Abstract
The transcription factors encoded by the E2A gene are known to be essential for B lymphocyte development, and ectopic expression or gene inactivation studies have revealed several potential lineage-specific E2A target genes. However, it remains unknown whether these target genes are directly regulated by E2A at the transcriptional level. We therefore generated mice carrying an affinity-tagged E2A knock-in allele to provide a system for the direct elucidation of E2A target genes based on E2A binding to target regulatory regions. Abelson-transformed pre-B cell lines derived from these mice were used in chromatin immunoprecipitation experiments to identify regulatory sequences bound by E2A in the context of an early B lymphocyte environment. Significant E2A binding was detected at the promoters and enhancers of several essential B-lineage genes, including the Igκ intronic and 3′ enhancers, λ5 and VpreB surrogate light chain promoters, the EBF locus promoter region, and the mb-1 (Igα) promoter. Low levels of E2A binding were observed at several other lymphoid-restricted regulatory regions including the Ig heavy chain (IgH) intronic enhancer, the IgH 3′ enhancers hs3b/hs4, the RAG-2 enhancer, and the 5′ regions of the B29 and TdT loci. An E2A target gene, the predicted butyrophilin-like gene NG9 (BTL-II), was also identified by using a chromatin immunoprecipitation-based cloning strategy. In summary, our studies have provided evidence that E2A is directly involved in the transcriptional regulation of a number of early B-lineage genes.
The development of lymphocytes from hematopoietic stem cells involves a series of highly regulated differentiation events that depend on the collaborative efforts of a number of transcription factors. These transcription factors coordinately regulate lymphopoiesis through the initiation, maintenance, and restriction of lineage-specific gene expression programs. The transcription factors encoded by the E2A gene are known to play critical roles in the regulation of lymphocyte development. E2A proteins are highly expressed in developing lymphoid cells (1, 2), and gene targeting studies have shown that E2A proteins are required for the initiation of B cell development in the bone marrow. Mice deficient in E2A demonstrate a complete and persistent block at the earliest stage of B cell development before the initiation of Ig heavy chain gene rearrangements, as well as a dramatic reduction in thymocyte number (3, 4).
The mammalian E2A gene encodes two major products, E12 and E47, which are members of the basic helix–loop–helix (bHLH) family of transcription factors (5, 6). bHLH proteins are characterized by a conserved HLH dimerization domain and an adjacent basic region that mediates DNA binding (7, 8). E12 and E47 are generated by alternative splicing of adjacent exons encoding their bHLH domains and bind the consensus E-box sequence CANNTG as homodimers or as heterodimers with other bHLH transcription factors (5–7, 9, 10). As E-box binding factors, E12 and E47 are members of a subfamily of bHLH transcription factors denoted the E proteins (9). The E protein family also includes the transcription factors E2-2 and HeLa E-box binding protein (HEB), which play important roles in lymphocyte development as well (11–14).
E2A proteins were initially characterized for their binding activity at the Ig heavy and light chain enhancers, which were subsequently shown to contain several functionally relevant E2A binding sites (10, 15–18). Ectopic expression of E2A in non-B cells was also found to induce germ-line Ig transcription and rearrangement, suggesting that E2A might play a key role in Ig gene regulation (19–21). More recent studies demonstrated that transfection of the kidney cell line BOSC23 with E2A and the RAG recombination machinery was sufficient to drive diverse recombination events at the Ig heavy and light chain loci (22, 23). Overexpression studies have also implicated E2A in the regulation of other genes involved in the initiation and maintenance of the B cell developmental program. Ectopic expression of E12 in a macrophage line was shown to result in the induction of the B-lineage transcription factors EBF and Pax5/BSAP, as well as IL-7Rα and RAG-1 (24). Other potential E2A targets include several components of the pre-B cell and B cell receptor (BCR) complexes, including the surrogate light chains λ5 and VpreB and the BCR signaling molecules mb-1 (Igα) and B29 (Igβ) (25, 26). However, it remains unclear whether the lineage and stage-specific expression of these genes is directly regulated by E2A at the transcriptional level or, alternatively, is activated by other factors downstream of E2A.
Chromatin immunoprecipitation (ChIP) strategies have recently been used for more direct assessments of target gene regulation based on transcription factor binding to suspected regulatory regions. For example, ChIP has been successfully used in the verification and cloning of several E2F target genes in human cell lines (27). In yeast, ChIP-based analysis of transcription factor binding to relevant target sequences has been facilitated by the introduction of affinity tags to endogenous proteins by homologous recombination (28). We have therefore combined an in vivo murine gene-tagging approach with a ChIP assay to investigate both suspected and novel E2A target genes in lymphoid cells. Mice carrying a dual affinity-tagged E2A knock-in allele have been generated and used to derive pre-B cell lines that express a functional E2A fusion protein. This fusion protein provides a means for highly efficient immunoprecipitation of E2A-bound DNA fragments, which have been screened for the presence of several suspected E2A target sequences. We have used this approach to identify a subset of B-lineage genes whose regulatory regions are bound by E2A under physiological conditions, providing evidence for direct transcriptional regulation of these genes by E2A. The pre-B cell lines expressing affinity-tagged E2A have also been used in ChIP-based cloning of a previously uncharacterized E2A target gene.
Materials and Methods
E2AFH Knock-In Mice.
The E2AFH gene-targeting construct was generated by using an E2A genomic sequence isolated from the 129/sv strain. A 2-kb genomic fragment of homologous sequence was used in subcloning to create an in-frame fusion of the E2A carboxyl terminus with oligonucleotides encoding the following amino acid sequence: Ala-Gly-Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys-Ala-Gly-His-His-His-His-His-His-Stop. A phosphoglycerate kinase promoter-driven neomycin resistance cassette (PGK-Neo) was inserted into a unique XbaI site downstream of the E2A poly(A) signal and between the homologous segments for positive selection of clones undergoing homologous recombination. A PGK promoter-driven thymidine kinase (PGK-TK) cassette was placed outside of the homologous targeting sequences to allow for negative selection against random nonhomologous recombination events. The final targeting construct was linearized and transfected into embryonic stem cells by electroporation. Transfected clones were cultured under double selection with gancyclovir and G418, and correct targeting of 27 of 95 clones was determined by PCR screening. Germ-line transmission was obtained from one of two injected clones, and mice carrying the E2AFH allele were maintained in a specific pathogen-free environment at Duke University's animal facility.
Derivation of Abelson Pre-B Cell Lines.
Abelson murine leukemia virus (AMLV) was prepared as supernatant from ABO10 cells, pretreated with polybrene, and added to E2AFH/FH or E2Aloxp/loxp bone marrow cells cultured in RPMI 1640 media supplemented with 10% FBS, penicillin/streptomycin, β-ME, and gentamycin. After 3 wk, bulk cultures were subcloned to yield the Abelson lines E2AFH1B, E2AFH4, and E2Aloxp 1AB1.
Chromatin Extracts and Immunoprecipitations.
Cells were fixed and washed for preparation of chromatin extracts essentially as reported by Fernandez et al. (29). Fixed cells were then sonicated (Fisher Scientific 550 tapered microtip probe, setting 4) for 20–25 cycles of 25 s on a cold block with 15 s cooling between each burst, to obtain 0.5- to 1.0-kb DNA fragments. Sonicates were centrifuged at 14,000 × g for 5 min, and supernatants were harvested and stored at −80°C. ChIP procedures were adapted from those described by Fernandez et al. (29). Chromatin extracts (1–2 mg) from the E2AFH1B pre-B cell line or the untagged control cell line AMLV-3B (derived from C57BL/6 bone marrow) were diluted 1:10 in ChIP buffer (140 mM NaCl/100 μg/ml BSA/100 μg/ml yeast tRNA/1% Triton X-100/1 mM PMSF) and incubated with 50 μl of anti-FLAG agarose for 2 h at 4°C, rotating slowly. The bound agarose beads were harvested by centrifugation (14,000 × g for 15 s) and washed three times in 1 ml of IP buffer, twice in 1 ml of IP buffer containing 500 mM NaCl, twice in 1 ml of wash buffer (10 mM Tris⋅HCl, pH 8.0/250 mM LiCl/1 mM EDTA), and three times in 1 ml of TE buffer (10 mM Tris⋅HCl, pH 8.0/1 mM EDTA). Bound DNA was eluted, incubated at 65°C overnight to reverse cross-links, and then RNase-treated, deproteinized, and precipitated as described by Fernandez et al. (29). Processed DNA fragments were resuspended in 100 μl of distilled water. Cloning of immunoprecipitated DNA fragments was performed essentially as described by Weinmann et al. (27).
PCR Screening of Suspected Target Genes.
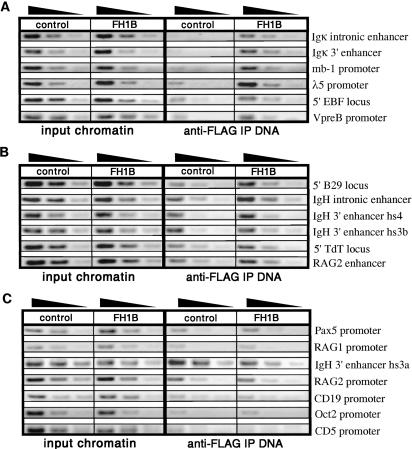
A series of 4-fold dilutions of input chromatin and immunoprecipitated DNA from E2AFH and control cell lines was PCR-amplified for 29–32 cycles (94°C, 1 min; 58°C, 1 min; 72°C, 1min; with 2 min final extension at 72°C) in 20 μl PCR buffer containing 3 mM MgCl2 and Platinum Taq polymerase (Invitrogen). The entire PCR samples were then resolved on 1.5% agarose gels and visualized by ethidium bromide staining. Relative band intensity was used in qualitative comparisons of target sequence enrichment from ChIPs of E2AFH vs. untagged control cell lines. All oligonucleotides used in PCR screening of immunoprecipitated DNA were 23-mers (IDT DNA) designed to amplify 150- to 200-bp regions surrounding potential E2A-binding sites within the regulatory regions of suspected target genes (Table 1, Regulatory region and Clone). In some cases, multiple primer sets were used to cover larger genomic sequences and potential regulatory regions; representative results for each of these regions are presented in Fig. 4.
Table 1.
Oligonucleotide primer pairs used for ChIP-PCR screening of putative E2A target genes (Regulatory region), ChIP-PCR enrichment confirmation of clones isolated as E2A-bound sequences (Clone), and RT-PCR analysis of Abelson cell line transcripts (Transcript)
| Gene locus | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| Regulatory region | ||
| Igκ intronic enhancer | CAG AGG GGA CTT TCC GAG AGG CC | ACC CTG GTC TAA TGG TTT GTA AC |
| Igκ 3′ enhancer | ATA GCA ACT GTC ATA GCT ACC GT | GCA GGT GTA TGA GGC TTT GGA AA |
| mb-1 promoter | CCA CGC ACT AGA GAG AGA CTC AA | CCG CCT CAC TTC CTG TTC AGC CG |
| λ5 promoter | GGG TTA AGA CAG GCA GCT GTG AG | CAA ACC CCA GGC TGT CTC TAG TT |
| 5′ EBF locus | GTG GGG TAC CAG CTG AAC TCC AC | CAG CTG AGC ATG TGT TTT AAT TG |
| Pax5 promoter | ATG GAG GTT GCA ATT GAG TTG GG | ACA ATT CTG CCA AGC AAG TGG TT |
| IgH intronic enhancer | TCA GAA CCA GAA CAC CTG CAG CA | GGT GGG GCT GGA CAG AGT GTT TC |
| IgH 3′ enhancer HS3a | GCT CTG GTT TGG GGC ACC TGT GC | GGG TAG GGC AGG GAT GCT CAC AT |
| IgH 3′ enhancer HS3b | TGG TTT GGG GCA CCT GTG CTG AG | GGG TAG GGC AGG GAT GTT CAC AT |
| IgH 3′ enhancer HS4 | GGG TAG ATG CAG CCT GTG TTC CG | GGA GTG TAG AGG AGA GCT GTG GC |
| RAG-1 promoter | AGC CAG GTG CAG CTG GAG CTG GG | CAA CAT ATG CTG TCT ACT CTC TC |
| RAG-2 promoter | TGA CTG GTA TCT CGG GAC TTA AC | GTG CCT ACA GAT GTT CCA GTG AG |
| RAG-2 enhancer | GTC ACT TGG AAA CCA CGT GGT TA | TAG TGC ACA TGC TGC TGC TTA TA |
| 5′ B29 locus | ACA TGC TGC CCA GGG TAG AGA TT | ATC TGG GAG CCC CTT AAA CAA CT |
| 5′ TdT locus | TGA GCT GGT TTT GTA ATT ATT AG | GAC TCA GGA AGA CGC ACA TAC CA |
| CD19 promoter | CCT AAT GCT ATC CCC AGA TGA TA | TAA ATA TTT TTC AGA TGA GTG GG |
| Oct-2 promoter | TGG CCT GAT AGT GTG GAA CTG TA | CGG CCA TTT GCA CAC AGC CAC GG |
| VpreB promoter | TGC CAA GCT GGC CAT GTG AAC AC | GAT GTT CCT CTA CCA TAT GTG AG |
| CD5 promoter | CAT GTG AAT GGC CAG TGG GCA CT | CAG GCA GTG TGG GCC TGT GTC AC |
| Clone | ||
| ChIP-1 | CAC TTC TAA ACG GGC AGA CTT TA | GGA GGA AAT GCC AAG GCA CAT CC |
| ChIP-5 | AAG CCA CAT GGG GAT CTC TCC CC | TCA CAA CTA ATC TGT TAC AAG TG |
| ChIP-6 | ATT GTA GTG TCC CTG GGT GAG CA | AGA GAA TTG TCT TAG GAG TCA GA |
| ChIP-9 | GGA GTG GCA GAG GTT AAG TAC CT | TTA CAG GGA ACC TGA GCC ACA AC |
| ChIP-11 | ATG TTC CAA AGC CAA CAT GCA AC | TCA CAC ATG CAC ATA CAC GAA AA |
| ChIP-13 | ATC TCT CTA GCT GAA ATT TAG AT | TTT CTA CCA CAA GCA TTA TGT TA |
| ChIP-17 | GCA TAG GCA CTT ATG TAG TGT CC | TGC TTG AAT TAG CTC TTA TAG AA |
| ChIP-18 | TGA GAG CTA GCA CTA CAG GCT TA | AGC ATT TAG GAA GAA GCA AAG GC |
| ChIP-20 | ATG CCC TCT CCT GGT CTT TAT GG | CAG AAA TTT TCC TAA TAG GAC CT |
| ChIP-24 | ATT TAT TGC TAA GAA GAC ATG CA | CAA ACA CAG TCT TTT GGC ACA TG |
| Transcript | ||
| 5′ NG9 | AAA GTC GGG GAA GAT GCC CTG CT | CGG CCT CCG TAC CCC TCC ATC GG |
| 3′ NG9 | CTG GGC CAG GAG AAA ACA GCC CG | GGA AGT CGC CGC CTG TGG TTT TC |
| Igk | TCC ATC TTC CCA CCA TCC AG | GAT GTC TTG TGA GTG GCC TC |
| λ5 | TGT GAA GTT CTC CTC CTG CTG | ACC ACC AAA GTA CCT GGG TAG |
| Iμ | GGT GGC TTT GAA GGA ACA ATT CCA C | TCT GAA CCT TCA AGG ATG CTC TTG |
| EF1α | AGT TTG AGA AGG AGG CTG CT | CAA CAA TCA GGA CAG CAC AGT C |
Fig 4.
ChIP-PCR screen for E2A binding to regulatory regions of potential target genes. A series of 4-fold dilutions of input chromatin and immunoprecipitated DNA from E2AFH1B or untagged control pre-B cell lines were PCR-amplified by using primers specific for suspected E2A target regions (see Table 1, Regulatory region). Sequence enrichment in immunoprecipitated DNA from E2AFH1B vs. control chromatin indicated E2A binding within genomic region. Each experiment was repeated at least three times, and results shown were reproducible for all targets. (A) Significant enrichment of the Igκ enhancers, λ5 and VpreB promoters, 5′ EBF locus, and mb-1 promoter was observed. (B) Low levels of enrichment were seen for the IgH intronic and hs3b/hs4 enhancers, 5′ B29 and TdT loci, and RAG-2 enhancer. (C) No enrichment was indicated for the Pax5, CD19, Oct-2, RAG-1, RAG-2, and CD5 promoters and the IgH hs3a enhancer.
Results
Generation of Affinity-Tagged E2AFH Mice.
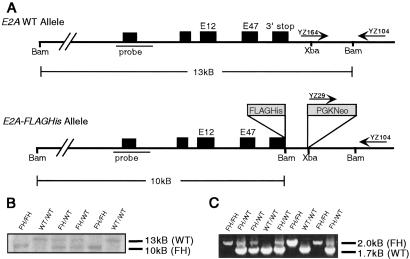
We wanted to use a ChIP strategy to elucidate target genes that are directly regulated by E2A during lymphocyte development. Because the efficiency of immunoprecipitation-based studies can be limited by antibody specificity or affinity, we used an in vivo gene-tagging approach in generating an affinity-tagged E2A knock-in mouse model to facilitate the isolation of endogenous E2A-bound DNA sequences. Oligonucleotides encoding dual affinity tags were cloned into the 3′ exon of the genomic E2A locus such that both E12 and E47 would be expressed as fusion proteins carrying carboxyl-terminal FLAG epitope and hexahistidine sequences (Fig. 1A). Proper targeting of the locus with the knock-in allele was confirmed by Southern blotting (Fig. 1B) and PCR genotyping (Fig. 1C). Bands representing the expected wild-type and E2AFH alleles were detected, and normal distributions of each genotype were observed, indicating that the knock-in allele did not have an effect on neonatal survival.
Fig 1.
Generation of dual affinity-tagged E2A knock-in allele. (A) Wild-type E2A locus and E2A-FLAGHis knock-in allele containing in-frame fusion of 3′ E2A exon sequence with FLAG and hexahistidine affinity tags. (B) Southern blot genotyping confirmation of BamHI-digested tail DNA from E2AFH/+ intercross pups. A 1.3-kb PCR-generated genomic E2A probe was used to detect the wild-type and knock-in alleles. (C) PCR genotyping of intercross pups by using YZ-104 (E2A antisense), YZ-29 (Neo sense), and YZ-164 (E2A sense) primers (31).
E2AFH Fusion Protein Supports Normal Lymphocyte Development.
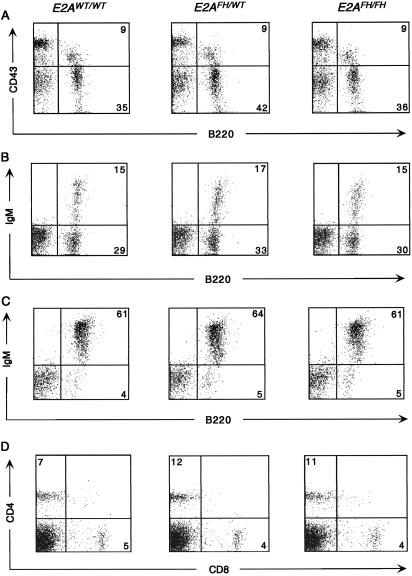
B lymphocyte development is highly dependent on E2A protein dosage, such that even a 50% reduction in E2A protein activity translates into altered B cell development in the bone marrow (4, 13, 30). We therefore wanted to investigate B-lymphopoiesis in E2AFH mice as a sensitive indicator of E2A fusion protein expression and function. Fluorescence-activated cell sorter (FACS; Becton Dickinson) analysis of bone marrow and splenocytes from wild-type, E2AFH/+ heterozygous, and E2AFH/FH homozygous mice showed that lymphocyte development was normal in the knock-in mice. The E2AFH allele led to no significant alterations in the relative percentages or numbers of pro- and pre-B cell populations (Fig. 2A) and mature IgM+ B cell populations in the bone marrow (Fig. 2B). Normal B and T lymphocyte profiles were also observed in the spleen (Fig. 2 C and D) and thymus, and other hematopoietic populations in the bone marrow and spleen were also normal (data not shown). The fusion protein was therefore expressed from the targeted locus and was functional in supporting normal lymphocyte development.
Fig 2.
Normal lymphocyte development in E2AFH mice. Bone marrow (A and B) and splenocytes (C and D) from 3-wk-old wild-type (E2AWT/WT), heterozygous (E2AFH/WT), and homozygous (E2AFH/FH) littermates were analyzed by FACS for expression of lymphocyte lineage surface markers by using the following fluorescent antibody conjugates: IgM-FITC, B220-APC, CD19-PE, CD4-FITC, and CD8-APC (Caltag, South San Francisco, CA); and CD43-PE (PharMingen). (A) Bone marrow staining for B220 and CD43. (B) Bone marrow staining for B220 and surface IgM. (C) Splenocyte staining for B220 and surface IgM. (D) Splenocyte staining for CD4 and CD8.
Abelson-Transformed Pre-B Cell Lines Express Functional E2AFH Fusion Protein.
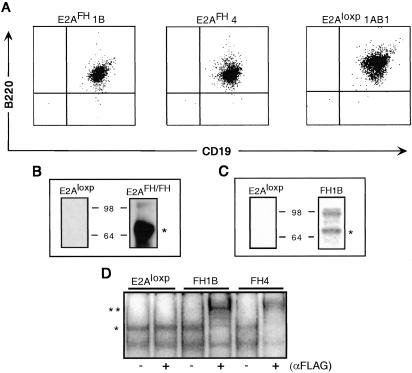
To establish a culture system for studying E2A target sequences in the context of an early B cell environment, bone marrow was isolated from E2AFH/FH mice and transformed with AMLV. The Abelson virus encodes the v-Abl oncogene and selectively targets developing pre-B cells for transformation. Primary transformants were established from E2AFH/FH bone marrow cultures, and two subcloned lines, E2AFH1B and E2AFH4, were then derived for subsequent studies. These lines, as well as an untagged control Abelson line (E2Aloxp 1AB1) generated from the bone marrow of mice carrying a loxp-flanked E2A knock-in allele (E2Aloxp/loxp), displayed a typical B220+CD19+ pre-B cell phenotype (Fig. 3A; ref. 31). Anti-FLAG immunoprecipitations of E2A from nuclear extracts from E2AFH1B or E2AloxP control cells were then performed to verify the expression and functional utility of the E2AFH fusion proteins. An anti-E2A Western blot of the immunoprecipitated extract confirmed that the fusion proteins were expressed and that affinity purifications could be performed by using the FLAG epitope tag (Fig. 3B). A band corresponding to the molecular mass of the immunoprecipitated E2A fusion protein (74 kDa) was also detected by colloidal staining of an SDS/PAGE gel (Fig. 3C). Electrophoretic mobility-shift analysis of nuclear extracts from the three Abelson clones (E2AFH1B, E2AFH4, and E2AloxP) indicated that the E2A fusion protein bound to a μE5 E-box oligonucleotide probe as expected and that the E2AFH-DNA complex could be mobility-shifted with anti-FLAG antibody (Fig. 3D).
Fig 3.
Characterization of Abelson-transformed pre-B cell lines. (A) FACS analysis for expression of B-lineage markers CD19 and B220 on E2AFH clone 1B, E2AFH clone 4, and E2Aloxp control clone 1AB1. (B) Anti-E2A (G193-86, PharMingen) Western blot of elution fractions from anti-FLAG agarose affinity-purification of primary E2Aloxp and E2AFH/FH transformant nuclear extracts (*, E2A monomer). (C) Colloidal Coomassie staining of FLAG peptide elution fractions from affinity-purification of subcloned Abelson lines. E2A monomer (*) is indicated, as well as a specific higher-molecular-weight band representing a commonly observed alternative isoform or modification of E2A. (D) Electrophoretic mobility-shift analysis on Abelson pre-B nuclear extracts for binding to radiolabeled μE5 oligonucleotide (10). E2A in μE5-binding complexes unshifted (*) or supershifted (**) with 1 μl of anti-FLAG antibody.
E2A Is Bound to a Subset of Lineage-Restricted Regulatory Regions.
Phenotypic and biochemical analysis of the E2AFH Abelson lines demonstrated that the E2A fusion protein was expressed, displayed normal DNA-binding characteristics in vitro, and could be immunoprecipitated with anti-FLAG antibody. The affinity-tagged protein should facilitate efficient immunoprecipitation of E2A-bound DNA sequences from cross-linked chromatin fragments. Chromatin extracts were therefore prepared from the E2AFH1B or untagged control cell lines, and E2A-bound DNA fragments were obtained by anti-FLAG immunoprecipitation. These sequences were then screened for enrichment of promoter and enhancer regions within several potential lymphoid-restricted E2A target genes. Unfractionated input chromatin and immunoprecipitated DNA from the E2AFH1B cell line and an untagged control cell line (AMLV-3B) were PCR-amplified by using primers surrounding the E-box sites within the regulatory elements of these genes (Table 1, Regulatory Region). Comparisons of the relative signals observed after amplification of immunoprecipitated DNA from tagged vs. control chromatin extracts allowed for qualitative determinations of whether each potential target sequence was enriched due to E2A binding.
Significant enrichment was observed for several suspected E2A targets, including the Igκ intronic and 3′ enhancers, the mb-1 (Igα) promoter, the λ5 and VpreB promoters, and the 5′ region of the EBF locus (Fig. 4A). Selective enrichment of these regulatory regions indicated E2A protein binding and suggested that E2A is likely to play a direct role in the transcriptional regulation of these genes during B cell development. Interestingly, several other potential E2A target sequences appeared to be only slightly enriched in the ChIP-PCR screen, including the IgH intronic and hs3b/hs4 3′ enhancers, the RAG-2 enhancer, and the 5′ regions of the B29 and TdT loci (Fig. 4B). Meanwhile, no enrichment was observed for several other potential E2A targets, including the IgH 3′ enhancer hs3a and the promoter sequences for Pax5, RAG-1, RAG-2, Oct-2, CD5, and CD19 (Fig. 4C). The observation that only a subset of putative E2A targets was actually enriched provided support for both the selectivity and relevance of the assay because enrichment required more than just an accessible locus containing potential E2A binding sites.
Identification of Novel E2A Target Genes by ChIP-Based Cloning.
Comparative PCR analysis provided a means for qualitative assessment of enrichment (and thus E2A binding) at a suspected target sequence. However, the regulatory regions identified as E2A targets by ChIP-PCR screening probably represent only a small subset of all genes subject to transcriptional regulation by E2A during B cell development. A more general approach was required to broaden the scope of the study to include the identification of novel E2A target genes. ChIP-based cloning strategies have recently been used in identifying novel target genes regulated by ubiquitous transcription factors such as E2F (27). We therefore used our E2AFH pre-B cell ChIP system for the cloning and characterization of novel E2A target sequences. Immunoprecipitated DNA fragments from E2AFH1B chromatin extracts were blunt-ended by polymerase treatment and ligated into pBluescript vector. The transformed clones were then screened by restriction digest, and 13 of 24 clones were found to contain inserts representing immunoprecipitated DNA fragments. These inserts were sequenced by using vector-specific primers, and the sequences obtained were then identified by blast or ensemble database searches for mouse genome matches. Ten of 13 sequences had single murine genome matches, whereas the remaining 3 yielded either no match or multiple genomic matches (Table 2). Sequence analysis of the 10 genomic clones indicated that 9 of 10 inserts contained at least one E box representing a potential E2A binding site. Importantly, eight of these nine sequences were subsequently found to be enriched by ChIP-PCR screening, providing strong confirmation that the majority of DNA fragments isolated by ChIP-based cloning represented actual E2A-bound sequences (Table 1, Clone; Table 2). Interestingly, seven of the eight sequences found to be enriched by ChIP-PCR were also located within predicted or known gene loci (Table 2).
Table 2.
Summary of inserts isolated as E2A-bound sequences from ChIP-based cloning of novel E2A target genes
| Clone | E boxes | Enriched | Chromosome | Gene | Family |
|---|---|---|---|---|---|
| ChIP-1 | 2 | Yes | 17 | NG9 | Butyrophilin-like |
| ChIP-2 | 0 | Multiple hits | |||
| ChIP-5 | 3 | Yes | 10 | None | N/A |
| ChIP-6 | 0 | No | 9 | Novel | PWWP domain |
| ChIP-8 | 0 | Multiple hits | |||
| ChIP-9 | 1 | Yes | 17 | Novel | Zn/PHD finger |
| ChIP-11 | 3 | Yes | 4 | Novel | TEF S-II homology |
| ChIP-13 | 1 | Yes | 6 | Novel | Similar to K-ras |
| ChIP-14 | 2 | No match | |||
| ChIP-17 | 1 | Yes | 15 | CDH7 | Cadherin precursor |
| ChIP-18 | 2 | Yes | 5 | Novel | Unknown |
| ChIP-20 | 2 | Yes | 8 | CSMD2 | C4B binding |
| ChIP-24 | 1 | No | X | None | N/A |
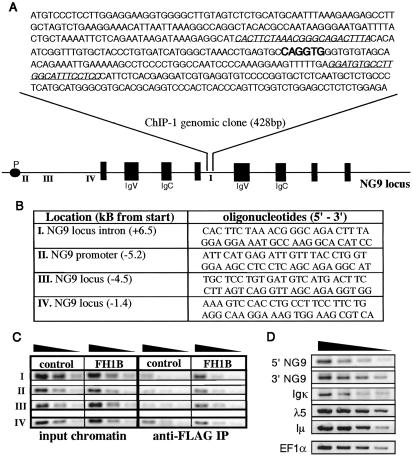
The ChIP-1 insert yielded a perfect match to an intronic sequence of the putative mouse butyrophilin-like gene NG9 (BTL-II), located within the extended MHC II locus (GenBank accession no. AF050157.1). The 428-bp sequence also contained two E-box sites, including one canonical E2A binding site, CAGGTG (Fig. 5A). The genomic region represented by the ChIP-1 clone was therefore selected for further analysis. Primers (I) specific for the sequences flanking the prominent ChIP-1 E box were used to confirm E2A-dependent enrichment of the region by PCR (Fig. 5 A and B). Because the clone represented intronic sequence between central exons of the gene, two additional primer sets (II, III) were also designed to screen for enrichment of the putative promoter region of NG9. A fourth primer pair (IV) covering what was speculated to be a nonregulatory region upstream of the first NG9 exon was used in ChIP-PCR screening as well. Interestingly, the genomic regions corresponding to both the ChIP-1 clone and the putative NG9 promoter region were moderately enriched in immunoprecipitated DNA from E2AFH1B chromatin (Fig. 5C). No significant enrichment of the nonregulatory region represented by primer pair IV was observed, supporting the specificity of the enrichment seen with the other primers.
Fig 5.
E2A target gene isolated by ChIP-based cloning. Immunoprecipitated DNA fragments from E2AFH1B chromatin extract were cloned, sequenced, and identified by mouse genome database search (see Table 2). (A) ChIP-1 clone insert with an exact match to an intronic region within the putative butyrophilin-like gene NG9. Clone-specific primers (I; in italics) surrounding the consensus E box (CAGGTG) were used to confirm E2A binding by ChIP-PCR. (B) Primers to the ChIP-1 insert (I), the predicted promoter region (II and III) of the NG9 locus, and a region upstream of the transcription start site (IV). (C) ChIP-PCR screen for E2A binding at the ChIP-1 insert genomic sequence and NG9 5′ regulatory regions. (D) Semiquantitative RT-PCR analysis of four 3-fold dilutions of E2AFH1B cDNA for the predicted NG9 transcript, along with relevant B-lineage genes (Igκ, λ5, Iμ) and a loading control (EF1α) (see Table 1, Clone). Thirty cycles (21 cycles for EF1α) of PCR amplification (94°C, 30 s; 57°C, 30 s; 72°C, 30 s, with 1 min extension at 72°C) were performed in PCR buffer containing 3 mM MgCl2 and Platinum Taq polymerase (Invitrogen).
The NG9 gene contains several conserved domains characteristic of the butyrophilin gene family, including two Ig variable-like (IgV) domains and two Ig constant region-like (IgC) domains (Fig. 5A) (32). No functional information on the gene product is available, however, and little is known about its expression in lymphoid cells. Because E2A was shown to bind to putative regulatory regions of NG9 and typically functions as a positive regulator of transcription, we wanted to evaluate NG9 transcription in the cells from which the ChIP clone was isolated. cDNA from E2AFH1B cells was therefore PCR-amplified by using primers specific for the 5′ and 3′ regions of the predicted NG9 transcript, as well as primers to several other B-lineage genes (Table 1, Clone). NG9 transcripts were detectable in E2AFH1B cells along with Igκ, λ5, and Iμ transcripts, further supporting a role for E2A in the regulation of NG9 expression (Fig. 5D).
Discussion
To our knowledge, there have been no previous studies in which an in vivo murine gene-tagging approach has been used to facilitate analysis of target genes regulated by an endogenously expressed transcription factor. The gene-tagging system provides a distinctive alternative to the use of ectopically expressed proteins and also bypasses the challenges of low antibody affinity and specificity that can sometimes interfere with traditional protein isolation approaches. Similarly, to our knowledge there have been no previous studies to directly characterize E2A binding to a number of lymphoid lineage-restricted regulatory regions at the chromatin level. Here, we have demonstrated E2A binding to regulatory regions within a select subset of genes required for B lymphocyte development. Previous ectopic expression and promoter-analysis studies have implicated E2A in the regulation of many of these target genes, including the Ig heavy and κ light chains, surrogate light chains λ5 and VpreB, and the transcription factor EBF (22, 24, 26). Significant enrichment of these sequences by E2A-ChIP provides more conclusive evidence for direct transcriptional regulation by E2A. Binding of E2A to 5′ regulatory regions within the EBF locus is also in agreement with recent studies by Smith et al. (33), who have characterized a potential E2A binding site within the newly identified EBF promoter region. We were also intrigued to see strong enrichment of the mb-1 (Igα) promoter region, because this observation provided direct evidence for the regulation of yet another component of the B cell receptor complex by E2A. Although mb-1 has been implicated as a potential E2A target in genetic studies, it has generally been considered a primary target for other transcription factors, such as Pax5/BSAP, Ets, and Oct-2 (34, 35). However, our data on E2A binding to the mb-1 promoter supports unpublished work by Hagman et al. demonstrating occupation of an mb-1 promoter E-box site by DNA footprinting analysis (J. Hagman, personal communication). These studies provide further insight into the collaborative regulation of gene expression by E2A and other transcription factors during B-lymphopoiesis.
We consistently observed that certain regulatory regions (Igκ enhancers, mb-1 promoter, 5′ EBF locus, and λ5 and VpreB promoters) are significantly enriched after E2A-DNA immunoprecipitations, whereas other putative targets (IgH intronic and hs3b/hs4 enhancers, 5′ TdT and B29 loci, and RAG-2 enhancer) show only very low levels of enrichment. We have also evaluated a group of lymphoid lineage-restricted genes that show no detectable enrichment in our ChIP-PCR assay, including the IgH 3′ enhancer hs3a and the promoters for Pax5, RAG-1, RAG-2, CD19, Oct-2, and CD5. The lack of enrichment of these sequences further substantiates the specificity of the ChIP-PCR system as a means for elucidation of direct vs. indirect E2A target genes. Although the transcription of these genes has been shown to be affected by E2A activity, previous studies have provided no significant evidence for direct transcriptional regulation by E2A. Our data suggests that Pax5/BSAP, Oct-2, CD19, and RAG-1 may be targets for transcriptional regulation by other genes downstream of E2A. This possibility is supported by previous observations that ectopic expression of EBF in a macrophage line leads to activation of a subset of E2A-responsive genes including Pax5 (24). E2A-mediated activation of the EBF locus may therefore play a role in establishing the hierarchy of transcription factors at the earliest stages of B-lymphopoiesis, with EBF subsequently contributing to the induction of Pax5 and other downstream genes.
Perhaps the most surprising data obtained from the ChIP-PCR screen were the minimal enrichment observed for the IgH enhancers. The presence of abundant Iμ transcripts in the pre-B cell lines indicates that the IgH locus is accessible and transcriptionally active. The IgH enhancers also contain well-characterized E-box sites, which are required for normal Ig gene transcription and rearrangement (15, 18). However, previous transfection and DNase hypersensitivity studies have shown that only one of the 3′ IgH enhancers, hs4, is thought to be active at the early stages of B cell development (36, 37). Interestingly, hs4 was the most prominently enriched of the 3′ enhancers, with its most proximal enhancer, hs3b, showing slightly lower enrichment. No detectable enrichment of IgH enhancer hs3a was observed, which is in agreement with previous data suggesting that this enhancer is only functional in activated mature B cells (reviewed in ref. 37). Here, we must also emphasize that a lack of significant E2A binding at any given regulatory region in the pre-B cell lines does not exclude the possibility of direct transcriptional regulation by E2A. Alternatively, certain regulatory regions may be bound by E2A only at restricted developmental stages during B cell development and/or activation. This caveat may be particularly relevant in the analysis of genes involved in Ig rearrangement, such as RAG-1 and RAG-2, TdT, and the Ig genes themselves. The tight and highly ordered developmental regulation of these loci during B-lymphopoiesis may involve significant E2A binding only during specific phases of Ig gene rearrangement, assembly, and expression. Future studies on E2A-bound regulatory regions in primary cells at different stages of development should provide further insight on these dynamic regulatory processes.
A ChIP-based cloning strategy was used to isolate several novel E2A-bound sequences, including an intronic region within the novel E2A target gene NG9 (also known as BTL-II for butyrophilin-like MHC class II-associated). E2A binding at E-box-containing genomic regions across this locus was subsequently investigated by sequence analysis and ChIP-PCR screening. The NG9 locus lies downstream from the Eα locus and upstream from a cluster of other butyrophilin-like genes within the extended MHC II locus (32). As with other butyrophilin genes, each conserved domain of NG9 is encoded by a separate exon, providing support for the notion that the butyrophilin gene clusters arose through exon shuffling and duplication. However, NG9 lacks both the transmembrane domain and the conserved carboxyl-terminal B30.2 domain of the traditional butyrophilins. Interestingly, sequencing data from multiple cell lines indicates that the NG9/BTL-II locus is also highly polymorphic with respect to HLA haplotype (32). These polymorphisms translate into at least five different NG9 alleles and may have been generated and maintained during the diversification of the MHC/HLA loci. Although NG9 transcripts have previously been detected in skeletal muscle and a number of gut tissues, to our knowledge no previous work has evaluated potential regulatory regions and characterized NG9 transcription specifically in lymphoid cells (32). A number of other potential target sequences were also isolated by ChIP-based cloning, most of which appear to be bona fide E2A-bound targets based on their enrichment and their location within known or predicted gene loci. The ChIP-PCR screening strategy used in characterizing E2A binding at the NG9 locus should also prove quite useful in evaluating these and other large genomic regions for transcription factor binding in vivo. In future studies, we will be combining the ChIP-based cloning system with gene array analysis on E2A-deficient cell lines to characterize additional E2A target genes. This powerful two-tiered approach will allow for the elucidation of target genes based on both E2A binding and the requirement for E2A in the normal expression of these genes.
Acknowledgments
We are grateful to Cheryl Bock at the Duke University Transgenic Mouse Facility for her work in generating the E2AFH mice. We also thank Michael Krangel at Duke University for equipment use and critical review of the manuscript, William Forrester at Harvard University for providing the ABO10 viral producer line, and Marco Davila at Duke University for donating the control Abelson line AMLV-3B. This work was supported by a research grant from the National Cancer Institute (RO1 CA72433) and a scholarship from the Leukemia and Lymphoma Society to Y.Z.
Abbreviations
bHLH, basic helix–loop–helix
ChIP, chromatin immunoprecipitation
AMLV, Abelson murine leukemia virus
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Jacobs Y., Vierra, C. & Nelson, C. (1993) Mol. Cell. Biol. 13, 7321-7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xin X., Nelson, C., Collins, L. & Dorshkind, K. (1993) J. Immunol. 151, 5398-5407. [PubMed] [Google Scholar]
- 3.Bain G., Maandag, E. R., Izon, D. J., Amsen, D., Kruisbeek, A. M., Weintraub, B. C., Krop, I., Schlissel, M. S., Feeney, A. J., van Roon, M., et al. (1994) Cell 79, 885-892. [DOI] [PubMed] [Google Scholar]
- 4.Zhuang Y., Soriano, P. & Weintraub, H. (1994) Cell 79, 875-884. [DOI] [PubMed] [Google Scholar]
- 5.Murre C., McCaw, P. S. & Baltimore, D. (1989) Cell 56, 777-783. [DOI] [PubMed] [Google Scholar]
- 6.Sun X. H. & Baltimore, D. (1991) Cell 64, 459-470. [DOI] [PubMed] [Google Scholar]
- 7.Murre C., McCaw, P. S., Vaessin, H., Caudy, M., Jan, L. Y., Jan, Y. N., Cabrera, C. V., Buskin, J. N., Hauschka, S. D., Lassar, A. B., et al. (1989) Cell 58, 537-544. [DOI] [PubMed] [Google Scholar]
- 8.Voronova A. & Baltimore, D. (1990) Proc. Natl. Acad. Sci. USA 87, 4722-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ephrussi A., Church, G. M., Tonegawa, S. & Gilbert, W. (1985) Science 227, 134-140. [DOI] [PubMed] [Google Scholar]
- 10.Murre C., Voronova, A. & Baltimore, D. (1991) Mol. Cell. Biol. 11, 1156-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bain G., Gruenwald, S. & Murre, C. (1993) Mol. Cell. Biol. 13, 3522-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J. S., Olsen, E. N. & Kingston, R. E. (1992) Mol. Cell. Biol. 12, 1031-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhuang Y., Cheng, P. & Weintraub, H. (1996) Mol. Cell. Biol. 16, 2898-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergqvist I., Erikksson, M., Saarikettu, J., Erikksson, B., Corneliussen, B., Grundstrom, T. & Holmberg, D. (2000) Eur. J. Immunol. 30, 2857-2863. [DOI] [PubMed] [Google Scholar]
- 15.Fernex C., Capone, M. & Ferrier, P. (1995) Mol. Cell. Biol. 15, 3217-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henthorn P., Kiledjian, M. & Kadesch, T. (1990) Science 247, 467-470. [DOI] [PubMed] [Google Scholar]
- 17.Kiledjian M., Su, L. K. & Kadesch, T. (1988) Mol. Cell. Biol. 8, 145-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenardo M., Pierce, J. W. & Baltimore, D. (1987) Science 236, 1573-1577. [DOI] [PubMed] [Google Scholar]
- 19.Schlissel M., Voronova, A. & Baltimore, D. (1991) Genes Dev. 5, 1367-1376. [DOI] [PubMed] [Google Scholar]
- 20.Choi J. K., Shen, C. P., Radomska, H. S., Eckhardt, L. A. & Kadesch, T. (1996) EMBO J. 15, 5014-5021. [PMC free article] [PubMed] [Google Scholar]
- 21.Sigvardsson M., O'Riordan, M. & Grosschedl, R. (1997) Immunity 7, 25-36. [DOI] [PubMed] [Google Scholar]
- 22.Romanow W. J., Langerak, A. W., Goebel, P., Wolvers-Tettero, I., van Dongen, J., Feeney, A. J. & Murre, C. (2000) Mol. Cell 5, 343-353. [DOI] [PubMed] [Google Scholar]
- 23.Goebel P., Janney, N., Valenzuela, J. R., Romanow, W. J., Murre, C. & Feeney, A. J. (2001) J. Exp. Med. 194, 645-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kee B. L. & Murre, C. (1998) J. Exp. Med. 188, 699-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Riordan M. & Grosschedl, R. (1999) Immunity 11, 21-31. [DOI] [PubMed] [Google Scholar]
- 26.Sigvardsson M. (2000) Mol. Cell. Biol. 20, 3640-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinmann A. S., Bartley, S. M., Zhang, T., Zhang, M. Q. & Farnham, P. J. (2001) Mol. Cell. Biol. 21, 6820-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cosma M. P., Tanaka, T. & Nasmyth, K. (1999) Cell 97, 299-311. [DOI] [PubMed] [Google Scholar]
- 29.Fernandez L., Winkler, M. & Grosschedl, R. (2001) Mol. Cell. Biol. 21, 196-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhuang Y., Barndt, R. J., Pan, L., Kelley, R. & Dai, M. F. (1998) Mol. Cell. Biol. 18, 3340-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan L., Hanrahan, J., Li, J., Hale, L. P. & Zhuang, Y. (2002) J. Immunol. 168, 3923-3932. [DOI] [PubMed] [Google Scholar]
- 32.Stammers M., Rowen, L., Rhodes, D., Trowsdale, J. & Beck, S. (2000) Immunogenetics 51, 373-382. [DOI] [PubMed] [Google Scholar]
- 33.Smith E. M., Gisler, R. & Sigvardsson, M. (2002) J. Immunol. 169, 261-270. [DOI] [PubMed] [Google Scholar]
- 34.Fitzsimmons D., Hodsdon, W., Wheat, W., Maira, S. M., Wasylyk, B. & Hagman, J. (1996) Genes Dev. 10, 2198-2211. [DOI] [PubMed] [Google Scholar]
- 35.Malone C. S., Patrone, L. & Wall, R. (2000) Mol. Immunol. 37, 321-328. [DOI] [PubMed] [Google Scholar]
- 36.Madisen L. & Groudine, M. (1994) Genes Dev. 8, 2212-2226. [DOI] [PubMed] [Google Scholar]
- 37.Arulampalam V., Eckhardt, L. & Pettersson, S. (1997) Immunol. Today 18, 549-554. [DOI] [PubMed] [Google Scholar]