia cell lines, the same approach was used as the one we described previously (4, 5). The KS6 clone was isolated from K562 cells and the US4.2 clone from U937 cells. The KS6 clone differs from KS cells (4) by having a significantly more suppressed malignant phenotype and by lack of reexpression of p53. The US4.2 clone has a malignant suppressed phenotype similar to the US cells (5). For the three solid tumors, we chose breast carcinoma cell lines BT20, T47D, and MDA-MB231. One should note that all these cell lines, whether hematopoietic or of breast cancer origin, have a mutant p53, and that as a consequence, the revertant cells were obtained in a p53 mutant background. K562/KS6, BT20/BT20S, and T47D/T47DS were tested in soft agar, whereas the U937 and MDA-MB231 cell lines do not grow in these conditions. As shown in Fig. 1A, both the number and size of the colonies are significantly lower for the revertant clones.
Fig 1.
Characterization of the revertant cells. (A) In vitro soft agar assay measuring, respectively, the number of colonies (Upper) and mean colony diameter (Lower). (B) In vivo tumorigenicity after injection of 10⋅106 cells per site. (C) PCR analysis specific for a region of 254 base pairs of the H-1 parvovirus in NBE cells uninfected (CTR−), in parvovirus infected (CTR+), and in the studied cells and their revertants.
In vivo tumorigenicity in scid/scid mice was tested for the K562/KS6, U937/US4.2, BT20/BT20S, and MDA-MB231/MDA-MB231S, whereas T47D did not grow at all in the scid/scid animals. For all of the revertants tested, the in vivo tumorigenicity was highly decreased (Fig. 1B). BT20S revertants do not form any tumors when injected into scid/scid mice; KS6 form four tumors of 20 injections; US4.2 form two tumors on 20 injections; and MDA-MB231S form 19 tumors on 20 injections, but the size of the tumors is significantly lower. Our previous results indicated that both the KS revertants from K562 cells and US revertants from U937 cells continue to produce functional H-1 parvovirus for years after the initial infection (4, 5). This is the reason we isolated the new clones, KS6 and US4.2, to check whether they would also continue to produce the parvovirus. The newly isolated KS6 and US4.2 revertants continue to produce parvovirus (Fig. 1C). However, for the breast cancer cells, only the revertants derived from the MDA-MB231 produced parvovirus, whereas BT20S and T47DS did not produce any viral particles detectable by PCR (Fig. 1C) or in functional assays examining the supernatant for viral cytopathic activity (data not shown). For further molecular analysis of tumor reversion, we also used the U937 (8) and the MCF7 cells (9) stably transfected with the human SIAH-1 gene. We have, in fact, previously described the inhibition of tumorigenicity by SIAH-1 in U937 cells (8), and Fig. 1 shows the inhibition of tumorigenicity in the MCF7 cells when transfected with SIAH-1. These MCF7 SIAH-1 transfected cells did not grow when injected into scid/scid mice (data not shown).
Differential Gene Expression Analysis in Tumor Reversion.
This study was performed in two parts. First, all of the expression analysis was done on the U937/US4.2 system, then on the six other systems, including the K562/KS6, BT20/BT20S, T47D/T47DS, MDA-MB231/MDA-MB231S, U937/U937-SIAH-1, and MCF7/MCF7-SIAH-1 (illustrated as two separate columns in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org; yellow is used for the first part and blue and light blue for the second part). All of the data presented for the U937/US4.2 system in Table 1 were confirmed by Northern blot analysis or real-time PCR. The gene expression pattern for the U937/US4.2 was first analyzed by differential display (24). We identified 600 bands whose expression varied between the U937 and US4.2. One hundred seven genes were isolated, 35 of which showed differentially expressed mRNAs (listed in Table 1). Most of the other 72 genes were either undetectable by Northern blot (59) or showed a difference in expression that was considered as nonsignificant (13 were less than 1.5-fold increased or decreased). We further performed the screening by megasort (25), which generates as results cDNA sequences, and mpss (26), which generates signatures. Only the most significant differences as selected by the highest scores for differential expression combined with the most significant P value (<0.001) were further taken into consideration. The megasort analysis indicated 855 sequences, of which 185 were clusters (the most differentially expressed genes). Of these clusters, 148 encoding for human genes were included in Table 1 after confirmation by Northern blot analysis. mpss generated 3,246 signatures that were differentially expressed, of which 1,638 correspond to genes, 1,558 have no match in the nonredundant database (GenBank, European Molecular Biology Laboratory, DNA databank of Japan, Protein Data Bank), and 50 correspond to ALU sequences. Of these 1,638 signatures, 134 corresponded to human genes that were differentially expressed by Northern blot analysis or real-time PCR and thus are listed in Table 1). The three technologies together generated 263 human genes that were differentially expressed in the U937/US4.2 system and listed in the left part of Table 1. Discrepancies were found between the results of the Northern blots or real-time PCR and those generated by the megasort (47 genes) and mpss (34 genes). These inconsistencies are either due to the fact that the probe did not detect the specific gene whose expression was too low to be seen on the Northern blot, or because of errors in sampling for megasort and mpss.
For the second part of the study, we used only the mpss method for the analysis of gene expression in the six model systems. Table 1 (second part, blue and light blue) contains genes that are differentially expressed but match already with a gene that was differentially expressed in the U937/US4.2 system. This part of the study was aimed at investigating whether there are common effector genes between the different biological model systems.
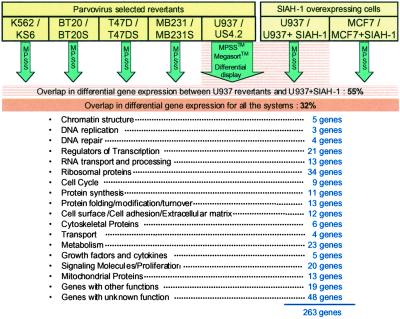
As illustrated in Table 1, almost all of the vital processes of the cell participate in tumor reversion. More than 12% of the genes identified encode for ribosomal proteins. Besides providing a global view of tumor reversion, the most striking and surprising finding was that 32% of all of the genes are differentially expressed in all of the biological models, whether parvovirus selected, hematological, or breast malignancies, or transfectants of U937 and MCF7 cells with the SIAH-1. When one is comparing only the gene differences between the U937/US4.2 system and U937-SIAH-1 transfectants, one finds 144 on the 263 genes (55%) (summary in schematic Fig. 2) to be differentially expressed in both systems. Besides the genes that form the “common core” of reversion, it was striking to see that they fluctuate in their expression from one biological model to the other and form a “variable ensemble.” The biological models selected by parvovirus share also a large part of their repertoire with the SIAH-1 transfectants. We did not include in Table 1 the 25 genes that we previously isolated by differential screening analysis (8, 28). Classification for function was performed by using the Online Mendelian Inheritance in Man, GeneOntology, and Golden Path databases and further extensively checked by reviewing the literature. By no means do the 263 genes that we identified form an exhaustive list of genes involved in tumor reversion; they are meant only to provide a global view of gene variations that can occur in the tumor reversion process.
Fig 2.
Schematic representation of the differential gene expression analysis in tumor reversion. (The complete data are in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org.)
Inhibition of tpt1 Expression by Anti-Sense cDNA or Knock Down by siRNA.
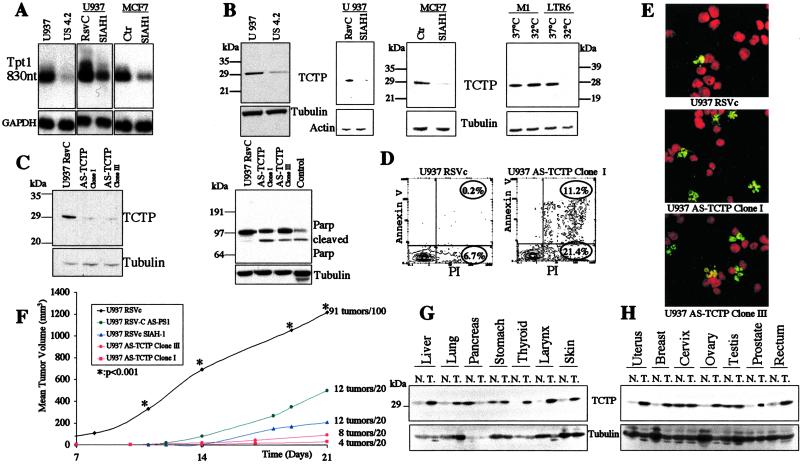
The most differentially expressed gene, using first the megasort technology between the U937 and US4.2 cells, was tpt1 for having detected the signal 124 times in the malignant cells versus once in the revertant US4.2 cells (numbers for the megasort not shown in Table 1). This strong differential expression was also confirmed by Northern and Western blot analyses (Fig. 3 A and B).
Fig 3.
tpt1/TCTP expression and functional analysis. (A) Northern blot analysis of U937 cells, US4.2 revertants (first gel), U937 cells and U937 SIAH-1 stable transfectants (second gel), MCF7 cells, and MCF7 cells stably transfected with SIAH-1 (third gel). (B) Western blot analysis for the expression of tpt1/TCTP protein in U937 cells, US4.2 revertants (first gel), U937 cells and U937 SIAH-1 stable transfectants (second gel), MCF7 cells, and MCF7 cells stably transfected with SIAH-1 (third gel). The fourth gel is a Western blot analysis of tpt1/TCTP in the M1 cells and LTR6 cells stably transfected with the temperature-sensitive p53 val135 mutant. (C) Western blot analysis of tpt1/TCTP in U937 cells stably transfected with the vector alone (first lane) or anti-sense tpt1/TCTP cDNA (second and third lanes). Second gel: analysis of Parp cleavage in U937 cells transfected with the vector alone or anti-sense tpt1/TCTP cDNA. Positive control, last lane. (D) Percentage of annexin V in U937 cells transfected with the vector alone or anti-sense tpt1/TCTP. (E) Terminal deoxynucleotidyltransferase-mediated dUTP end labeling (TUNEL) assay in U937 transfected with the vector alone or anti-sense tpt1/TCTP. TUNEL-positive cells are in green. (F) In vivo tumorigenicity assay after injection of 10⋅106 cells per site into scid/scid mice of U937 cells, U937 cells stably transfected with anti-sense PS1, U937 cells stably transfected with SIAH-1, U937 cells stably transfected with anti-sense tpt1/TCTP. (G and H) Western blot analysis of tpt1/TCTP in human tumor tissues. The expression at the protein level was analyzed in different normal and tumor tissues, as indicated. Tubulin was used as a control for equal loading.
tpt1/TCTP is also inhibited in its expression in the U937 and MCF7 cells transfected with SIAH-1, which also have a suppressed malignant phenotype (Fig. 3 A and B). Although this inhibition of tpt1/TCTP expression is already clearly present, at the transcriptional level we tested whether SIAH-1 could bind and degrade TCTP via the ubiquitination pathway, which is not the case (L.S., data not shown). Interestingly, in the LTR6 system (29) after activation of wild-type p53 function, expression of tpt1/TCTP is drastically inhibited (Fig. 3B, last gel). To mimic this inhibition of tpt1/TCTP as shown in the biological systems above, U937 cells were stably transfected with tpt1/TCTP anti-sense cDNA. As shown in Fig. 3C, Western blot analysis of two transfected clones shows a significant decrease in the tpt1/TCTP protein. This decrease in TCTP has as consequence an increased rate of apoptosis as confirmed by PARP cleavage (Fig. 3C, second gel), annexin V (Fig. 3D), and terminal deoxynucleotidyltransferase-mediated dUTP end labeling staining (Fig. 3E). This increase in apoptosis, although moderate (15%), was reproducible. When these transfectants were injected into scid/scid mice, they showed a robust inhibition in tumorigenicity (Fig. 3F). After 3 weeks of 20 injections of 10⋅106 cells, clone I formed four tumors, and clone III formed eight tumors, but extremely small ones at the limit of being palpable, whereas the parental U937 cells formed tumors in 90% of the injections, which were so large that the animals had to be killed. This inhibition of tumorigenicity was also more profound than in the U937 cells stably transfected with SIAH-1 or anti-sense PS1 (Fig. 3F); for both genes, we previously described their effect on tumorigenicity (8, 23). Importantly, tpt1/TCTP is also up-regulated at the protein level in most of the tumor samples we have tested (Fig. 3 G and H).
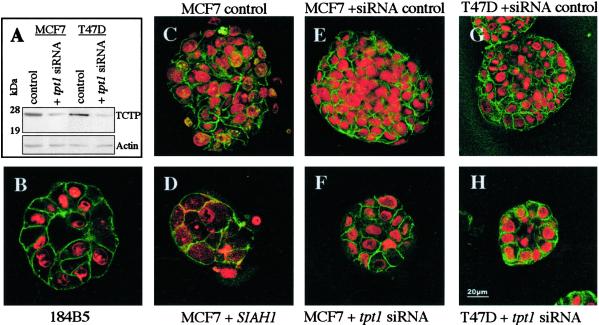
To exclude that the difference in tumorigenicity of the stable U937 tpt1/TCTP transfectants was due to a clonal difference, we investigated changes in 3D reconstituted basement membrane (3DrB-M) (30) matrigel cultures of MCF7 and T47D by using knock-down tpt1/TCTP by siRNA (27) (Fig. 4). The siRNA inhibited the expression of tpt1/TCTP (Fig. 4A) in both the MCF7 and T47D cells, as compared with the clones transfected with control mouse trt/TCTP siRNA, which differs in 4 bp from the human one. Both MCF7 and T47D cells transfected with a specific tpt1/TCTP siRNA (Fig. 4 F and H) showed drastic changes in the matrigel and formed structures reminiscent now of the growth of 184 B5 cells (Fig. 4B), which are considered as the normal control. Because MCF7 cells stably transfected with SIAH-1 show a strong down-regulation of tpt1/TCTP (Fig. 3 A and B), and because the SIAH-1 pathway shares 55% of differentially expressed genes with the U937/US4.2 cells (Table 1), we investigated whether these SIAH-1 stables have a modified growth in 3DrB-M matrigel cultures. As shown in Fig. 4D, the MCF7–SIAH-1 transfectants are reorganized in structures very similar to the MCF7 cells transfected with tpt1/TCTP siRNA.
Fig 4.
Three-dimensional reconstituted basement membrane matrigel cultures. (A) Western blot analysis of tpt1/TCTP expression in MCF7 and T47D cells transfected with a specific anti-human tpt1/TCTP siRNA. (B) 184B5 cells. (C) MCF7 in standard growth medium. (D) MCF7 cells stably transfected with SIAH-1 cDNA. (E) MCF7 cells transfected with control trt/TCTP siRNA. (F) MCF7 cells transfected with tpt1/TCTP siRNA. (G) T47D with the same control siRNA. (H) T47D transfected with tpt1/TCTP siRNA.
Discussion
Two fundamental questions in cancer research are: (i) how does a normal cell become malignant, and (ii) how does a malignant cell revert and lose its aggressive phenotype? There have been only a few attempts to answer the second question, and these were oriented mainly to the analysis of flat revertants (2, 3). A different aspect of tumor reversion was discovered by using blocking antibodies against integrin (30) and by analyzing the phenotype of breast cells in a three-dimensional culture. These studies suggested that the phenotype generated by reversion is dominant over the genotype.
The most direct way to approach reversion is to identify how it occurs in a way dictated by nature, like reversion of bacterial strains, yet in an extremely rare fashion. The process of reversion is so rare that one could not simply take a tumor from a patient and just look for revertants, because they could be mistaken for any normal cells, such as those forming blood vessels and the blood cells. One feasible approach is to obtain from a cancer sample a cell line representative of the specific tumor, and only then to derive the revertants (4).
By using the H-1 parvovirus as a tool to kill the malignant cells, we isolated clones resistant to this cytopathic effect. As demonstrated here, these resistant cells have a suppressed malignant phenotype. Revertants derived from K562 and U937 cells continued to produce the H-1 parvovirus, raising the possibility that the virus itself could play a dominant role in the reversion. By expanding the study to other systems like breast cancer cells, we found that the BT20S and T47DS revertants do not harbor the virus, and that only the MDA-MB231S continue to produce it. These results indicate that the BT20S and T47DS revertants are not induced but selected by the H-1 parvovirus. However, even in these cells, the parvovirus selection process may have initiated changes that we were unable to detect, but that are part of the tumor reversion process. This is why we compared in the gene expression analysis U937 or MCF7 cells to their counterparts, stably transfected with the SIAH-1 gene. This gene was previously described as playing a role in programmed cell death and in controlling the fate of cancer cells by promoting, among others, β catenin and numb degradation (5, 8, 11–13, 15–17, 28). These two stable SIAH-1 transfectants also show inhibition of their tumorigenicity. In addition, the MCF7-SIAH-1 transfectant cells have a wild-type p53, indicating that the reversion process can override the oncogenic events, even in the presence of wild-type p53, which is of interest for the further study of tumors that do not have a mutation in this gene.
By applying a large-scale screening analysis for variation in gene expression, 263 genes were identified as differentially expressed when confirmed by Northern blot analysis or real-time PCR. Although every compartment of the cell seems to participate in the reversion, it is noteworthy to observe how the mRNAs coding for the ribosomal proteins are reorganized in their expression. Some of the oncogenes, like myb, are down-regulated during the reversion process. The expression of the amyloid precursor protein (APP) also varies in the reversion. However, these changes may equally be a consequence of the reversion program and not a contributor to this program. A cause–effect conclusion can be made only on the basis of further functional studies, although previous data on Presenilin1 (a predisposition gene for a familial form of Alzheimer's disease that cleaves the APP) already indicated that it participates in the regulation of programmed cell death and tumor suppression (23, 31). Most striking is the fact that 32% of the genes we identified are differentially expressed in all of the biological systems analyzed. Also, the stable U937 transfectants overexpressing SIAH-1 have 55% of their changes in gene expression, matching those obtained by selecting for U937 revertants with H-1 parvovirus. These findings could indicate the presence of a “common core” of genes intervening in all situations of suppression we tested. The differential expression of this core of genes does not always follow the same direction (activated or inhibited) in all of the models presented here, which may indicate that leukemia cells or different types of breast tumor cells do not require the same modifications to revert. Thus, these data suggest the presence of a “variable ensemble” of genes that are responsible for reversion. Every cell system would choose among these genes the most adequate ones to perform the definitive task.
The most differentially expressed gene is tpt1/TCTP. The magnitude in differential gene expression for tpt1/TCTP that we initially found in the U937/US4.2 system, and that was confirmed to be strongly decreased by Northern and Western blot in different systems, including after induction of wild-type p53 function in the LTR6 system (29) and after overexpression of the SIAH 1 gene in U937 and MCF7 cells, prompted us to further study tpt1/TCTP to validate the experimental strategy that unravels effector genes of tumor reversion. The mainstream research for the role of tpt1/TCTP is in allergic response (21). tpt1/TCTP was identified as the human histamine releasing factor. It has been shown to be one of the first proteins to be induced in Ehrlich ascites tumor cells after mitotic stimulation (20). tpt1/TCTP was also described as binding in a yeast two-hybrid assay MCL1, a Bcl-2 homologue, and identified as an antiapoptotic protein (32).
We demonstrate that inhibition of tpt1/TCTP expression by anti-sense cDNA and siRNA suppresses the malignant phenotype. U937 leukemic cells stably transfected with anti-sense tpt1/TCTP generate significantly less tumors when injected in scid/scid mice. We observed phenotypic changes in MCF7 or T47D cells transfected with tpt1/TCTP siRNA by using three-dimensional reconstituted basement membrane matrigel cultures. More organized ductal-like structures similar to those generated by down-regulation of β1 integrin were evident (30). When stably transfected with SIAH-1, these MCF7 cells also show this striking change in morphology. These SIAH-1 transfected cells have a strong down-regulation of tpt1/TCTP expression. The above-described results indicate that lowering of tpt1/TCTP expression in different types of cancer cell lines, such as a leukemic and breast cancer cell lines, is a critical factor to allow the cell to revert to a suppressed malignant phenotype. They further provide the argument toward a common mechanism of reversion underlying activation of SIAH-1 pathway or reduction of tpt1/TCTP. However, one suspects that the complexity of the process may be higher than presently envisaged, and that other genes described in Table 1 may be as important in controlling reversion as the particular genes targeted in the present study: SIAH 1 and tpt1/TCTP.
In conclusion, by using the H-1 parvovirus as a sift to isolate single revertants among millions of tumor cells, we provided a tool to study global tumor reversion. The gene expression profile suggests that it is not the processes per se of cell cycle arrest, apoptosis, and terminal differentiation, that matter here, and that provide by themselves the framework for reversion. It is rather a “reorganizing” function of all these processes as a form of rerouting and trigger of the whole machinery that enables the tumor cells to quit the malignant pathway, even bypassing mutant or wild-type p53. It is possible that activation of SIAH-1 or inhibition of tpt1 expression is related not only to a pro- or antiapoptotic process but rather an organizing function such as seen in the development of the ommatidia in Drosophila. We suggest that reversion as described here operates through at least three mechanisms. The first one would involve the inhibition of mRNA synthesis for genes encoding ribosomal proteins. The second mechanism is powerfully inhibiting expression of tpt1/TCTP. The third one would involve the SIAH-1 pathway, as presented in this study, the result of a more specific protein targeting toward proteasomal degradation. All three mechanisms combined would reprogram the cancer cell to recover some of its normal functions, such as for breast cells to form ductal-like structures. There is no evidence to rule out that the three mechanisms are at least partially redundant. In fact, the data do suggest that SIAH-1 is dominant over tpt1/TCTP and can reduce its expression, consistent with SIAH-1 being upstream of tpt1/TCTP in the same pathway. This mechanism of reversion could be an alternative pathway that forms the “intracellular defense system against cancer,” a function inhibited in the tumor. Such a system could override the genetic changes in cancer without necessarily correcting the causal mutation, deletion, or translocation but by bypassing them.
Supplementary Material
Acknowledgments
A.T. and R.A. dedicate this study to Sydney Brenner for a discussion we started in Cambridge (U.K.) 14 years ago and for providing the megasort and mpss technologies. We thank Anne Géant, Françoise Rohfritsch, Florence Lethrosne, Dominique Duflaut, and Emmanuelle Mas for excellent technical support; Jennifer Richardson for initial collaboration; Nicolas Privat for microscopy; Laurie Goodman and colleagues at Lynx for their efficiency; and Philippe Genne at Oncodesign for the animal studies.
Abbreviations
siRNA, small interfering RNA molecules
References
- 1.Sager R. (1997) Proc. Natl. Acad. Sci. USA 94, 952-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noda M., Selinger, Z., Scolnick, E. M. & Bassin, R. H. (1983) Proc. Natl. Acad. Sci. USA 80, 5602-5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitayama H., Sugimoto, Y., Matsuzaki, T., Ikawa, Y. & Noda, M. (1989) Cell 56, 77-84. [DOI] [PubMed] [Google Scholar]
- 4.Telerman A., Tuynder, M., Dupressoir, T., Robaye, B., Sigaux, F., Shaulian, E., Oren, M., Rommelaere, J. & Amson, R. (1993) Proc. Natl. Acad. Sci. USA 90, 8702-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nemani M., Linares-Cruz, G., Bruzzoni-Giovanelli, H., Roperch, J. P., Tuynder, M., Bougueleret, L., Cherif, D., Medhioub, M., Pasturaud, P., Alvaro, V., et al. (1996) Proc. Natl. Acad. Sci. USA 93, 9039-9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toolan H. W. (1967) Nature 214, 1036. [DOI] [PubMed] [Google Scholar]
- 7.Mousset S. & Rommelaere, J. (1982) Nature 300, 537-539. [DOI] [PubMed] [Google Scholar]
- 8.Roperch J. P., Lethrone, F., Prieur, S., Piouffre, L., Israeli, D., Tuynder, M., Nemani, M., Pasturaud, P., Gendron, M. C., Dausset, J., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 8070-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruzzoni-Giovanelli H., Faille, A., Linares-Cruz, G., Nemani, M., Le Deist, F., Germani, A., Chassoux, D., Millot, G., Roperch, J. P., Amson, R., et al. (1999) Oncogene 18, 7101-7109. [DOI] [PubMed] [Google Scholar]
- 10.Carthew R. W. & Rubin, G. M. (1990) Cell 63, 561-577. [DOI] [PubMed] [Google Scholar]
- 11.Della N. G., Senior, P. V. & Bowtell, D. D. (1993) Development (Cambridge, U.K.) 117, 1333-1343. [DOI] [PubMed] [Google Scholar]
- 12.Hu G., Zhang, S., Vidal, M., Baer, J. L., Xu, T. & Fearon, E. R. (1997) Genes Dev. 11, 2701-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuzawa S., Takayama, S., Froesch, B. A., Zapata, J. M. & Reed, J. C. (1998) EMBO J. 17, 2736-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Relaix F., Wei, X., Li, W., Pan, J., Lin, Y., Bowtell, D. D., Sassoon, D. A. & Wu, X. (2000) Proc. Natl. Acad. Sci. USA 97, 2105-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuzawa S. I. & Reed, J. C. (2001) Mol. Cell 7, 915-926. [DOI] [PubMed] [Google Scholar]
- 16.Liu J., Stevens, J., Rote, C. A., Yost, H. J., Hu, Y., Neufeld, K. L., White, R. L. & Matsunami, N. (2001) Mol. Cell 7, 927-936. [DOI] [PubMed] [Google Scholar]
- 17.Susini L., Passer, B. J., Amzallag-Elbaz, N., Juven-Gershon, T., Prieur, S., Privat, N., Tuynder, M., Gendron, M. C., Israel, A., Amson, R., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 15067-15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnsen S. A., Subramaniam, M., Monroe, D. G., Janknecht, R. & Spelsberg, T. C. (2002) J. Biol. Chem. 277, 30754-30759. [DOI] [PubMed] [Google Scholar]
- 19.Yenofsky R., Cereghini, S., Krowczynska, A. & Brawerman, G. (1983) Mol. Cell. Biol. 3, 1197-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohm H., Benndorf, R., Gaestel, M., Gross, B., Nurnberg, P., Kraft, R., Otto, A. & Bielka, H. (1989) Biochem. Int. 19, 277-286. [PubMed] [Google Scholar]
- 21.MacDonald S. M., Rafnar, T., Langdon, J. & Lichtenstein, L. M. (1995) Science 269, 688-690. [DOI] [PubMed] [Google Scholar]
- 22.Thaw P., Baxter, N. J., Hounslow, A. M., Price, C., Waltho, J. P. & Craven, C. J. (2001) Nat. Struct. Biol. 8, 701-704. [DOI] [PubMed] [Google Scholar]
- 23.Roperch J. P., Alvaro, V., Prieur, S., Tuynder, M., Nemani, M., Lethrosne, F., Piouffre, L., Gendron, M. C., Israeli, D., Dausset, J., et al. (1998) Nat. Med. 4, 835-838. [DOI] [PubMed] [Google Scholar]
- 24.Liang P. & Pardee, A. B. (1992) Science 257, 967-971. [DOI] [PubMed] [Google Scholar]
- 25.Brenner S., Williams, S. R., Vermaas, E. H., Storck, T., Moon, K., McCollum, C., Mao, J. I., Luo, S., Kirchner, J. J., Eletr, S., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 1665-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brenner S., Johnson, M., Bridgham, J., Golda, G., Lloyd, D. H., Johnson, D., Luo, S., McCurdy, S., Foy, M., Ewan, M., et al. (2000) Nat. Biotechnol. 18, 630-634. [DOI] [PubMed] [Google Scholar]
- 27.Elbashir S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- 28.Amson R. B., Nemani, M., Roperch, J. P., Israeli, D., Bougueleret, L., Le Gall, I., Medhioub, M., Linares-Cruz, G., Lethrosne, F., Pasturaud, P., et al. (1996) Proc. Natl. Acad. Sci. USA 93, 3953-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yonish-Rouach E., Resnitzky, D., Lotem, J., Sachs, L., Kimchi, A. & Oren, M. (1991) Nature 352, 345-347. [DOI] [PubMed] [Google Scholar]
- 30.Weaver V. M., Petersen, O. W., Wang, F., Larabell, C. A., Briand, P., Damsky, C. & Bissell, M. J. (1997) J. Cell Biol. 137, 231-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amson R., Lassalle, J. M., Halley, H., Prieur, S., Lethrosne, F., Roperch, J. P., Israeli, D., Gendron, M. C., Duyckaerts, C., Checler, F., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 5346-5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F., Zhang, D. & Fujise, K. (2001) J. Biol. Chem. 276, 47542-47549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.