Abstract
Ubiquitylated protein aggregates are characteristic features of neurodegenerative disorders that are also found in acute pathological states of the brain such as stroke. Many of the proteins connected to neurodegenerative diseases play a role in the ubiquitin-proteasomal pathway. Mutation of one of these proteins, the E3 ubiquitin ligase parkin, is the cause of autosomal recessive juvenile Parkinson's disease. Here we show that transient focal cerebral ischemia of 1-h duration induces marked depletion of parkin protein levels, to 60%, 36%, 33%, and 25% of controls after 1, 3, 6, and 24 h of reperfusion, but that ischemia does not cause lower protein levels of E2 ubiquitin-conjugating enzymes Ubc6, Ubc7, or Ubc9. After 3 h of reperfusion, when parkin protein levels were already reduced to <40% of control, ATP levels were almost completely recovered from ischemia and we did not observe DNA fragmentation, suggesting that parkin depletion preceded development of neuronal cell death. Up-regulation of the expression of parkin has been shown to protect cells from injury induced by endoplasmic reticulum (ER) dysfunction, and this form of cellular stress is also triggered by transient cerebral ischemia. However, in contrast to observations in neuroblastoma cells, we saw no up-regulation of parkin expression in primary neuronal cell cultures after induction of ER dysfunction. Our data thus suggest that ischemia-induced depletion of parkin protein may contribute to the pathological process resulting in cell injury by increasing the sensitivity of neurons to ER dysfunction and the aggregation of ubiquitylated proteins during the reperfusion period.
A key feature of transient cerebral ischemia is the blocking of translation at the initiation step, as indicated by severe suppression of the incorporation of amino acids into proteins, increased phosphorylation of the eukaryotic initiation factor 2α (eIF2α), and disaggregation of polyribosomes (1–3). To date, the only eIF2α kinase shown to be activated after transient ischemia is the double-stranded RNA-dependent protein kinase-like endoplasmic reticulum (ER) kinase (4), an enzyme specifically activated under conditions associated with ER stress (5). This finding suggests that transient ischemia induces ER dysfunction (6, 7). Ischemia-induced suppression of protein synthesis is reversible in resistant neurons, but is irreversible in vulnerable cells (8, 9). The inability of affected neurons to synthesize new proteins has far-reaching consequences, because such neurons cannot react to the severe form of stress elicited by transient ischemia with an up-regulation of the expression of stress genes coding for neuroprotective proteins like HSP70 or Bcl-2. Transcription of various stress genes is indeed markedly activated after ischemia (10), but the respective proteins are not synthesized (10–13). Experiments on the stable neuroblastoma cell line SH-SY5Y (14) showed that transcription and translation of the E3 ubiquitin ligase parkin is considerably up-regulated under conditions associated with ER dysfunction. ER stress-induced up-regulation of parkin expression is a defensive response as this protein confers protection on cells from toxicity elicited by ER dysfunction (14). To elucidate the mechanisms underlying neuronal cell injury triggered by transient cerebral ischemia, we sought to investigate parkin protein levels in transient focal cerebral ischemia, a pathological process associated with severe suppression of protein synthesis (8, 9), ER dysfunction (4), and disturbance of the ubiquitin-proteasomal pathway (15).
Materials and Methods
Animal Experiments.
Experiments were carried out according to the National Institutes of Health guidelines for the care and use of laboratory animals and approved by local authorities. Experiments were performed with adult male C57Black mice weighing 22–26 g. Animals were anaesthetized with 1.5% halothane in 70% N2O and 30% O2, and focal cerebral ischemia was induced by occluding the left middle cerebral artery (MCA) by using the thread occlusion technique, essentially as described (16). MCA occusion-induced changes in cortical blood flow and the extent of reperfusion were verified by laser-Doppler flowmetry. Body temperature of animals was kept constant at 37 ± 0.5°C by using a feedback-controlled heating system. At the end of the experiments, animals were reanesthetized and brains were frozen in situ. Frozen brains were removed in a low-temperature cabinet at −20°C, and tissue samples were dissected from the MCA territory of both hemispheres.
Primary Neuronal Culture.
Primary neuronal cell cultures were prepared from cortices of embryonic rat brains at days 15–17 of gestation, essentially as described (17). Cells were kept in MEM supplemented with 5% horse serum, 30 mM glucose, 2 mM glutamine, 10 units/ml penicillin, and 10 ng/ml streptomycin for 10 days before being used for experiments. ER stress was produced by exposing cells to 1 μM thapsigargin (Tg), an irreversible inhibitor of ER Ca2+-ATPase (18). After 30 min of exposure to Tg or the solvent DMSO (0.1%), cells were transferred to Tg-free medium. The half-life of parkin protein was evaluated by exposing cells to the protein synthesis blocker cycloheximide (20 μg/ml) for 1–72 h. This cycloheximide concentration suppressed protein synthesis to <3% of control and reduced cell viability to 97%, 56%, and 17% of control after 24, 48, and 72 h of exposure, respectively (not shown). At the end of the experiments, cultures were washed with PBS solution.
PCR.
Changes in parkin mRNA levels of cultures exposed to Tg were evaluated by quantitative PCR as described (19, 20). The following parkin primers were used: (sequence taken from GenBank, accession no. AF257234), upper-strand primer, 5′-GTGGGGCTGGCAGTCATTCTGG-3′; and lower-strand primer, 5′-GGGATGCTGCGGCTGTTGTTGG-3′; amplification product, 362 bp. PCRs were run in the presence of defined amounts of an internal standard amplified with the same set of primers but yielding a smaller amplification product of 272 bp. PCR products were separated on a 2% agarose gel, bands were photographed, and changes in parkin mRNA levels were quantified by image analysis using an appropriate standard curve (19). The standard curve was produced in the following way: PCRs were run with different concentration ratios of plasmids containing standard cDNA or parkin cDNA as inserts. The OD ratios of bands (OD parkin band/OD internal standard band) was then related to the concentration ratios of plasmids.
Immunoblotting.
Brain and tissue culture samples were homogenized in lysis buffer, which was composed of 50 mM β-glycerophosphate, pH 7.4, supplemented with EGTA and EDTA (1 mM each), 0.5 mM Na3V04, 1% Triton X-100, 1 mM DTT, the protease inhibitors aprotinin, pepstatin, leupeptine, and benzamidine (1 μg/ml each), 10 mg/ml PMSF, and the calpain inhibitors calpeptin and calpastatin (1 μM each). Homogenates were centrifuged at 14,000 × g, and supernatant fractions were used for immunoblotting. Equal amounts of protein from each sample were loaded onto 8% Tris⋅HCl SDS/PAGE gels, as evaluated by eIF2α immunoblotting. Two different parkin antibodies (PAR-N1 and T160) were used. The PAR-N1 was produced by immunizing rabbits with the synthetic peptide EVDSDTSIFQLKEVVAKC corresponding to amino acid residues 16–32 of human parkin, with a cysteine residue in the C-terminal position. The peptide was coupled to keyhole limpet hemocyanin before immunization. The serum was affinity-purified on a column with the synthetic peptide coupled via its C-terminal cysteine to thiopropyl-Sepharose (Amersham Pharmacia). The T160 antibody was produced by immunizing rabbits with partially purified recombinant hexa-histidine-tagged human parkin. IgG fractions from both sera were isolated by protein A-Sepharose chromatography (Amersham Pharmacia). Anti-rabbit/goat horseradish peroxidase (HRP) conjugates were taken as secondary antibody (Amersham Pharmacia). Monoclonal antibodies directed against ubiquitin-conjugating enzymes Ubc6, Ubc7, and Ubc9 were from Transduction Laboratories (Lexington, KY), and goat/anti-eIF2α antibody was from Santa Cruz Biotechnology. Membranes were incubated with the following secondary antibodies: anti-rabbit/goat HRP conjugates (Pharmacia Biotech), anti-goat/donkey HRP conjugates (Santa Cruz Biotechnology), and anti-mouse/sheep HRP conjugates (Pharmacia Biotech). Bands were visualized by using the ECL Western Blot analysis system (Amersham Pharmacia). OD of bands was quantified by image analysis.
Immunohistochemistry.
Ischemia-induced changes in levels and distribution of parkin protein were evaluated by immunohistochemistry with cryostat sections and the PAR-N antibody. Sections were fixed by incubation in 20 mM Tris⋅HCl (TBS) pH 7.2, supplemented with 50 mM NaCl and 4% paraformaldehyde at 4°C for 5 min, followed by washing with TBS. Endogenous peroxidase was blocked with methanol/H2O2 (0.3%) solution for 20 min, followed by washing with TBS. To optimize reactivity of parkin antibody with parkin protein in the tissue, sections were exposed for 1 min to TBS heated to 95°C. Slices were then incubated in blocking solution for 3 h (TBS supplemented with 0.3% Triton X-100 and 10% normal goat serum), followed by exposure to the PAR-N antibody (1:200) in blocking solution overnight at 4°C. The second antibody [biotinylated goat anti-rabbit (1:100); Santa Cruz Biotechnology] was left on slices for 2 h at room temperature. Finally, slices were incubated with avidin/biotin/peroxidase complex (Vectastatin, Vector Laboratories), followed by visualization of peroxidase with diaminobenzidine/peroxide solution.
Detection of DNA Fragmentation.
DNA fragmentation was investigated by using the deoxynucleotidyl transferase-mediated UTP nick end labeling (TUNEL) techniques as described (21). In short, after immersion in terminal deoxynucleotidyl transferase buffer, sections were incubated with terminal deoxynucleotidyl transferase (Life Technologies, Eggenstein, Germany). The reaction was stopped after 60 min, and sections were treated with avidin-biotin-HRP (ABC kit, Vector Laboratories) to visualize incorporated biotin-dUTP.
Analysis of ATP Levels.
Ischemia-induced changes in ATP levels were measured by HPLC as described (22). The MCA territory of both hemispheres was sampled from frozen brains, ATP was extracted with perchloric acid, and extracts were neutralized. ATP levels in neutralized extracts were measured by HPLC using a reverse-phase ODS-3 column connected to a UV detector. Separation was achieved by using a stepwise methanol gradient.
Data Presentation.
Quantitative data are presented as means ± SD, with n = 4 for cell culture studies, n = 5 for evaluation of parkin and ATP levels, and n = 3 for measurement of DNA fragmentation. Statistically significant differences between parkin mRNA levels in experimental groups and controls or between parkin protein levels in the right and left MCA territory of animals subjected to transient left MCA occlusion were evaluated by ANOVA, followed by Fisher's protected least-significant difference test.
Results and Discussion
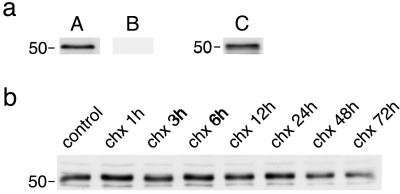
We used two different parkin antibodies to evaluate ischemia-induced changes in parkin protein levels. In samples derived from control brains, immunoblotting produced a major band at ≈52 kDa with the PAR-N1 antibody (Fig. 1aA). The band disappeared completely when the Par-N1 antibody was preincubated with the PAR-N1 peptide used for immunization (Fig. 1aB), and the band also appeared at immunoblots prepared with the T160 antibody (Fig. 1aC), suggesting that it represents parkin protein. Experiments performed with primary neuronal cell cultures exposed to the protein synthesis inhibitor cycloheximide showed that parkin protein had a long half-life (Fig. 1b). Cells were exposed to 20 μg/ml cycloheximide for 1–72 h. With this cycloheximide concentration, protein synthesis was reduced to below 3% of control, and cell viability was reduced to 96.7%, 55.6%, and 17.2% of control after 24, 48, or 72 h of exposure, respectively (not shown). We did not observe any major changes in the band intensity of the 52-kDa parkin band in cultures exposed to cycloheximide for up to 24 h, indicating a long half-life of parkin protein.
Fig 1.
Immunoblotting of parkin protein. Protein extracts from control brains were used to evaluate parkin immunoblotting with the PAR-N1 and T160 antibody. (a) A protein band of ≈52 kDa was present in immunoblots produced with the PAR-N1 antibody (A). Appearance of the band was blocked when the PAR-N1 antibody was preincubated with the PAR-N1 peptide (B), and the band also appeared in immunoblots prepared with the T160 antibody (C). (b) Changes in 52-kDa parkin band after blocking protein synthesis by exposing primary neuronal cell cultures to cycloheximide (chx; 20 μg/ml) for the indicated periods of time.
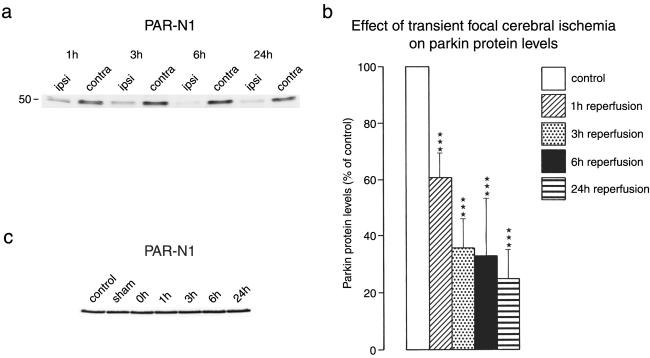
A well-characterized experimental model of transient focal cerebral ischemia (16, 23) was used to investigate ischemia-induced changes in parkin protein levels. With this model, we previously found a long-lasting suppression of protein synthesis during reperfusion (24) despite a transient almost complete recovery of energy metabolism (25). The parkin band started to disappear at 1 h of reperfusion after 1 h left MCA occlusion, and band intensity decreased further with increasing recovery times (Fig. 2 a and b). In the contralateral nonischemic hemisphere, parkin protein levels did not change during or after ischemia (Fig. 2c). A similar pattern of postischemic depletion of parkin protein levels was observed in immunoblots prepared with the T160 antibody (not shown). In samples taken from postischemic brains, we did not find any new band of smaller molecular mass, which might have been indicative of a degradation product of 52-kDa parkin protein (not shown), suggesting that ischemia-induced parkin degradation was a fast process. In additional experiments, mice were subjected to 3 h of permanent left MCA occlusion. In samples derived from the ischemic tissue, parkin protein levels were decreased to 36.8 ± 5.5% of the nonaffected tissue (P < 0.001), indicating that depletion of parkin protein was induced by both permanent ischemia and reperfusion after shorter periods of ischemia.
Fig 2.
Ischemia-induced changes in parkin protein levels. Mice were subjected to 1 h of left MCA occlusion, followed by varying periods of reperfusion. In samples derived from the ischemic left MCA territory, the parkin band started to disappear after 1 h of reperfusion (a). Quantification of ODs of the parkin band revealed a marked decrease of parkin protein levels (b; ***, P > 0.001, left versus right hemisphere). In the contralateral nonischemic hemisphere, parkin protein levels did not change during ischemia or after reperfusion (c). Western blots were run with samples from the MCA territory of plain control animals, sham-operated animals, and the nonischemic MCA territory of the right hemisphere of animals subjected to left MCA occlusion without recovery (0 h) or with 1–24 h of reperfusion.
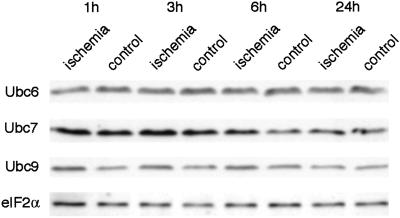
To investigate whether the depletion of parkin protein observed after transient focal cerebral ischemia was specific to the ubiquitin E3 ligase parkin, we evaluated ischemia-induced changes in protein levels of ubiquitin-conjugating enzymes Ubc6, Ubc7, and Ubc9 (Fig. 3). Protein levels of Ubc7 were not decreased but transiently increased in both hemispheres after ischemia. Levels of Ubc9 were transiently increased in the ischemic MCA territory. Protein levels of Ubc6 did not change during or after transient focal cerebral ischemia. Because the ubiquitin system may be up-regulated after transient cerebral ischemia, we also measured protein levels of eIF2α, a soluble cytosolic protein playing a central role in the initiation step of translation. As we have shown (24), eIF2α protein levels did not change during or after transient focal cerebral ischemia (Fig. 3), suggesting that depletion of parkin protein levels did not result from an unspecific gross leakage of soluble proteins from severely damaged neurons.
Fig 3.
Ischemia-induced changes in Ubc6, Ubc7, and Ubc9 protein levels and levels of eIF2α. Ischemia-induced changes in Ubc6, Ubc7, Ubc9, and eIF2α protein levels were evaluated after 1 h of MCA occlusion followed by the indicated reperfusion times. Transient focal cerebral ischemia did not cause a decrease but caused a transient increase in protein levels of ubiquitin-conjugating enzymes Ubc7 and Ubc9. Protein levels of Ubc6 and eIF2α did not change after transient focal cerebral ischemia.
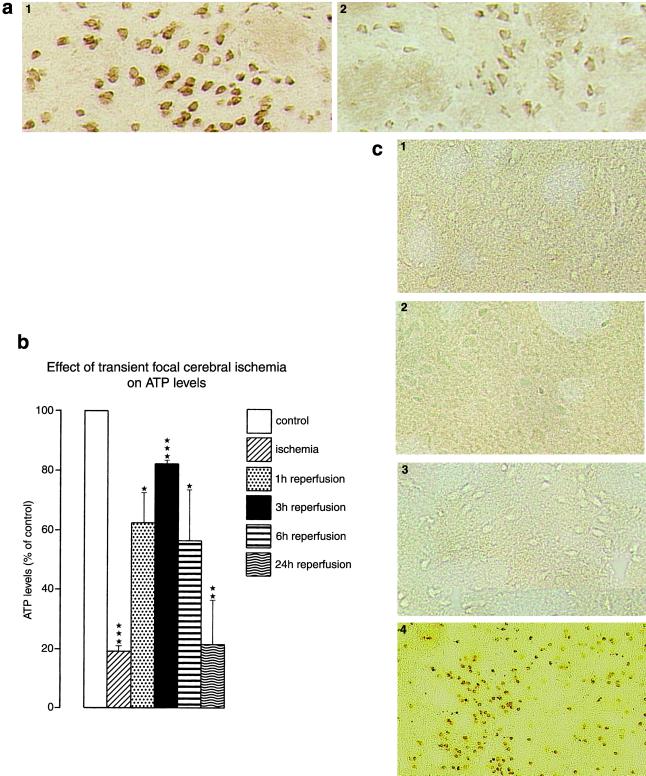
We also evaluated changes in parkin protein levels by immunohistochemistry using the Par-N antibody (Fig. 4a). It has been shown that in the brain parkin protein is largely confined to the cell body of neurons (26, 27). Parkin immunohistochemistry revealed a massive decrease of neuronal parkin protein levels in the ipsilateral striatum compared with the contralateral tissue and less decrease in the cerebral cortex (not shown). Immunohistochemistry was performed with sections of brains subjected to 1 h of MCA occlusion followed by 1 h (not shown) or 3 h (Fig. 4a) of recovery. Three hours is a reperfusion time when the energy state of cells has maximally recovered from interruption of blood supply (Fig. 4b). We also measured the temporal profile of severe cell injury induced in this model, by analyzing DNA fragmentation with the TUNEL technique (Fig. 4c). After 72 h of reperfusion, most of the cells of the ipsilateral striatum (Fig. 4c) and the ipisilateral cortex (not shown) had massive brown deposits, indicating DNA double-strand breaks. After 1 or 3 h of reperfusion, however, we did not observe TUNEL-positive cells in the striatum (Fig. 4c) and the cerebral cortex (not shown). Thus, 3 h after focal ischemia, when parkin protein levels were already decreased to clearly <40% of control (see Fig. 2b), neurons were still not damaged, indicating that ischemia-induced depletion of parkin protein preceded neuronal cell death.
Fig 4.
Ischemia-induced changes in neuronal parkin protein levels (a, immunohistochemistry, 3 h of reperfusion), ATP levels in the territory supplied by the left MCA (b), and DNA fragmentation (c). (a) Parkin immunohistochemistry was performed by using the PAR-N antibody: 1, contralateral striatum; 2, ipsilateral striatum (×200). (c) DNA fragmentation was evaluated by using the TUNEL technique: 1, control; 2, 1 h of reperfusion; 3, 3 h of reperfusion; 4, 72 h of reperfusion. Focal cerebral ischemia caused an almost complete depletion of ATP levels. ATP levels recovered transiently on onset of reperfusion, peaking at 3 h of restoration of blood flow. After 72 h of reperfusion, most of the neurons of the ipsilateral MCA territory showed massive brown deposits, indicating DNA double-strand breaks. After 1 or 3 h of reperfusion, however, we did not observe TUNEL-positive cells, indicating the absence of DNA double-strand breaks (×200).
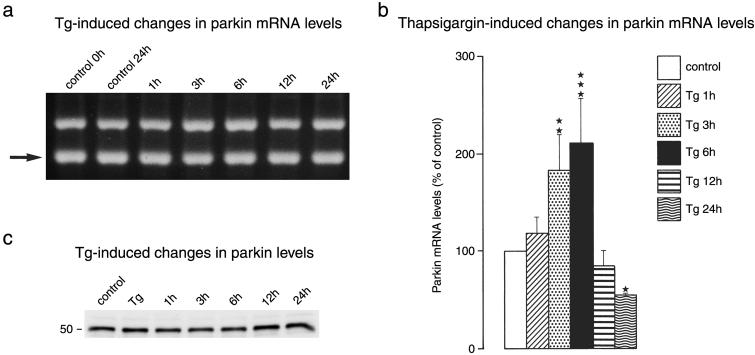
In further experiments, we exposed primary neuronal cell cultures to Tg, an irreversible inhibitor of the ER calcium pump, to investigate the effect of ER stress on parkin expression. We observed an only small transient increase in parkin mRNA levels in cultures exposed to Tg (Fig. 5 a and b). After 6 h of exposure, parkin mRNA levels increased about 2-fold. The moderate rise in parkin mRNA levels was not paralleled by a corresponding increase in parkin protein levels (Fig. 5c). We did not observe any major changes in the intensity of the 52-kDa parkin band.
Fig 5.
Tg-induced changes in parkin expression. Primary neuronal cell cultures were exposed to Tg (1 μM) or the Tg solvent DMSO (0.1%) for 30 min, followed by varying recovery times in Tg-free medium. PCR products were separated on 2% agarose gels (a). The internal standard band is highlighted by an arrow. Gels were photographed and OD of bands was analyzed by image analysis (b). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (ANOVA, followed by Fisher's protected least-significant difference test). (c) Tg-induced changes in parkin protein levels.
The results of the Tg experiments suggest that primary neuronal cell cultures respond to conditions associated with ER stress in a different way to stable cell lines. In experiments performed on the neuroblastoma SH-SY5Y cell line, Imai and colleagues (14) observed an ≈10-fold increase in parkin mRNA and parkin protein levels several times higher than controls after induction of ER stress, indicating that ER stress causes an activation of the ubiquitin-proteasomal pathway. The special response of primary neurons to conditions associated with ER dysfunction, which differs from that of neuroblastoma cells, may have far-reaching consequences for the affected cells, because increased parkin protein levels appear to have protected cells from toxicity elicited under conditions associated with ER stress (14). Recently, we confirmed that our primary neuronal cell cultures react to ER stress with an activation of the unfolded protein response (UPR), a severe suppression of the initiation step of protein synthesis and a 100- to 200-fold increase in mRNA levels of genes coding for ER stress proteins, including grp78, grp94, and gadd153 (28). In contrast, the expression of genes such as hsp70, which do not react specifically to ER stress, were only slightly activated (28). We found their degree of activation to be similar to that of parkin. Our observations thus suggest that, in neuronal cell cultures, parkin does not behave like a gene responding specifically to ER dysfunction.
Besides the moderate increase in neuronal parkin mRNA levels observed in cultures exposed to ER stress, activation of the translation of parkin may be hindered and the extent of this effect is related to the level of suppression of global protein synthesis under our experimental conditions (28). Evidence is indeed accumulating to suggest that the normal stress response of cells is suppressed in neurons. Besides the inability of these cells to react to ER stress with an up-regulation of parkin protein synthesis, as observed in this study, we previously found that an activation of the synthesis of ER calcium pump protein SERCA 2b observed in PC12 cells exposed to Tg (29) is absent in neurons (28). In both cell types, Tg exposure induced an ≈4-fold increase in SERCA 2b mRNA levels (28, 29). However, in contrast to PC12 cells where protein synthesis recovered completely after Tg-induced blocking of the ER calcium pump (29) and where the rise in SERCA 2b mRNA levels was paralleled by a marked increase in SERCA 2b protein levels, SERCA 2b protein levels did not increase in neurons after Tg exposure, most probably because protein synthesis remained markedly suppressed in these cells (28).
Our observations of parkin protein depletion after transient focal cerebral ischemia suggest that the pathological processes culminating in neuronal cell injury in stroke and degenerative diseases are linked, and that parkin may be a key to this link. The mechanisms underlying neuronal cell injury in stroke and degenerative diseases are usually investigated separately for the various pathological states of the brain. However, stroke and degenerative diseases may have common links (6, 7, 30). Additional evidence corroborating this view is indeed accumulating. First, diminished or impaired cerebral perfusion is a risk factor for Alzheimer's disease (31–33); second, in animals expressing mutant presenilin-1, the extent of brain injury induced by focal ischemia and the behavioral outcome is worsened (34); and third, transient ischemia induces β-amyloid peptide formation (35, 36). The observation that ubiquitylated protein aggregates, characteristic features of degenerative diseases (37), are also found in acute pathological states of the brain such as stroke (15) further corroborates the notion that common denominators for cell injury exist in acute and degenerative diseases of the brain (6, 7).
The causal relationship between protein aggregates and cell death remains to be established. Neuronal cell injury may be caused by a direct toxic effect of protein aggregates. Alternatively, appearance of protein aggregates may indicate that ER function and/or the ubiquitin-proteasomal pathway is disturbed, a pathological process that would result in ER stress and an up-regulation of the UPR (38). A regulatory link between ER-associated protein degradation and induction of UPR has indeed been observed (39, 40), and it has recently been shown that pathogenic expansion of CAG trinucleotide repeats, the underlying mechanism of at least nine inherited human neurodegenerative disorders including Huntington's disease, induces ER stress through proteasomal dysfunction (41). In additional experiments, we have indeed shown that blocking the ubiquitin-proteasomal pathway in neurons induces a marked increase in processed xbp1 mRNA levels (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org), indicative of ER stress (42). We have observed a similar increase in processed xbp1 mRNA levels after transient cerebral sichemia (unpublished observation). It can be concluded therefore that depletion of parkin protein observed here after transient focal cerebral ischemia and thus a dysfunction of the ubiquitin-proteasomal pathway may contribute to the induction of ER stress triggered by transient ischemia (6, 7).
UPR is a highly conserved stress response of cells to conditions associated with ER dysfunction (38). Activation of UPR induced by an accumulation of unfolded proteins in the ER lumen causes an up-regulation of the expression of genes coding for ER chaperons, thus increasing folding capacity and finally normalizing protein synthesis and ER function. This process seems to be hindered in vulnerable neurons in which protein synthesis does not completely recover after a severe form of stress. This long-lasting down-regulation of translation in neurons exposed to a transient energy crisis, which prevents cells from responding to transient ischemia with an up-regulation of the synthesis of neuroprotective proteins such as parkin, HSP70, or Bcl-2, may play a key role in the pathological process culminating in neuronal cell injury. This view is corroborated by the observation that in preconditioned, as opposed to nonpreconditioned, animals ischemia-induced suppression of protein synthesis is less pronounced and reversible (43), and that in these animals translation of neuroprotective proteins is markedly activated after transient ischemia (12, 44, 45). Thus, knowledge of the molecular mechanisms underlying the inability of vulnerable neurons to restore normal protein synthesis rate after a severe form of stress would help to elucidate why these cells are particularly sensitive to stressful conditions.
Supplementary Material
Acknowledgments
The excellent technical assistence of Ursula Gerster, Änne Pribliszki, and Cordula Strecker is gratefully acknowledged. This work was supported by Deutsche Forschungsgemeinschaft Grant Pa 266/13.
Abbreviations
eIF2α, eukaryotic initiation factor 2α
ER, endoplasmic reticulum
MCA, middle cerebral artery
Tg, thapsigargin
TUNEL, transferase-mediated UTP nick end labeling
UPR, unfolded protein response
HRP, horseradish peroxidase
References
- 1.Kleihues P., Hossmann, K.-A., Pegg, A. E., Kobayashi, K. & Zimmermann, V. (1975) Brain Res. 95, 61-73. [DOI] [PubMed] [Google Scholar]
- 2.Cooper H. K., Zalewska, T., Kawakami, S. & Hossmann, K.-A. (1977) J. Neurochem. 28, 929-934. [DOI] [PubMed] [Google Scholar]
- 3.Burda J., Martin, M. E., García, A., Alcázar, A., Fando, J. L. & Salinas, M. (1994) Biochem. J. 302, 335-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar R., Azam, S., Sullivan, J. M., Owen, C., Cavener, D. R. C., Zhang, P., Ron, D., Harding, H. P., Chen, J.-J., Han, A., et al. (2001) J. Neurochem. 77, 1418-1421. [DOI] [PubMed] [Google Scholar]
- 5.Harding H. P., Zhang, Y. & Ron, D. (1999) Nature 397, 271-274. [DOI] [PubMed] [Google Scholar]
- 6.Paschen W. & Doutheil, J. (1999) J. Cereb. Blood Flow Metab. 19, 1-18. [DOI] [PubMed] [Google Scholar]
- 7.Paschen W. & Frandsen, A. (2001) J. Neurochem. 79, 719-725. [DOI] [PubMed] [Google Scholar]
- 8.Bodsch W., Takahashi, K., Barbier, A., Grosse Ophoff, B. & Hossmann, K.-A. (1985) Prog. Brain Res. 63, 197-210. [DOI] [PubMed] [Google Scholar]
- 9.Thilmann R., Xie, Y., Kleihues, P. & Kiessling, M. (1986) Acta Neuropathol. 71, 88-93. [DOI] [PubMed] [Google Scholar]
- 10.Massa S. M., Swanson, R. A. & Sharp, F. R. (1996) Cerebrovasc. Brain Metabol. Rev. 8, 95-158. [PubMed] [Google Scholar]
- 11.Nowak T. S. (1990) Cerebrovasc. Brain Metabol. Rev. 2, 345-366. [PubMed] [Google Scholar]
- 12.Chen J., Graham, S. H., Nakayama, M., Zhu, R. L., Jin, K., Stetler, R. A. & Simon, R. P. (1997) J. Cereb. Blood Flow Metab. 17, 2-10. [DOI] [PubMed] [Google Scholar]
- 13.Hara A., Iwai, T., Niwa, M., Uematsu, T., Yoshimi, N., Tanaka, T. & Hori, H. (1996) Brain Res. 711, 249-253. [DOI] [PubMed] [Google Scholar]
- 14.Imai Y., Soda, M. & Takahashi, R. (2000) J. Biol. Chem. 275, 35661-35664. [DOI] [PubMed] [Google Scholar]
- 15.Hu B.-R., Janelidze, S., Ginsberg, M. D., Busto, R., Perez-Pinzon, M., Sick, T. J., Siesjö, B. K. & Liu, C. L. (2001) J. Cereb. Blood Flow Metab. 21, 865-875. [DOI] [PubMed] [Google Scholar]
- 16.Hata R., Mies, G., Wiessner, C., Fritze, K., Hesselbarth, D., Brinker, G. & Hossmann, K.-A. (1998) J. Cereb. Blood Flow Metab. 18, 367-375. [DOI] [PubMed] [Google Scholar]
- 17.Paschen W., Doutheil, J., Gissel, C. & Treiman, M. (1996) J. Neurochem. 67, 1735-1743. [DOI] [PubMed] [Google Scholar]
- 18.Inesi G. & Sagara, Y. (1994) J. Membr. Biol. 141, 1-6. [DOI] [PubMed] [Google Scholar]
- 19.Gissel C., Doutheil, J. & Paschen, W. (1997) J. Neurochem. 69, 2538-2545. [DOI] [PubMed] [Google Scholar]
- 20.Paschen W., Mengesdorf, T., Althausen, S. & Hotop, S. (2001) J. Neurochem. 76, 1916-1924. [DOI] [PubMed] [Google Scholar]
- 21.Wiessner C., Brink, I., Lorenz, P., Neumann-Haefelin, T., Vogel, P. & Yamashita, K. (1996) Neuroscience 72, 947-958. [DOI] [PubMed] [Google Scholar]
- 22.Djuricic B., Röhn, G., Paschen, W. & Hossmann, K.-A. (1994) Metab. Brain Dis. 9, 235-247. [DOI] [PubMed] [Google Scholar]
- 23.Hata R., Maeda, K., Hermann, D., Mies, G. & Hossmann, K.-A. (2000) J. Cereb. Blood Flow Metab. 20, 937-946. [DOI] [PubMed] [Google Scholar]
- 24.Althausen S., Mengesdorf, T., Mies, G., Oláh, L., Nairn, A. C., Proud, C. G. & Paschen, W. (2002) J. Neurochem. 78, 779-787. [DOI] [PubMed] [Google Scholar]
- 25.Paschen W., Oláh, L. & Mies, G. (2000) J. Neurochem. 75, 1675-1680. [DOI] [PubMed] [Google Scholar]
- 26.D'Agata V., Grimaldi, M., Pascale, A. & Cavallaro, S. (2000) Eur. J. Neurosci. 12, 3583-3588. [DOI] [PubMed] [Google Scholar]
- 27.Stichel C. C., Augustin, M., Kuhn, K., Zhu, X. R., Engels, P., Ullmer, C. & Lubbert, H. (2000) Eur. J. Neurosci. 12, 4181-4194. [PubMed] [Google Scholar]
- 28.Mengesdorf T., Althausen, S., Oberndorfer, I. & Paschen, W. (2001) Biochem. J. 356, 805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caspersen C., Pedersen, P. S. & Treiman, M. (2000) J. Biol. Chem. 275, 22363-22372. [DOI] [PubMed] [Google Scholar]
- 30.Mattson M. P., Duan, W., Pedersen, W. A. & Culmsee, C. (2001) Apoptosis 6, 69-81. [DOI] [PubMed] [Google Scholar]
- 31.de la Torre J. C. (2000) Neurobiol. Aging 21, 331-342. [DOI] [PubMed] [Google Scholar]
- 32.de la Torre J. C. & Stefano, G. B. (2000) Brain Res. Rev. 34, 119-136. [DOI] [PubMed] [Google Scholar]
- 33.Kudo T., Imaizumi, K., Tanimukai, H., Katayama, T., Sato, N., Nakamura, Y., Tanaka, T., Kashiwagi, Y., Jinno, Y., Tohyama, M. & Takeda, M. (2000) Neurobiol. Aging 21, 215-224. [DOI] [PubMed] [Google Scholar]
- 34.Mattson M. P., Zhu, H., Yu, J. & Kindy, M. S. (2000) J. Neurosci. 20, 1358-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nihasi T., Inao, S., Kajita, T., Sugimoto, T., Niwa, M., Kabeya, R., Hata, N., Hayashi, S. & Yoshida, J. (2001) Acta Neurochir. 143, 287-295. [DOI] [PubMed] [Google Scholar]
- 36.Popawagner A., Schroder, E., Walker, L. C. & Kessler, C. (1998) Stroke 29, 2196-2202. [DOI] [PubMed] [Google Scholar]
- 37.Chung K. K. K., Dawson, V. L. & Dawson, T. M. (2001) Trends Neurosci. 24,Suppl., S7-S14. [DOI] [PubMed] [Google Scholar]
- 38.Kaufman R. J. (1999) Genes Dev. 13, 1211-1233. [DOI] [PubMed] [Google Scholar]
- 39.Friedlander R., Jarosch, E., Urban, J., Volkwein, C. & Sommer, T. (2000) Nat. Cell Biol. 2, 379-384. [DOI] [PubMed] [Google Scholar]
- 40.Travers K. J., Patil, C. K., Wodicka, L., Lockhart, D. J., Weissman, J. S. & Walter, P. (2000) Cell 101, 249-258. [DOI] [PubMed] [Google Scholar]
- 41.Nishitoh H., Matsuzawa, A., Tobiume, K., Saegusa, K., Takeda, K., Inoue, K., Hori, S., Kakizuka, A. & Ichijo, H. (2002) Genes Dev. 16, 1345-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calfon M., Zeng, H., Urano, F., Till, J. H., Hubbart, S. R., Harding, H. P., Clark, S. G. & Ron, D. (2002) Nature 415, 92-96. [DOI] [PubMed] [Google Scholar]
- 43.Kato H., Kogure, K., Nakata, N., Araki, T. & Itoyama, Y. (1995) Brain Res. Bull. 36, 205-208. [DOI] [PubMed] [Google Scholar]
- 44.Shimazaki K., Ishida, A. & Kawai, N. (1994) Neurosci. Res. 20, 95-99. [DOI] [PubMed] [Google Scholar]
- 45.Nakatsuka H., Ohta, S., Tanaka, J., Toku, K., Kumon, Y., Maeda, N., Sakanaka, M. & Sakaki, S. (2000) Neurosci. Lett. 278, 53-56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.