Abstract
Deafness in spontaneously occurring mouse mutants is often associated with defects in cochlea sensory hair cells, opening an avenue to systematically identify genes critical for hair cell structure and function. The classical semidominant mouse mutant varitint-waddler (Va) exhibits early-onset hearing loss, vestibular defects, pigmentation abnormalities, and perinatal lethality. A second allele, VaJ, which arose in a cross segregating for Va, shows a less severe phenotype. By using a positional cloning strategy, we identify two additional members of the mucolipin gene family (Mcoln2 and Mcoln3) in the 350-kb VaJ minimal interval and provide evidence for Mcoln3 as the gene mutated in varitint-waddler. Mcoln3 encodes a putative six-transmembrane-domain protein with sequence and motif similarities to the family of nonselective transient-receptor-potential (TRP) ion channels. In the Va allele an Ala419Pro substitution occurs in the fifth transmembrane domain of Mcoln3, and in VaJ, a second sequence alteration (Ile362Thr) occurring in cis partially rescues the Va allele. Mcoln3 localizes to cytoplasmic compartments of hair cells and plasma membrane of stereocilia. Hair cell defects are apparent by embryonic day 17.5, assigning Mcoln3 an essential role during early hair cell maturation. Our data suggest that Mcoln3 is involved in ion homeostasis and acts cell-autonomously. Hence, we identify a molecular link between hair cell physiology and melanocyte function. Last, MCOLN2 and MCOLN3 are candidate genes for hereditary and/or sporadic forms of neurosensory disorders in humans.
Perception and transmission of acoustic stimuli in the mammalian cochlea is a stratified process involving the establishment of the endolymph through melanocytes located in the stria vascularis; deflection of stereocilia situated on the apical surface of sensory hair cells in the organ of Corti (OC); opening of mechanosensitive transduction channels, nonselectively permeable for cations, including Ca2+; and transmission of the electrical impulse onto the eighth cranial nerve for further processing in the auditory brainstem and cortex. Mutations affecting these processes are often associated with circling behavior, ataxic movements, and pigmentation anomalies. Circling and waltzing phenotypes are usually caused by mutations primarily affecting structure and function of hair cells, which, directly or indirectly, lead to stereocilia disorganization and hair cell degeneration. Genes critical for hair cells encode a functionally heterogenous group of proteins that include structural proteins (myosins, cadherins, and spectrins), extracellular matrix proteins (tectorins), transmembrane proteins (channels, pumps, ion pores, and exchangers), and others (for review see ref. 1). Among the many coat color and spotting mutants, dominant spotting (KitW), steel (KitlSt), microphthalmia (Mitfmi), and varitint-waddler (Va), exhibit deafness in combination with pigmentation defects. The genes underlying W, St, and mi have been shown to encode the c-Kit receptor tyrosine kinase (2, 3), its ligand Kitl (4, 5), and the Mitf transcription factor (6), respectively. Mutations in Kit and Mitf impair the survival of melanocytes in the stria vascularis, resulting in the loss of the endochochlea potential, and deafness (7, 8).
To identify genes important for hair cell function, we adopted a positional cloning approach of mouse mutations with known defects in the OC. Two mutations, Va and VaJ, arose spontaneously at the varitint-waddler locus, causing a distinct allele-specific set of phenotypes (9–11). Most severely affected are Va/Va mice, which exhibit deafness, circling behavior, sterility, an almost entirely white coat color, and reduced viability (9). The mildest phenotype is seen in +/VaJ mice, which are viable, show normal vestibular function, display only limited variegation and coat color dilution, and have some residual hearing (10, 12, 13). VaJ/VaJ and +/Va mice show intermediate and similar phenotypes. Hearing tests and anatomical studies associated hearing loss in VaJ mice with a primary defect in the sensory epithelium and reduced pigmentation in melanocytes located in the stria vascularis (12). These defects were found to occur independently, suggesting that Va acts as a cell-autonomous factor. Thus, unlike other spotted deaf mouse mutants (Mitfmi, KitW, and KitlSl), in which neuroepithelial degeneration develops secondary to the stria vascularis defects, Va might reveal an interesting cellular and molecular principle shared by inner ear hair cells and melanocytes.
Materials and Methods
Mice and DNA.
Mutant and common inbred mouse strains were obtained from The Jackson Laboratory. B6Fe-a/a-Hoxa13Hd VaJ mice were backcrossed to the C57BL/6J strain for10 generations and outcrossed to C3HeB/FeJ, and the resulting progeny were intercrossed. RSV/Le-Va/+ mice were outcrossed to C3HeB/FeJ and then intercrossed. Intercrossed offspring from Va and VaJ strains were used for phalloidin staining and immunocytochemistry. Genomic DNA was obtained from The Jackson Laboratory. Animal care and procedures were in accordance with National Institutes of Health guidelines (Animal Study Protocol 821/97).
Physical Map.
The physical map was constructed by screening the ES-129/SvJ I (Incyte Genomics, Palo Alto, CA) and the C57BL/6J RPCI-23 (Roswell Park Cancer Institute) mouse bacterial artificial chromosome (BAC) libraries. We screened the ES-129/SvJ I BAC library by PCR with the flanking recombinant markers (D3Mit85, D3Mit260, D3Mit259, and D3Mit292). Probes from the BAC ends of the ES-129/SvJ I BAC clones were used to screen the RPCI-23 library by hybridization. BAC clones were sized by restriction endonuclease digestion with NotI (NEB) followed by pulsed-field gel electrophoresis (PFGE) on a Chef-DRII apparatus (Bio-Rad). We sequenced BAC ends with vector-specific primers by using BigDye terminator chemistry (Applied Biosystems) and an ABI 377 sequencer (Applied Biosystems). To confirm the chromosomal location of each identified clone and to establish the orientation of Sp6 and T7 ends, we developed sequence-tagged sites (STSs), primer sets, and probes from the end sequences of BAC clones and mapped them to the predicted VaJ physical interval by using PCR cross-screening experiments and Southern blot hybridizations. Overlaps among individual BAC clones were confirmed by fingerprinting with HindIII and/or EcoRI single or double digestions.
Gene Identification and Mutation Analysis.
Draft sequences from RP23-108E10 and RP23-121J1 BAC clones were generated by the Department of Molecular Genetics, Albert Einstein College of Medicine Genome Center, through the transnih BAC Sequencing Program (www.nih.gov/science/models/bacsequencing) and released into GenBank (accession nos. AC068947 and AC079941). blast searches of the EST database with these genomic sequences identified a cluster of mouse ESTs with high similarity to the full-length human cDNA clone AK001868. To obtain the homologous mouse sequence, we sequenced the longest EST clones AI787597 and AA756265. We designed primers and performed RT-PCR on C57BL/6J-derived brain cDNA to establish the mouse Mcoln3 cDNA sequence (GenBank accession no. AY083531). We deduced its genomic structure by aligning the mouse and human cDNA against genomic sequences by using blastn searches and DNA-Pustell matrix alignments (macvector Version 6.0, Oxford Molecular). To identify mutations, genomic DNA from wild-type (C57BL/10J and C57BR/cdJ) and mutant (Va/Va and VaJ/VaJ) strains was used to amplify by PCR exon-specific products by using primer pairs complementary to flanking intronic sequences. PCR was carried out by using the Advantage cDNA polymerase mix (CLONTECH) according to the manufacturer's instructions. PCR products were gel purified following the Qiagen (Chatsworth, CA) gel extraction kit protocol and sequenced by using the BigDye chemistry on an ABI 377 automated sequencer. Wild-type and mutant sequences were compared by using sequencher Version 3.0 (Gene Codes, Ann Arbor, MI) software. Primers to amplify across the Va 1255G → C mutation are as follows: 5′-1138-TCAACTATGCTCGTGTGGC-3′ (forward), 5′-1312-GGTAAGGGCCCAGCACAATCC-3′ (reverse). Primers to amplify across the VaJ 1085T → C mutation are as follows: 5′-970-CACTACAAGAAGGAAGTTTCGG-3′ (forward), 5′-intronic-GCCCAGAATTTTTTCACAGTTTGG-3′ (reverse). To identify mutations in +/VaJ and VaJ/VaJ -derived cDNA, poly(A)+ RNA was isolated from the brain and spleen according to the Oligotex Direct mRNA kit protocol (Qiagen), and reverse transcribed with an oligo(dT) primer by using the SuperScript Preamplification System (Invitrogen). RT-PCR was carried out and resulting products were directly sequenced.
Phalloidin Staining.
Cochlear ducts were dissected in Leibovitz's L-15 medium (Invitrogen) from wild-type, VaJ, and Va mice. Ducts were fixed in 4% paraformaldehyde for 2 h at room temperature and microdissections were performed to isolate the OC. Tissues were permeabilized in 0.5% Triton X-100 in PBS for 30 min, washed twice in PBS for 10 min, and then stained with 2 μg/ml fluorescein-conjugated phalloidin (Sigma) in PBS for 20 min. After three 10-min washes in PBS, samples were mounted by using the ProLong Antifade kit (Molecular Probes) and examined with a laser scanning confocal microscope (LSM 510, Zeiss).
Immunocytochemistry.
To obtain antibodies against Mcoln3 (GenBank accession no. AY083531), rabbits were immunized (Covance Research Products, Denver, PA) against synthetic peptide 1, NH2-446-RVSECLFSLINGDDMFS-COOH, and peptide 2, NH2-529-KDLPNSGKYRLEDDPPGSLL-COOH (Princeton Biomolecules, Langhorne, PA). Immunocytochemistry was performed as described (14). OCs were dissected as described above. After permeabilization in 0.5% Triton X-100 for 30 min, samples were washed three times in PBS for 10 min and incubated in 5% normal goat serum (Life Technologies, Grand Island, NY) and 2% BSA (ICN) in PBS for 2 h to block nonspecific binding sites. After incubation with primary antibodies at 3–6 μg/ml in blocking solution for 2 h at room temperature, samples were washed several times in PBS and incubated in a 1:200 dilution of the fluorescein-conjugated anti-rabbit IgG secondary antibody (Amersham Pharmacia Biotech, Uppsala) for 40 min. Samples were mounted by using the ProLong Antifade kit and examined with a laser scanning confocal microscope (LSM 510, Zeiss). Preimmune sera and preincubation of primary antibody with an excess of immunogenic peptide were used as negative controls. Some samples were stained for actin with rhodamine-conjugated phalloidin. We obtained the same results (Fig. 4) with affinity-purified (Covance Research Products) antisera PB221 and PB220 raised against peptide 1, and antisera HL4559 and HL4560 raised against peptide 2 (Fig. 4).
Fig 4.
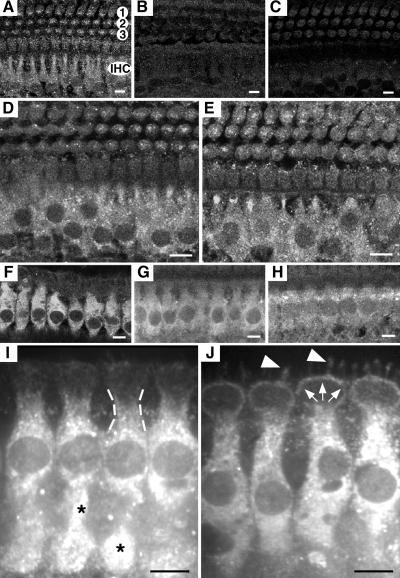
Localization of Mcoln3 in mouse and rat OC by immunofluorescence. Shown are cross-sections through the OC showing the labeling of IHCs for Mcoln3. (A–E) Mouse OC at P11, and optical sections at the level of the cell body of IHCs. In A a single row of IHCs and three rows of OHCs (1, 2, and 3) are presented. (A, D, and E) PB221 labeling for Mcoln3. The cytoplasm of IHCs is intensively labeled in wild type (A), +/VaJ (D), and VaJ/VaJ (E); staining is absent after incubation with preimmune serum (B) and excess specific peptide (C). Some labeling is also observed in the cytoplasm of OHCs (A, D, and E), and under the cuticular plate (not shown). (F–J) Rat OC at 4 weeks of age, and optical sections at the level of cell bodies (F–H) and the cuticular plate (I and J) of IHCs. All four anti-Mcoln3 antisera, PB220 (F and I), PB221 (J), HL4459 (G), and HL4460 (H) showed immunoreactivity in the cytoplasm of IHCs. In the cytoplasm, labeling appears as a punctuate pattern (*, I) throughout the IHC bodies, and it is also detected in the subcuticular region (dashed lines, I), the pericuticular necklace (white arrows, J), and in the IHC bundles (white arrowheads, J). Specificity of all four antisera was confirmed by immunoblot after bacterial expression of a partial Mcoln3 fusion protein (data not shown). Stereocilia staining was also observed on mouse OC (data not shown). (Bars, 80 μm.)
GenBank Accession Numbers.
Mouse mucolipin-1 (Mcoln1), AF302009; human mucolipin-1 (MCOLN1), AF249319; mouse mucolipin-2 (Mcoln2), AY083532; human mucolipin-2 (MCOLN2), AY083533; mouse mucolipin-3 (Mcoln3), AY083531; human mucolipin-3 (MCOLN3), AF475085; Drosophila CG8743, AAF49118; Caenorhabditis elegans CUP-5, AF338583; Pkd2, AF014010; and TRP, JU0092. Genomic clones containing Mcoln2 and Mcoln3, AC068947 (RP23-108E10), and AC079941 (RP23-121J1).
Results
Perinatal Cochlea Hair Cell Defects in Va and VaJ Mutants.
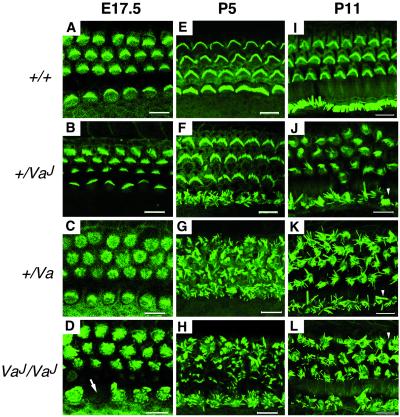
Previous ultrastructural studies showed hair cell degeneration in 14-day-old [postnatal day 14 (P14)] VaJ mutants (12). To determine the onset and extent of the hair cell defects in both alleles, we examined phalloidin-stained whole mount OC preparations with scanning laser confocal microscopy. The first signs of abnormalities were detected in VaJ/VaJ and as early as embryonic day 17.5 (E17.5), when the growing microvilli of both inner hair cells (IHCs) and outer hair cells (OHCs) show an irregular arrangement (Fig. 1D). Occasionally, gaps were observed in the row of IHCs, indicating signs of degeneration (Fig. 1D, arrow). At P5, stereocilia bundles of IHCs and OHCs seem to have grown to their normal length but they appear in small clumps at the top of some hair cells, showing a disorganized pattern (Fig. 1H) rather than the normal arrangement observed in littermate controls (Fig. 1E). Stereocilia disorganization in both IHCs and OHCs continued to progress from P5 to P11 when many of them appear missing through the length of the OC, and extensive fusion and clumping are apparent (Fig. 3L, arrowhead). We also examined HCs at the level of the cuticular plate but no irregularities were detected; occasionally, however, we noticed an extrusion of hair cell bodies through the reticular lamina and an abnormal cytoplasmic architecture of the hair cell body (data not shown). In +/Va we observed a similar progressive disorganization, with stereociliar bundles of both IHCs and OHCs becoming equally disorganized at all time points (Fig. 1 C, G, and K). The least severe pathology is seen in +/VaJ, in which the general organization of the microvilli appears mostly normal at E17.5 (Fig. 1B). However, disruption of the stereocilia bundle is apparent by birth and mainly in IHCs, where disorganization of the bundle remains severe throughout development (Fig. 1 F and J). Only some minor signs of disorganization are noticeable in the OHC bundles at P5 (Fig. 1F) but stereocilia disorganization continues to progress, becoming more pronounced by P11 (Fig. 1J). The progression of hair cell defects from base to apex, along its developmental gradient, and the perinatal expression of the phenotype attributes to Va a critical role during early steps of hair cell differentiation.
Fig 1.
Development of cochlear hair cells in varitint-waddler mutants. Labeling of cochlear hair cells F-actin by FITC-phalloidin at E17.5 (A–D), P5 (E–H), and P11 (I–L). Shown are optical sections through the OC at the level of the stereocilia. Normal hair cells can be distinguished by E17.5 as a regular array of microvilli from which ordered rows of stereocilia bundles develop; in wild-type mice, hair cell stereocilia are V-shaped in OHCs (top three rows), and straight in IHCs (bottom row) (A, E, and I). In +/VaJ mutants hair cell bundles appear mostly normal at E17.5 (B); disorganization of IHC stereocilia is severe by birth and continues to progress from P5 (F) to P11 (J); only minor signs of disorganization are noticeable in OHC bundles by P5 (F), but disorganization of the OHC bundle continues to progress, becoming more pronounced by P11 (J). In +/Va mutants, IHC and OHC stereociliar bundles are equally disorganized at all time points (C, G, and K). In VaJ/VaJ mutants, stereocilia disorganization is apparent in both IHCs and OHCs at E17.5 (D), and progresses from P5 (H) to P11 (L). In all mutant genotypes, by P11 many stereocilia appear missing and extensive fusion and clumping are apparent (J–L, arrowheads). The spatial organization of hair cells in the OC in all mutants remains unaltered with three rows of OHCs and one row of IHCs. (Bars, 10 μm.)
Fig 3.
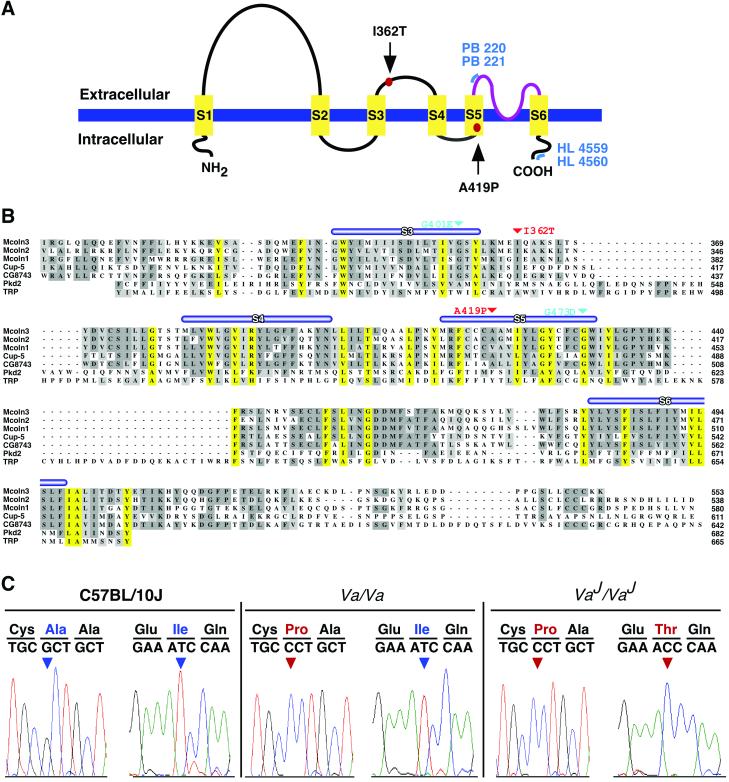
Sequence analysis of Mcoln3. (A) Schematic representation of predicted structure of Mcoln3. Six putative transmembrane domains (S1–S6) and a putative pore region (amino acids 480–505; in purple) are shown. Mutations in Mcoln3 are indicated by red dots. Blue lines represent regions of Mcoln3 containing polyclonal antibody recognition sites: PB221 and PB220 antisera were raised against amino acids 446–462, HL4559, and HL4560 against amino acids 529–548. (B) clustalw alignment of the mucolipin family. Multiple sequence alignment of Mcoln3, Mcoln1, Mcoln2, C. elegans CUP-5, and Drosophila CG743 is shown with Pkd2 (amino acids 478–682) and TRP (amino acids 430–665). Putative transmembrane domains (S1–S6) are indicated by thick blue lines. Predicted ion transport domain and TRPL motif (PS50272) of Mcoln3 are located between amino acids 337–501 and amino acids 317–505, respectively. Conserved amino acids across the Mcoln3 TRPL region are shaded yellow. Sites of mutations in Mcoln3 are shown in red and in CUP-5 in teal. Amino acid positions are given on the right. (C) Mutation analysis. Sequence chromatographs showing nucleotide sequence and translation across the sites of mutations in C57BL/10J, Va/Va, and VaJ/VaJ genomic DNA. Nucleotide changes are shown in red.
The VaJ Critical Interval Spans 350 kb and Contains Three Genes.
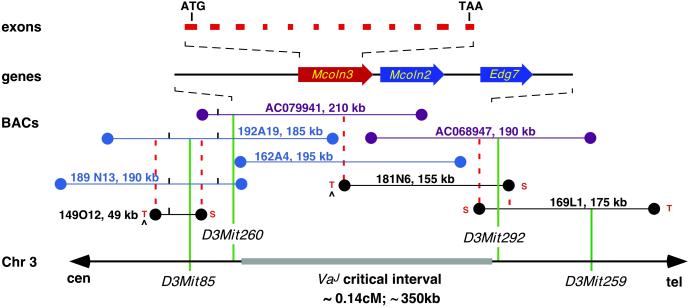
Previous linkage analyses placed Va to distal chromosome 3 (15), and we recently refined the VaJ map position to a 0.14-centimorgan interval (13). To clone Va, we established a physical map with BAC clones, spanning an estimated physical distance of not more than 350 kb, across the VaJ recombinant interval (Fig. 2). Two overlapping BAC clones (RP23–121J1 and RP23–108E10), positive for the recombinant flanking markers, were chosen for sample sequencing. Homology searches with genomic sequences from these clones identified the previously known lysophosphatidic acid receptor gene, Edg7 (16), a full-length mouse mRNA AK014467, and several overlapping mouse ESTs representing a cDNA not previously described. AK014467 and the mouse transcript shared significant amino acid similarities (67% and 73%, respectively) to human mucolipin-1 (MCOLN1, AF249319) and were subsequently designated Mcoln2 (AY083532) and Mcoln3 (AY083531), respectively.
Fig 2.
Physical map of the VaJ critical region. The thick horizontal gray line (Bottom) represents the VaJ critical interval defined by the polymorphic Mit markers on mouse chromosome 3. cM, centimorgan. A contig of 15 overlapping BAC clones is represented by horizontal lines; RPCI-23 BAC clones are shown in blue, Incyte Genomics BAC clones are in black, and BAC clones used for sample sequencing are in purple. Coordinates and corresponding sizes appear next to each BAC clone. SP6 and T7 ends of BACs are indicated by S or T, respectively. BAC ends used to derive probes for screening of the RPCI-23 BAC library are indicated by ^. Dashed lines in red indicate anchored SP6 and T7 BAC ends that have been used in PCR cross-screening and/or Southern blot experiments to confirm overlaps among BAC clones. Genes that map within the candidate region are shown with an arrow indicating transcription orientation. Mcoln3 exons are indicated. Genomic clones containing Mcoln2 and Mcoln3, AC068947 (RP23-108E10), and AC079941 (RP23-121J1).
Mucolipin-3 (Mcoln3) Encodes a Predicted Ion Channel.
Mcoln3 has an ORF of 1,662 bp and encodes a protein of 553 aa with a predicted molecular mass of ≈64 kDa (Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). The deduced amino acid sequence shares the highest degree of similarity with Mcoln2 (77%) and Mcoln1 (74%). Secondary structure analyses predict that Mcoln3 contains six transmembrane domains (S1–S6) with short cytoplasmic amino and carboxy termini (Fig. 3A). Sequence motif analyses identify an ion-transport domain (Pfam00520) and a transient-receptor-potential-like (TRPL) motif (PS50272) between S3 and S6, as well as a putative pore region between S5 and S6 (PS50273) (Fig. 3B). Sequence similarities of Mcoln3 with Drosophila CG8743 (63%) and C. elegans CUP-5 (55%) homologues are significant, with the highest degree of conservation being observed across the TRPL motif (74% and 73%, respectively). Amino acid sequence comparison of Mcoln3 with Pkd2 and TRP reveals only limited but significant similarity (49% and 32%, respectively) across the TRPL region (Fig. 3B).
The Varitint-Waddler Phenotype Is Associated with Mutations in Mcoln3.
All 12 coding exons of Mcoln3 were analyzed for mutations by sequence analyses of PCR products amplified from the genomic DNA of mutant (Va/Va and VaJ/VaJ) and control strains (C57BL/10J and C57BL/6J; Fig. 3C). The Va allele carries a 1255G → C transversion in exon 10 leading to an Ala419Pro substitution in the fifth predicted transmembrane domain of the protein (Fig. 3 A and C). The mutation in VaJ is a 1085T → C transition in exon 8 resulting in an Ile362Thr substitution in the second predicted extracellular loop (Fig. 3 A and C). Interestingly, we also detected the 1255G → C nucleotide change in VaJ, suggesting that VaJ arose in cis to Va (Fig. 3C). These nucleotide changes cosegregated with VaJ in the critical recombinants, and were not present in the parental or other representative inbred strains. No mutations were found in the coding regions of Edg7 and Mcoln2.
Mcoln3 Localizes to Vesicular Compartments and Stereocilia in Cochlea Hair Cells.
To gain insights into the cellular function of Mcoln3, we determined its subcellular location. By using polyclonal antibodies directed against either an extracellular or cytoplasmic epitope of Mcoln3 (Fig. 3A), the distribution of the protein in the OC was determined in wild-type (Fig. 4A) and mutant mice (Fig. 4 D and E), and adult rat (Fig. 4 F–J). In the mouse, Mcoln3 is highly expressed in the cytoplasm of IHCs and OHCs where labeling appears as a punctuate pattern throughout the hair cell bodies (Fig. 4A). A similar staining was also observed in the cytoplasm of IHCs and some OHCs of VaJ/VaJ and +/VaJ mice (Fig. 4 D and E). Immunoreactivity in the adult OC of the rat was specific and concentrated to cytoplasmic compartments of IHCs with all four anti-Mcoln3 antisera that we generated (Fig. 4 F–J). Fluorescence labeling was also detected in the subcuticular region (Fig. 4I) and the pericuticular necklace (Fig. 4J). In addition, modest immunoreactivity localizes to the plasma membrane of the stereocilia (Fig. 4J). Consistent with circling behavior in Va and VaJ mutants, we also detected Mcoln3 expression in vestibular hair cells (data not shown).
Discussion
A high-resolution genetic and physical mapping approach localized Va to a minimal region of 350 kb, in which three genes were identified. The only nucleotide changes were found in Mcoln3, and those were absent from parental controls (C57BR/cdJ, C57L/J, C58/J, C57BL/6J, C57BL/10J, C57BL/KS, C57BL/ScSnJ, and STOCK-a/a ma ft/ma ft) and in other strains that are representative members of different subgroups of inbred mouse strains (17) such as Castle's mice (CBA/CaJ, 129/SvJ, DBA/2J, BALB/cByJ, and AKR/J), Swiss mice (FVB/FnJ and NOD/LtJ) and wild-derived mice from different continents (PERC/Ei, MOLF/Ei, CZECHII/Ei, and CAST/Ei) suggesting that the sequence alterations are causative mutations. Moreover, similar pathogenic mutations were found in Cup-5 (G401E and G473D) and MCOLN1 (D362Y and V446L). The Ala419Pro substitution in Va in the fifth transmembrane domain, located near the predicted pore region, is likely to cause a gain-of-function or dominant-negative effect (18), although other mechanisms, such as the partial loss of functionality, cannot be entirely ruled out. VaJ was first recognized as a less variegated offspring from a Va linkage test cross, and subsequent breeding showed that the deviant was a somatic and germ-line mosaic and allelic to Va (10). The presence of two missense mutations in Mcoln3 in VaJ argues that the 1085T → C transition arose in cis to Va. Given the consistently milder phenotypes in VaJ, it suggests that Ile362Thr acts as an intragenic suppressor mutation. However, definitive proof of the causative nature of the missense mutations awaits further transgenic experiments and molecular functional studies.
Mucolipins constitute a newly recognized family of cation channel proteins with homologues in mouse (Mcoln1), D. melanogaster (CG8743), and C. elegans (CUP-5). Mutations in human MCOLN1 cause the neurodegenerative lysosomal disorder mucolipidosis type IV (OMIM 252650) (19–22), and loss-of-function mutations in C. elegans-CUP-5 lead to endocytosis defects, formation of large lysosomal vacuoles, and increased apoptosis (23, 24). Here, we describe two additional members, Mcoln2 and Mcoln3, of the mammalian gene family. The actual role of Mcoln2 is not known and may be defined by either cellular or genetic means. The motif and sequence similarities of Mcoln3 with C. elegans CUP-5 and human MCOLN1 as well as the punctuate staining of cytoplasmic compartments and the percuticular necklace suggest that Mcoln3 is associated with vesicular structures (25), and thus plays a critical role in vesicles. Defects in vesicle function often result in skin pigmentation abnormalities such as seen in the dilute, leaden, chocholate, or ashen mutants in which mutations in myosin Va (MyoVa), melanophilin (Mlph), Rab38, and Rab27a affect transport and trafficking of late-stage melanosomes (26–29). Based on the motif similarities with ion channels, we hypothesize that Mcoln3 is involved in vesicular and/or intracellular ion homeostasis in inner ear hair cells and melanocytes.
Va homozygotes were reported to be less viable (9), and we recently showed that VaJ homozygotes on the C57BL/6J background (after six backcross generations) have only a 34% chance of survival (13). Possibly, embryonic melanocytes fail to differentiate, migrate, or survive, and subsequently, downstream apoptotic events, similar to those observed in the null allele of CUP-5 (24), may cause the embryonic lethal phenotype. This observation and our generation of a C57BL/6J congenic strain (B6.VaJ) present an additional avenue to further characterize the cellular function of Mcoln3 and its dependence on the genetic background.
Mucolipins show sequence and motif similarities to the TRP ion channel family (30–32), including the polycystin-2 (PKD2) protein family of nonselective intracellular ion channels (33–35). TRP channels are characterized by a six-transmembrane-domain topology and their permeability to cations, including Ca2+ (31), a characteristic that is shared with the mechanosensitive transduction channel in vertebrate hair cells. Proteins of the TRP family are involved in the transduction of sensory stimuli in vision (36), olfaction (37), thermoreception (38), osmoregulation (39), and mechanosensation (40). Two members of the TRP gene family, NompC and TRPV4 (former Osm-9), were shown to be involved in mechanotransduction in D. melanogaster and C. elegans, respectively (37, 40), and more recently the murine homolog of TRPV4 was localized to inner ear hair cells (41). Our localization of Mcoln3 to the stereocilia, however, leads to the interesting hypothesis that this predicted channel might also be involved in sensory transduction.
Several lines of evidence argue for a cell-autonomous function of Mcoln3: (i) independent defects in the OC and stria vascularis as shown (12), (ii) erratic circling behavior indicative of vestibular hair cell defects, and (iii) localization of Mcoln3 to cochlea and vestibular hair cells. Thus, Va encodes an interesting molecular link between inner ear hair cells and melanocytes. Finally, human MCOLN2 and MCOLN3 are candidate genes for inherited and/or sporadic neurosensory disorders.
Supplementary Material
Acknowledgments
We thank Bill Pavan, Mark Fogg, Feng Qian, Emma Morton-Bours, Keith Vokey, and Susan Cole for discussions, Doris Wu, Dennis Drayna, Bob Wenthold, and Heinz Arnheiter for valuable comments on the manuscript, Alfredo Calderon for technical assistance, and Melissa Irby and Jimmy Fiallos for animal colony management. This work was supported by the intramural program of the National Institute on Deafness and Other Communication Disorders.
Abbreviations
TRP, transient receptor potential
BAC, bacterial artificial chromosome
IHCs, inner hair cells
OHCs, outer hair cells
OC, organ of Corti
Pn, postnatal day n
En, embryonic day n
References
- 1.Steel K. P. & Kros, C. J. (2001) Nat. Genet. 27 143-149. [DOI] [PubMed] [Google Scholar]
- 2.Geissler E. N., Ryan, M. A. & Housman, D. E. (1988) Cell 55 185-192. [DOI] [PubMed] [Google Scholar]
- 3.Chabot B., Stephenson, D. A., Chapman, V. M., Besmer, P. & Bernstein, A. (1988) Nature 335 88-89. [DOI] [PubMed] [Google Scholar]
- 4.Matsui Y., Zsebo, K. M. & Hogan, B. L. (1990) Nature 347 667-669. [DOI] [PubMed] [Google Scholar]
- 5.Huang E., Nocka, K., Beier, D. R., Chu, T. Y., Buck, J., Lahm, H. W., Wellner, D., Leder, P. & Besmer, P. (1990) Cell 63 225-233. [DOI] [PubMed] [Google Scholar]
- 6.Hodgkinson C. A., Moore, K. J., Nakayama, A., Steingrimsson, E., Copeland, N. G., Jenkins, N. A. & Arnheiter, H. (1993) Cell 74 395-404. [DOI] [PubMed] [Google Scholar]
- 7.Cable J., Huszar, D., Jaenisch, R. & Steel, K. P. (1994) Pigment Cell Res. 7 17-32. [DOI] [PubMed] [Google Scholar]
- 8.Cable J., Jackson, I. J. & Steel, K. P. (1995) Mech. Dev. 50 139-150. [DOI] [PubMed] [Google Scholar]
- 9.Cloudman A. M. & Bunker, L. E. (1945) J. Hered. 36 258-263. [Google Scholar]
- 10.Lane P. W. (1972) J. Hered. 63 135-140. [DOI] [PubMed] [Google Scholar]
- 11.Deol M. S. (1954) J. Genet. 52 562-588. [Google Scholar]
- 12.Cable J. & Steel, K. P. (1998) Hear. Res. 123 125-136. [DOI] [PubMed] [Google Scholar]
- 13.Kim, H. J., Jackson, T. & Noben-Trauth, K. (September 18, 2002) J. Assoc. Res. Otolaryngol., 10.1007/s10162-002-3011-0. [DOI] [PMC free article] [PubMed]
- 14.Belyantseva I. A., Adler, H. J., Curi, R., Frolenkov, G. I. & Kachar, B. (2000) J. Neurosci. 20 RC116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mobraaten L. E., Bunker, H. P., DeMaeyer-Guignard, J., DeMaeyer, E. & Bailey, D. W. (1984) J. Hered. 75 233-234. [DOI] [PubMed] [Google Scholar]
- 16.Contos J. J. & Chun, J. (2001) Gene 267 243-253. [DOI] [PubMed] [Google Scholar]
- 17.Beck J. A., Lloyd, S., Hafezparast, M., Lennon-Pierce, M., Eppig, J. T., Festing, M. F. & Fisher, E. M. (2000) Nat. Genet. 24 23-25. [DOI] [PubMed] [Google Scholar]
- 18.Lester H. A. & Karschin, A. (2000) Annu. Rev. Neurosci. 23 89-125. [DOI] [PubMed] [Google Scholar]
- 19.Bargal R. & Bach, G. (1997) J. Inherit. Metab. Dis. 20 625-632. [DOI] [PubMed] [Google Scholar]
- 20.Bargal R., Avidan, N., Ben-Asher, E., Olender, Z., Zeigler, M., Frumkin, A., Raas-Rothschild, A., Glusman, G., Lancet, D. & Bach, G. (2000) Nat. Genet. 26 118-123. [DOI] [PubMed] [Google Scholar]
- 21.Sun M., Goldin, E., Stahl, S., Falardeau, J. L., Kennedy, J. C., Acierno, J. S., Jr., Bove, C., Kaneski, C. R., Nagle, J., Bromley, M. C., et al. (2000) Hum. Mol. Genet. 9 2471-2478. [DOI] [PubMed] [Google Scholar]
- 22.Bassi M. T., Manzoni, M., Monti, E., Pizzo, M. T., Ballabio, A. & Borsani, G. (2000) Am. J. Hum. Genet. 67 1110-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fares H. & Greenwald, I. (2001) Nat. Genet. 28 64-68. [DOI] [PubMed] [Google Scholar]
- 24.Hersh B. M., Hartwieg, E. & Horvitz, H. R. (2002) Proc. Natl. Acad. Sci. USA 99 4355-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kachar B., Battaglia, A. & Fex, J. (1997) Hear. Res. 107 102-112. [DOI] [PubMed] [Google Scholar]
- 26.Loftus S. K., Larson, D. M., Baxter, L. L., Antonellis, A., Chen, Y., Wu, X., Jiang, Y., Bittner, M., Hammer, J. A., III & Pavan, W. J. (2002) Proc. Natl. Acad. Sci. USA 99 4471-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matesic L. E., Yip, R., Reuss, A. E., Swing, D. A., O'Sullivan, T. N., Fletcher, C. F., Copeland, N. G. & Jenkins, N. A. (2001) Proc. Natl. Acad. Sci. USA 98 10238-10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X. S., Rao, K., Zhang, H., Wang, F., Sellers, J. R., Matesic, L. E., Copeland, N. G., Jenkins, N. A. & Hammer, J. A., III (2002) Nat. Cell Biol. 4 271-278. [DOI] [PubMed] [Google Scholar]
- 29.Wilson S. M., Yip, R., Swing, D. A., O'Sullivan, T. N., Zhang, Y., Novak, E. K., Swank, R. T., Russell, L. B., Copeland, N. G. & Jenkins, N. A. (2000) Proc. Natl. Acad. Sci. USA 97 7933-7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Littleton J. T. & Ganetzky, B. (2000) Neuron 26 35-43. [DOI] [PubMed] [Google Scholar]
- 31.Clapham D. E., Runnels, L. W. & Strubing, C. (2001) Nat. Rev. Neurosci. 2 387-396. [DOI] [PubMed] [Google Scholar]
- 32.Montell C., (2001) Sci. STKE, http://stke.sciencemag.org/cgi/content/full/OC_sigtrans;2001/90/re1. [DOI] [PubMed]
- 33.Gonzalez-Perret S., Kim, K., Ibarra, C., Damiano, A. E., Zotta, E., Batelli, M., Harris, P. C., Reisin, I. L., Arnaout, M. A. & Cantiello, H. F. (2001) Proc. Natl. Acad. Sci. USA 98 1182-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanaoka K., Qian, F., Boletta, A., Bhunia, A. K., Piontek, K., Tsiokas, L., Sukhatme, V. P., Guggino, W. B. & Germino, G. G. (2000) Nature 408 990-994. [DOI] [PubMed] [Google Scholar]
- 35.Koulen P., Cai, Y., Geng, L., Maeda, Y., Nishimura, S., Witzgall, R., Ehrlich, B. E. & Somlo, S. (2002) Nat. Cell Biol. 4 191-197. [DOI] [PubMed] [Google Scholar]
- 36.Zhu X., Jiang, M., Peyton, M., Boulay, G., Hurst, R., Stefani, E. & Birnbaumer, L. (1996) Cell 85 661-671. [DOI] [PubMed] [Google Scholar]
- 37.Colbert H. A., Smith, T. L. & Bargmann, C. I. (1997) J. Neurosci. 17 8259-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peier A. M., Moqrich, A., Hergarden, A. C., Reeve, A. J., Andersson, D. A., Story, G. M., Earley, T. J., Dragoni, I., McIntyre, P., Bevan, S. & Patapoutian, A. (2002) Cell 108 705-715. [DOI] [PubMed] [Google Scholar]
- 39.Strotmann R., Harteneck, C., Nunnenmacher, K., Schultz, G. & Plant, T. D. (2000) Nat. Cell Biol. 2 695-702. [DOI] [PubMed] [Google Scholar]
- 40.Walker R. G., Willingham, A. T. & Zuker, C. S. (2000) Science 287 2229-2234. [DOI] [PubMed] [Google Scholar]
- 41.Liedtke W., Choe, Y., Marti-Renom, M. A., Bell, A. M., Denis, C. S., Sali, A., Hudspeth, A. J., Friedman, J. M. & Heller, S. (2000) Cell 103 525-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.