Abstract
The t(1;19) translocation yields a fusion between E2A and PBX1 genes and occurs in 5% of acute lymphoblastic leukemia in children and adults. We used chromosomal translocations and Ig heavy chain (IGH)/T cell antigen receptor (TCR) rearrangements to develop an understanding of the etiology and natural history of this subtype of leukemia. We sequenced the genomic fusion between E2A and PBX1 in 22 preB acute lymphoblastic leukemias and two cell lines. The prenatal origin of the leukemia was assessed in 15 pediatric patients by screening for the clonotypic E2A-PBX1 translocation in neonatal blood spots, or Guthrie cards, obtained from the children at the time of birth. Two patients were determined to be weakly positive for the fusion at the time of birth, in contrast to previously studied childhood leukemia fusions, t(12;21), t(8;21), and t(4;11), which were predominantly prenatal. The presence of extensive N-nucleotides at the point of fusion in the E2A-PBX1 translocation as well as specific characteristics of the IGH/TCR rearrangements provided additional evidence for a postnatal, preB cell origin. Intriguingly, 16 of 24 breakpoints on the 3.2-kb E2A intron 14 were located within 5 bp, providing evidence for a site-specific recombination mechanism. Breakpoints on the 232-kb PBX1 intron 1 were more dispersed but highly clustered proximal to exon 2. In sum, the translocation breakpoints displayed evidence of unique temporal, ontological, and mechanistic formation than the previously analyzed pediatric leukemia translocation breakpoints and emphasize the need to differentiate cytogenetic and molecular subgroups for studies of leukemia causality.
Pediatric leukemias are a group of diverse diseases at the chromosome level, with various subtypes recognizable by recurrent translocations and aneuploidies. Although these genetic abnormalities help to categorize leukemias for treatment strategy and prognosis, they also may delineate specific causal pathways to malignancy. The consideration of individual molecular subtypes is providing clarity to epidemiological and biological studies. The best characterized example of this approach is the infant leukemias with MLL translocations (MLL+), for which epidemiologic associations and molecular analysis of breakpoints point to an in utero origin of the translocations, with reactive metabolites of genotoxic chemicals playing a potential key role (1–5). The MLL+ leukemias, along with the hyperdiploid subtype (leukemia clones with >50 chromosomes), also display significant associations with variants in genes encoding functional metabolic enzymes, thus implying a risk of leukemia imparted by the enzymes' substrates or products (6–8). Childhood leukemias with TEL-AML1 translocations, representing ≈25% of common acute lymphoblastic leukemia (cALL), share the in utero timing of translocation formation with MLL+ leukemias (9); however, there is currently no etiological mechanism for the formation of this translocation, apart from the nonhomologous end-joining processes that rejoin broken DNA.
The second most common translocation in acute lymphoblastic leukemia (ALL) is t(1;19), which fuses the 5′ end of E2A with most of PBX1, yielding a chimeric protein that has cell transformation capability in both in vitro and in vivo models (reviewed in ref. 10). This leukemia subtype is morphologically and prognostically distinct in origin from the cALL subtype and represents ≈25% of the preB cell (CD10+, CD19+, CD34−, cIgμ+, sIgμ−) leukemia and 5% of childhood and adult ALL overall (11). The leukemia is a well established clinical and biologic entity, reinforced by its unique transcriptome (12); however, the timing, origin, and mechanism of the genetic rearrangements that create this leukemia are unclear. Like most B cell origin leukemias, E2A-PBX1 leukemias have germline-rearranged Ig heavy chain (IGH) rearrangements (13). The combinatorial and diversity characteristics in the leukemic IGH CDR3 region have been used to signify early (prenatal) from later (postnatal) development of normal lymphocytes as well as various pediatric leukemia subtypes (14–16). To date, no examples of the E2A-PBX1 translocation have been sequenced at the genomic level, nor has the natural history of this subtype been explored.
We developed methods to sequence E2A-PBX1 fusions rapidly at the genomic DNA level and have assessed whether this fusion arose prenatally by using archived neonatal blood spots, or Guthrie cards. Translocation breakpoints displayed a high degree of clustering and clear evidence of the involvement of a site-specific recombination mechanism. Little evidence for prenatal origin was found by direct assessment of the Guthrie spots, and indirect evidence from IGH and T cell antigen receptor (TCR) rearrangement patterns supported a postnatal origin with ontological specificity.
Materials and Methods
Cell Lines and Patient Samples.
Cell lines were obtained from American Type Culture Collection (cell line RCH-ACV) and cell line 697 was kindly provided by Michael Cleary (Stanford University, Stanford, CA). Patient samples were derived from the Northern California Childhood Leukemia study (#69, 71, 131, 348, 416, 398, and 498). Additional samples came from the Children's Oncology Group ALL cell bank (#058, 062, 228, 295, 482, 513, 633, 777, 780, and 913) and patients treated at University of Colorado Health Sciences Center (D1, D2, D3, and D4) or Stanford Medical Center (S1). Guthrie cards were obtained from the Genetic Diseases Branch of the California Department of Health Services.
Translocation Sequencing.
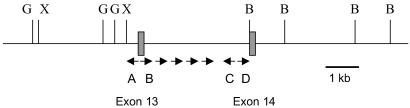
E2A-PBX1 fusions were sequenced by using Long Distance Inverse PCR (LDI-PCR) as described (17) with modifications. LDI-PCR was performed from the E2A sequence, given that the breakpoint cluster region is only 3.2 kb. The enzymes BglII and XbaI permitted amplification of E2A-PBX1 sequences, whereas enzymes BamH1, HpaI, and BsrDI were used to amplify the reciprocal PBX1-E2A (Fig. 1). PCRs were nested, using primers E2A A and B for the first round and E2A A-1 and B-1 for the second round, for amplifying E2A-PBX1 fusions. For the reciprocal translocation, C and D primers were used in turn (Fig. 1, and see primers in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). A rearrangement would be predicted to introduce a restriction site on the opposite side of the intron, allowing the preferential amplification of a smaller rearranged band over the larger wild type. After an initial successful nested PCR, tertiary reactions were performed by using E2A A-1 in all tubes with, in separate tubes, B-2, B-3, B-4, and B-5.
Fig 1.
Inverse PCR scheme. Locations of PCR primers and restriction sites around intron 13 of the E2A gene. G, BglII; X, XbaI; B, BamH1; A, location of invA and invA-1 primers; B, location of invB and invB-1 primers. InvB-2, 3, 4, and 5 are located just to the right of invB-1. C corresponds to invC and invC-1; D corresponds to invD and invD-1, respectively. InvC-2, 3, 4, and 5 are the complement of invB-5, 4, 3, and 2, respectively.
Guthrie card screening, or “backtracking,” was carried out by using Ampdirect methods, as described (9, 18), with two modifications. First, the presoaking in distilled water was omitted. Second, both primary and secondary reactions were performed by using Ampdirect buffers. The primary round was a 50-μl reaction with 1/16 segment of a 1.5-cm2 Guthrie card. The second round was a 25-μl reaction using 1 μl of the first round for template. The soaking step was omitted based on the observation that Guthrie cards stored in the freezer (as all California cards are) tend to leach DNA during this procedure. Primer sequences and product sizes can be found in Table 3, which is published as supporting information on the PNAS web site.
IGH/TCR Rearrangement Analysis.
IGH CDR3 region and TCRδ gene rearrangements were amplified by PCR as described (19). PCR products were separated on a 6% polyacrylamide gel and scanned and analyzed with a FluorImager 595 laser scanner and IMAGEQUANT software (Amersham Pharmacia). Clonal rearrangements were directly sequenced from agarose gel-purified PCR products as described (19).
Sequences were analyzed for VH, DH, and JH segments by using macvector 7.0 (Genetics Computer Group, Madison, WI). For DH identification (20), the criteria of a minimum of either seven uninterrupted DH germline nucleotides, or at least eight DH germline nucleotides with no more than one mismatch and at least two matching nucleotides at both the 5′ and 3′ ends, were used (21). No DIR segments, inverted D segments, or D-D recombinations were accepted (20).
Statistical Analysis.
Translocation breakpoint clustering was assessed by using a scan statistic as described (22). Prediction of heptamer-nonamer V(D)J recombinase site sequences (RSS; CACAGTG, 12 or 23 nucleotides, ACAAAAACC) was assessed by using the following rubric: the underlined sequence must be present, but any other nucleotide was allowable as long as there were less than 10 (E2A) or 5 (PBX) mismatches. A more stringent definition of RSS was chosen for the PBX1 gene because of the fact that the intron is much larger, although RSS distribution was similar regardless of stringency (data not shown). Both forward and complement sequences were scanned for RSS.
Results
E2A-PBX1 Genomic Fusions.
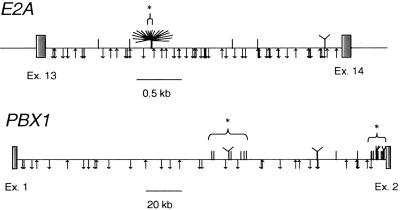
Sequences were obtained for 24 available patient or cell-line DNAs for E2A-PBX1. Sixteen breakpoints were located within 5 bp on the E2A side and two others within 12 bp, whereas breakpoints were more widely spaced on the PBX1 introns (Fig. 2). Overall assessment for clustering was highly significant on both introns. The single cluster of 18 breakpoints spanning 12 bp on E2A (see Figs. 2 and 3) signified a cluster by scan statistic (P < 0.0001), whereas two clusters were evident on PBX1: one proximal to exon 2 (12 breakpoints within 8,137 bp, P < 0.0001) and one in the right center of the intron (8 breakpoints within 21,400 bp, P = 0.056). A single base of E2A at which six E2A-PBX1 translocations were placed was also the site for two previously sequenced E2A-HLF and one base pair away from another E2A-HLF (Fig. 3; refs. 23 and 24). The PBX1 clusters were much more dispersed but still statistically clustered because of the lengthy intronic region of PBX1.
Fig 2.
Locations of E2A-PBX1 breakpoints and V(D)J recombinase sites. Exon sequences are indicated with boxes, and intronic sequences are indicated with horizontal lines. Patient breakpoints are identified with hash marks above the introns, and putative cryptic RSS are indicated below the introns. V(D)J sites are shown with an up arrow if they are on the sense strand and a down arrow if antisense. Eighteen breakpoints within a 12-bp segment of E2A are shown as a tree-like structure, and brackets above the introns (with a *) show the clusters indicated in Results.
Fig 3.
Fine structure of cluster region on E2A. Eighteen forward E2A-PBX1 breakpoints are indicated with lines and labels above the sequence segment of E2A intron 13, and six reciprocal (i.e., PBX1-E2A) breakpoints are indicated below the segment. The vertical lines delimit the E2A sequence to the left of the line for the E2A-PBX1 and delimit the E2A sequence to the right of the line in the case of the reciprocal. Six E2A-PBX1 translocations are designated at a single base, at which two E2A breakpoints from previously sequenced E2A-HLF+ patients were located, as shown with a dashed line (B, ref. 23). An additional E2A-HLF breakpoint is located 1 bp prior at A (24). Candidate RSS heptamers are boxed; nonamers are not shown for clarity.
Upon LDI-PCR and sequencing analysis, two unrelated patients (#69 and 131) exhibited the same E2A-PBX1 fusion sequence, including the presence of two nontemplate nucleotides “TT” at the point of fusion. To assess whether laboratory contamination may have played a role, we sequenced the IGH and TCR rearrangements of the patients as well as five polymorphisms that were found to be different (Table 1, supporting information, and data not shown). The breakpoints were confirmed by using a second diagnostic sample. The two patients were diagnosed at the same hospital, but because diagnosis and sample processing took place 18 months apart, it is unlikely that contamination was introduced within the laboratory or the hospital.
Table 1.
Age, gender, and genomic rearrangements in t(1;19) E2A-PBX1 patients
| Patient ID | Age, yr.mo | Gender | N-Nucl.E2A-PBX1 | IGH # | N-Nucl. IGH | FrameIGH | Vδ2-Dδ3 # | N-Nucl. Vδ2-Dδ3 |
|---|---|---|---|---|---|---|---|---|
| 398 | 1.8 | F | 13 | 1 | 28 | I | 0 | |
| 913 | 1.8 | M | 41 (0) | 1 | 9 | I | 0 | |
| 482 | 2.3 | F | 7 | 1 | 6 | O | 0 | |
| 777 | 2.5 | M | 6 | 1 | 19 | I | 0 | |
| 697 | 4 | F | 1 | 1 | 34 | S | 1 | 8 |
| 348 | 4.3 | M | 13 | 1 | 39 | I | 0 | |
| 131 | 4.4 | M | 2 | 0 | 2 | 3 | ||
| 228 | 4.8 | M | −1 | 1 | 4 | I | 0 | |
| 633 | 5.1 | F | 6 | 1 | 13 | I | 0 | |
| 295 | 5.3 | F | 4 | 1 | 14 | I | 0 | |
| 780 | 6.6 | F | 18 | 1 | 14 | I | 1 | 8 |
| 69 | 6.9 | F | 2 | 1 | 13 | I | 0 | |
| 498 | 7.3 | F | 15 (9) | 1 | 17 | I | 1 | 10 |
| 513 | 7.6 | F | 4 (15) | 1 | 17 | I | 0 | |
| RCH | 8 | F | 14 | 1 | 16 | O | 1 | 9 |
| 71 | 8.2 | F | 2 | 1 | 27 | I | 1 | 14 |
| 58 | 8.7 | F | 11 | 1 | 21 | I | 1 | 7 |
| D1 | 9.5 | F | 15 (5) | 1 | 14 | I | 1 | 6 |
| D2 | 10.2 | F | 16 | 0 | 1 | |||
| 416 | 11.2 | F | 19 | 1 | 14 | I | 1 | 3 |
| 62 | 12.5 | F | 17 | 1 | 22 | I | 0 | |
| D3 | 17.6 | M | 39 | 1 | 17 | I | 1 | 7 |
| D4 | 47.11 | F | 9 (6) | 1 | 15 | I | 0 | |
| S1 | NA | NA | 15 (16) | 1 | 25 | S | 0 |
N-Nucl. are nontemplate nucleotides. N-nucleotides for reciprocal fusions are in parentheses.
Pertains to IGH rearrangements. I, in frame and functional; O, out of frame and nonfunctional; S, in frame but stop codon.
NA, not available.
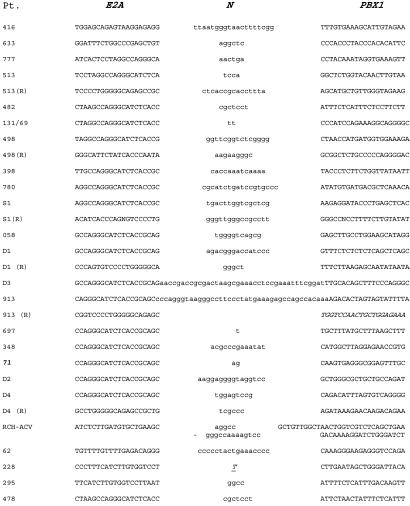
Twenty-three of 24 breakpoints (96%) contained the presence of 1–41 N-nucleotides. These are nontemplate nucleotides not present in either parent DNA sequence but probably introduced through the activity of terminal deoxynucleotidyltransferase (TdT) before ligation of DNA ends (Fig. 4). Only a single breakpoint did not contain N-nucleotides but displayed microhomology of one nucleotide at the fusion junction (#228). Another breakpoint exhibited a 30-bp duplication of PBX1 intronic sequence at the breakpoint along with 5 and 14 N-nucleotides at the E2A and PBX1 sides of the duplication, respectively (RCH-ACV, Fig. 4). Sequencing of reciprocal fusions (the 5′ end of the PBX1 gene fused to the 3′ end of E2A) was attempted in all cases but was successful in only five patients (Figs. 3 and 4). The rate of success (5 of 24 patients = 21%) is not far from the rate at which molecular and cytogenetically reciprocal translocations are identifiable in t(1;19)+ leukemia (25%; ref. 25). An additional patient exhibited a fusion between the intron 13 of E2A and intron 2 of the RODH 3-hydroxysteroid epimerase gene on chromosome 12 (#513, Fig. 4). The predicted fusion protein would not be in the proper reading frame but would result in the 3′ ends of each gene fused back-to-back, with no promoter (data not shown). Overall, when compared with other pediatric leukemia translocations, the reciprocal fusions were very conservative, demonstrating breakpoints in close proximity (average = 5 bp, excluding one with a 1-bp microhomology) to the primary fusion and also harboring similar presence of N-nucleotides.
Fig 4.
Fine structure of E2A-PBX1 genomic fusions. Patient number is displayed with the E2A and PBX1 sequences proximal to the fusions and N-nucleotides that were present between the E2A and PBX1 sequence (lowercase letters). The antisense strand of the reciprocal fusions (R) are displayed. Cell line RCH had two segments of N-nucleotides along with a tandem duplication of 30 nucleotides of PBX1 sequences shown in between, derived from sequence 7,398 bp 3′ of the breakpoint in reverse orientation. The “T” is a microhomology nucleotide (derived from E2A or PBX1, not an N-nucleotide). The reciprocal partner fusion to 913 is in italics, indicating a fusion of E2A to RODH intron 2.
Tight clustering of the breakpoints along E2A, along with the presence of N-nucleotides, suggests the activity of the V(D)J recombinase. Such a mechanism has one additional requirement: the presence of a canonical RSS consisting of a heptamer-nonamer sequence near the junction. This sequence would by rule be positioned in an orientation that would result in its excision from the E2A gene, because the N-nucleotides are added to the coding joint (26). Functional studies have identified basic requirements of functional RSS (CACAGTG, 12 or 23 nucleotides, ACAAAAACC), with the underlined nucleotides being present in close to 100% of functional RSS (27, 28). Breakpoints using the RSS yield a perfectly conserved pair of signal sequences linked to each other in reverse orientation and a modified coding end sequence which often has nucleotide loss (“nibbling”) as well as nucleotide gain (N-nucleotides) that contribute to the variable region of the IGH or TCR (reviewed in ref. 26). The slightly variable (within five nucleotides) nature of the E2A cluster as well as the presence of the N-nucleotides suggests that it may serve as a surrogate coding end in an aberrant V(D)J mechanism. However, the reciprocal fusions did not display signal end-like fusions, which typically consists of two abutting signal sequences with the lack of N-nucleotides, but instead showed similar characteristics of the forward fusion. We searched for potentially cryptic V(D)J RSS within the E2A and PBX1 introns and found no exact matches but many near matches that could conceivably serve as weak RSS and contribute to the translocation (28–30). In general, there were no apparent relationships between the locations of breakpoints and the generally even spreading of putative cryptic RSS sequences along the intronic regions (Fig. 2). Of course, this analysis does not take into account other aspects of DNA or chromatin structure that could influence accessibility of the DNA to recombinase activating RAG proteins. Some potential RSS in vicinity of the E2A cluster were found (Fig. 3). However, the closest putative RSS is in the orientation such that the E2A breakpoint in the E2A-PBX1 translocation would be the “coding end” fusion. If this were the case, the breakpoints should be oriented to the left side of the closest putative RSS sequence (Fig. 3), however 17 of 18 breakpoints are on the right side of the “CAC.” By these criteria there is poor evidence to support V(D)J-RSS to account for the breakpoint cluster on E2A. The PBX1 translocations were not site-specific, nor did they have potential V(D)J RSS at breakpoint fusions, and, thus, displayed little evidence of RSS-mediated recombination. However, the breakpoints were highly clustered at the 3′ end of the 232-kb intron 2 of this gene (Fig. 2).
Backtracking E2A-PBX1 to Birth.
Fifteen patients were assessed for prenatal origin of the clonotypic E2A-PBX1 translocation by PCR amplification of Guthrie cards from those individuals. Sensitivities of PCR reactions were initially determined by using a dilution series of patient diagnostic DNA that was derived from blood or bone marrow at the time of diagnosis, and each assay was run with the dilution series to assess sensitivity, which ranged from 5 to 500 pg. For each patient, a total of 12 segments (3/4 of one spot) were assayed in two PCR reactions (6 per reaction). Only one segment each from two patients (58 and 295) were identified to be “positive” for prenatal origin, indicated by the presence of leukemia clone-specific DNA sequence within the Guthrie segment in one assay (Fig. 5). This positive assay was not repeated in the second assessment of six segments for these patients. Sensitivity for patient #295 was 500 pg of DNA and 5 pg for patient #58. We cannot rule out lack of PCR sensitivity or lack of circulating E2A-PBX1 cells as a reason for the negative results in the other 13 cases; therefore, we recognize that those assays are indeterminate for prenatal origin of the clone.
Fig 5.
Guthrie card analysis of patient #295. The second-round PCR products are shown for the dilution series of diagnostic patient DNA (lanes 1–4, 1:10 dilution series, starting at 50 ng/μl patient DNA in lane 1). Guthrie card segments for control (C) cards are shown with segments from patient #295 (P) and a no-DNA blank (B). A single-lane positive shown was sequenced and determined to match the sequence from the patient. Results were similar for patient #58.
IGH and TCR Rearrangements.
The leukemic cells from 24 t(1;19) positive ALL patients were analyzed for clonal rearrangements of the IGH and TCR genes. Every patient was positive for at least one clonal rearrangement. One patient was positive for two Vδ2-Dδ3, and nine were positive for 1 IGH and 1 Vδ2-Dδ3 rearrangements. No clonal Dδ2-Dδ3 rearrangements were identified. Of the 22 IGH rearrangements identified, 20 were “in frame”; i.e., the triplet codon structure was maintained through the rearrangement. However, stop codons were introduced within two of these as a result of the rearrangement. Therefore, 18 of 22 (82%) would be predicted to translate into functional IGH heavy chains (Table 1 and Table 4, which is published as supporting information on the PNAS web site).
In normal IGH rearrangements, JH4 is the most commonly used JH segment at all stages of development, and JH2, JH3, and JH6 are the next most common JH segments during first trimester, third trimester, and adults, respectively (21). In our study, JH4 and JH6 were used in equal frequency (7/22 = 32% each), followed by JH5 (5/22) and JH1, JH2, and JH3 (one of each, see supporting information). Members of the DH3 family are normally observed in a higher percentage of IGH rearrangements as the age of the study population increases. For example, 5% of first trimester, 10% of second trimester, and 33% of adult sequences harbor DH3 segments (21). In our study, 9 of the 27 DH gene segments were identified in the clonal IGH rearrangements of the ALL patients. The DH3 family was represented most frequently (45%). The D7–27 gene segment is represented in 33% of fetal marrow B-lineage cells, in 17% of preterm infant mature B cells, and only in 1–2% of the adult DH segments (21, 31, 32). We observed no D7–27 segments in the 22 IGH rearrangements examined. In sum, the usages of JH and DH are consistent with a postnatal chronological age at which the blood cells that exhibit the leukemic phenotype passed through the developmental stage in which IGH rearrangement occurred.
The frequency of the Vδ2-Dδ3 rearrangement in childhood preB ALL reaches 32–35% (13, 33), and its presence is correlated with age; i.e., 86% of those younger than 2, 43% of those from 2 to 14, and 18% of those older than 15 were positive for Vδ2-Dδ3 (13). We identified Vδ2-Dδ3 rearrangements in 46% of the patients in our study, but identified no significant correlation between age and the presence of Vδ2-Dδ3 rearrangements (P > 0.9, Pearson's correlation). The frequency of the Dδ2-Dδ3 rearrangement in childhood preB ALL is historically ≈13% (34). We found no clonal Dδ2-Dδ3 rearrangements in the 24 t(1;19)+ patients in our study. A lower-than-expected frequency in our population may be because of a small sample size.
IGH N-Nucleotides.
The developmental regulation of nongermline (N) region addition has been well documented (21, 32, 35, 36). DHJH joining that lack N regions are found more frequently at the fetal stage of development. The proportion of sequences containing N additions increases from 25% during the second trimester to 94% in adults (21). In our study, all 22 clonal IGH gene rearrangements contained N-nucleotides (100%). The mean lengths of the N-regions on the 5′ and 3′ sides of the DH region were 10.4 and 7.3 nucleotides, respectively. The N-region lengths are consistent with those observed in adults, which average 8 and 6 nucleotides, respectively (32). There was no relationship between N-nucleotides at any of the three types of rearrangements assayed (i.e., IGH, TCRD, or E2A-PBX1; P > 0.4) and age at diagnosis.
Discussion
Childhood leukemia predominantly originates from B cell precursors, which are characterized by rapid developmental stage-specific proliferation as well as the activation of the V(D)J recombinase system. V(D)J and switch recombinases are the only mechanism in the body targeted to the somatic rearrangement of the genome and are well known to be involved in some translocation fusion breakpoints in another B cell cancer, the lymphomas (reviewed in ref. 37). It is reasonable to suppose that translocations in the B cell leukemias might arise from the aberrant activity of V(D)J recombinase; however, recent studies have failed to substantiate this hypothesis and, instead, suggest the activity of topoisomerase II, Alu-mediated homologous recombination, or unspecified damage-repair mechanisms involving DNA double-strand break repair without the involvement of recombinase-associated RSS sequences (38–40). This fact holds true for the most frequent translocation subtype of childhood ALL, TEL-AML1 fusions, which are generally widely spread over intron 5 of TEL and introns 1–2 of AML1. There is evidence of clustering of these breakpoints, but such clustering is multifocal and dispersed with cluster widths of hundreds of nucleotides rather than relegated to specific well defined recombination sites typical of V(D)J recombinase-induced genomic rearrangements (22, 41).
In contrast to other childhood leukemia breakpoints, E2A breakpoints were clustered in a site-specific manner. Sixteen E2A-PBX1 translocations of 24 were clustered within 5 bp (see Results). Three reported fusions between E2A and HLF also had breakpoints within the same cluster (Fig. 3; refs. 23 and 24). This clustering is the type evident in normal IGH and TCR V(D)J recombination, as well as putative V(D)J recombination events in other genes such as HPRT (42) and also in T cell oncogenic mutations at the TAL and MTS1 loci (43, 44) but not at any other B cell leukemia-associated breakpoints. The clustering exhibited on the E2A side thus suggests the involvement of the site-specific V(D)J recombinase. Indeed, the E2A gene itself produces the E12/E47 proteins that function as indispensable transcription factors for the induction of V(D)J recombination (45, 46), indicating that the chromatin configuration of the E2A gene itself is likely to be in an open conformation because of the fact that it is being transcribed, thus potentially accessible to recombinases. The ontological point at which E2A-PBX1 translocation occurs is likely to be later than that of other pediatric leukemia translocations that do not demonstrate site-specific clustering or the presence of extensive N-nucleotides. However, as explained in Results, there is no recombinase site sequence in the E2A gene in the correct orientation to account for typical V(D)J mechanisms. Although the presence of N-nucleotides clearly indicates the expression of TdT during the translocation process, further research will have to define the nature of the site-specific recombination and whether alternate mechanisms, such as transposase activity of RAG genes, might play a role in the mechanism of E2A-PBX1.
Breakpoints in the far larger exon 2 of PBX1 were clustered in a more dispersed manner compared with E2A. DNA breaks on the PBX1 side may be caused by factors in common with those inducing other pediatric chromosomal translocations, which are similarly clustered on a diffuse scale; for instance, TEL-AML1, AML1-ETO, and MLL-AF4 (38, 39, 41). DNA double-strand breaks induced by means other than RAG-induced recognition may be competent to participate in ligation to the E2A breakpoint. Interestingly, most PBX1 translocations were crowded next to exon 2, which suggests a contribution from chromatin, gene structure, or perhaps the presence of an open locus control region near the exon. The final ligation step between E2A and PBX1 is likely to be a nonhomologous end-joining event, in common with other translocations, as there is no evidence of homologous recombination.
The presence of nontemplate N-nucleotides at E2A-PBX1 fusions stands in marked contrast to the dearth of such nucleotides at TEL-AML1 fusions. It is known that normal V(D)J junctions formed during fetal development usually lack, or contain very few, N-nucleotides, whereas fusions in children and adults usually contain these nucleotides (21, 32, 35, 36). The lack of such nucleotides in TEL-AML1 fusions, and more importantly, the demonstration that clonotypic fusions present in leukemia cells at the time of diagnosis are present in the Guthrie cards of the children who later got disease (9), argues for the prenatal origin of these fusions. The presence of N-nucleotides at E2A-PBX1 fusions and IGH rearrangements as well as the JH and DH segment usage (see Results) does not date the timing of the fusion but supports a postnatal fusion event at a later ontological stage of blood cell development. In addition, the Guthrie card study did not support a prenatal origin for E2A-PBX1 translocations in the majority of cases, providing weak evidence in two. PCR analysis of clonotypic IGH sequences on Guthrie cards was not performed because of the limitations in the resource in relationship to the scientific value derived from backtracking IGH sequences.
Lymphomas are B cell tumors that are also caused, at least in part, by translocations that result in disregulated gene expression. The mechanisms involved in forming these oncogenic translocations have been linked to the IGH rearrangement processes that occur at the same maturation stage exhibited by the lymphoma (37, 47–49). Also, the normal antibody-producing IGH rearrangements in the lymphomas (not involved in oncogenic translocation) display characteristics typical of the phenotype of normal B cells from the organ from which the lymphoma is derived, providing evidence that the timing of transformation is linked in developmental stage with normal function of these B cells (reviewed in ref. 50). Pediatric leukemias, on the other hand, may harbor translocation before the cell-specific developmental stage at which the leukemia phenotype is clinically observed, given the lack of V(D)J evidence at translocation fusions despite rearranged IGH genes within the leukemia clones (15, 17, 51). In addition, the IGH genes are nonfunctional (i.e., out of frame) two thirds of the time in B cell leukemias, indicating that IGH rearrangement occurs without selection processes (15). This fact, along with the fact that B cell leukemias often include additional IGH subclones as well as cross-lineage TCR rearrangements (52), provides evidence that the transforming event for the cell precedes IGH rearrangement, because normal selection processes would result in deletion of these clones or further V(D)J recombination to produce a functional IGH gene. The exception to this could be t(1;19) E2A-PBX1+ leukemia, which may be derived from a preB cell transformation, post-IGH rearrangement, because 82% of the 22 rearranged IGH genes studied here are predicted to produce functional, in-frame genes (Table 1). This characteristic would argue that the t(1;19) E2A-PBX1 translocation as a transformation event occurred at the same time as or after IGH rearrangement and subsequent selection, linking transformation with the ontological stage of the leukemic cell. In addition, the pattern of germline segment usage and junctional diversity of the t(1;19)+ patients is consistent with rearranged IGH chain variable regions and TCR rearrangements typically seen in adults but rare during fetal development. In sum, these data suggest a more lymphoma-like etiology for t(1;19)+ leukemia than the other pediatric leukemias.
We conclude that the preB cells undergoing leukemic transformation that result in t(1;19)+ ALL typically undertake translocation as well as IGH gene rearrangement after birth in ontologically more mature cells than those involved in the majority of pediatric lymphocytic leukemias. Combined with its status as a unique clinical entity, t(1;19)+ ALL is a distinct subtype that should be stratified separately in etiology studies. The molecular analysis and natural history of gene rearrangements provide additional confidence in our strategy of stratification of this and other molecularly defined leukemia subtypes in the study of the causes of childhood leukemia.
Supplementary Material
Acknowledgments
We thank Andrew Carroll for cytogenetic analysis of COG patients and Cheryl Willman for maintenance of COG specimens. We thank Luoping Zhang and Weihong Guo (University of California, Berkeley), and ChangHong Yin and Qianxu Guo (New York Medical College) for technical assistance. We also thank Peggy Reynolds, Fred Lorey, and Michael Layefsky for coordinating Guthrie card retrieval, Institutional Review Board approval, and data management between the Children's Oncology Group and the California Department of Health Services. This work was supported by National Institutes of Health Grants CA89032 (to J.L.W.), ES09137, ES04705, and ES01896 (to M.T.S. and P.A.B.), Children's Oncology Group Grant CA30969, and the Children's Cancer Fund (to S.R.P.).
Abbreviations
ALL, acute lymphoblastic leukemia
IGH, Ig heavy chain
RSS, recombinase site sequences
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Strick R., Strissel, P. L., Borgers, S., Smith, S. L. & Rowley, J. D. (2000) Proc. Natl. Acad. Sci. USA 97, 4790-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander F. E., Patheal, S. L., Biondi, A., Brandalise, S., Cabrera, M. E., Chan, L. C., Chen, Z., Cimino, G., Cordoba, J. C., Gu, L. J., et al. (2001) Cancer Res. 61, 2542-2546. [PubMed] [Google Scholar]
- 3.Ford A. M., Ridge, S. A., Cabrera, M. E., Mahmoud, H., Steel, C. M., Chan, L. C. & Greaves, M. (1993) Nature 363, 358-360. [DOI] [PubMed] [Google Scholar]
- 4.Gale K. B., Ford, A. M., Repp, R., Borkhardt, A., Keller, C., Eden, O. B. & Greaves, M. F. (1997) Proc. Natl. Acad. Sci. USA 94, 13950-13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross J. A., Potter, J. D., Reaman, G. H., Pendergrass, T. W. & Robison, L. L. (1996) Cancer Causes Control 7, 581-590. [DOI] [PubMed] [Google Scholar]
- 6.Wiemels J. L., Pagnamenta, A., Taylor, G. M., Eden, O. B., Alexander, F. E. & Greaves, M. F. (1999) Cancer Res. 59, 4095-4099. [PubMed] [Google Scholar]
- 7.Wiemels J. L., Smith, R. N., Taylor, G. M., Eden, O. B., Alexander, F. E. & Greaves, M. F. (2001) Proc. Natl. Acad. Sci. USA 98, 4004-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith, M. T., Wang, Y., Skibola, C. F., Slater, D., Lo Nigro, L., Nowell, P. C., Lange, B. J. & Felix, C. A. (2002) Blood, in press. [DOI] [PubMed]
- 9.Wiemels J. L., Cazzaniga, G., Daniotti, M., Eden, O. B., Addison, G. M., Masera, G., Saha, V., Biondi, A. & Greaves, M. F. (1999) Lancet 354, 1499-1503. [DOI] [PubMed] [Google Scholar]
- 10.Aspland S. E., Bendall, H. H. & Murre, C. (2001) Oncogene 20, 5708-5717. [DOI] [PubMed] [Google Scholar]
- 11.Borowitz M. J., Hunger, S. P., Carroll, A. J., Shuster, J. J., Pullen, D. J., Steuber, C. P. & Cleary, M. L. (1993) Blood 82, 1086-1091. [PubMed] [Google Scholar]
- 12.Yeoh E. J., Ross, M. E., Shurtleff, S. A., Williams, W. K., Patel, D., Mahfouz, R., Behm, F. G., Raimondi, S. C., Relling, M. V., Patel, A., et al. (2002) Cancer Cells 1, 133-143. [DOI] [PubMed] [Google Scholar]
- 13.Brumpt C., Delabesse, E., Beldjord, K., Davi, F., Cayuela, J. M., Millien, C., Villarese, P., Quartier, P., Buzyn, A., Valensi, F. & Macintyre, E. (2000) Blood 96, 2254-2261. [PubMed] [Google Scholar]
- 14.Wasserman R., Galili, N., Ito, Y., Reichard, B. A., Shane, S. & Rovera, G. (1992) J. Exp. Med. 176, 1577-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steenbergen E. J., Verhagen, O. J., van Leeuwen, E. F., Behrendt, H., Merle, P. A., Wester, M. R., von dem Borne, A. E. & van der Schoot, C. E. (1994) Eur. J. Immunol. 24, 900-908. [DOI] [PubMed] [Google Scholar]
- 16.Fasching K., Panzer, S., Haas, O. A., Borkhardt, A., Marschalek, R., Griesinger, F. & Panzer-Grumayer, E. R. (2001) Blood 98, 2272-2274. [DOI] [PubMed] [Google Scholar]
- 17.Wiemels J. L. & Greaves, M. (1999) Cancer Res. 59, 4075-4082. [PubMed] [Google Scholar]
- 18.Wiemels J. L., Xiao, Z., Buffler, P. A., Maia, A. T., Ma, X., Dicks, B. M., Smith, M. T., Zhang, L., Feusner, J., Wiencke, J., et al. (2002) Blood 99, 3801-3805. [DOI] [PubMed] [Google Scholar]
- 19.Mayer S., Giamelli, J., Sandoval, C., Roach, A., Ozkaynak, M. F., Tugal, O., Rovera, G. & Jayabose, S. (1999) Leukemia 13, 1843-1852. [DOI] [PubMed] [Google Scholar]
- 20.Corbett S. J., Tomlinson, I. M., Sonnhammer, E. L., Buck, D. & Winter, G. (1997) J. Mol. Biol. 270, 587-597. [DOI] [PubMed] [Google Scholar]
- 21.Shiokawa S., Mortari, F., Lima, J. O., Nunez, C., Bertrand, F. E., III, Kirkham, P. M., Zhu, S., Dasanayake, A. P. & Schroeder, H. W., Jr. (1999) J. Immunol. 162, 6060-6070. [PubMed] [Google Scholar]
- 22.Segal M. R. & Wiemels, J. L. (2002) J. Am. Stat. Assoc. 97, 66-76. [Google Scholar]
- 23.Inaba T., Roberts, W. M., Shapiro, L. H., Jolly, K. W., Raimondi, S. C., Smith, S. D. & Look, A. T. (1992) Science 257, 531-534. [DOI] [PubMed] [Google Scholar]
- 24.Hunger S. P., Ohyashiki, K., Toyama, K. & Cleary, M. L. (1992) Genes Dev. 6, 1608-1620. [DOI] [PubMed] [Google Scholar]
- 25.van Dongen J. J., Macintyre, E. A., Gabert, J. A., Delabesse, E., Rossi, V., Saglio, G., Gottardi, E., Rambaldi, A., Dotti, G., Griesinger, F., et al. (1999) Leukemia 13, 1901-1928. [DOI] [PubMed] [Google Scholar]
- 26.Lewis S. M. (1994) Adv. Immunol. 56, 27-150. [DOI] [PubMed] [Google Scholar]
- 27.Hesse J. E., Lieber, M. R., Mizuuchi, K. & Gellert, M. (1989) Genes Dev. 3, 1053-1061. [DOI] [PubMed] [Google Scholar]
- 28.Lewis S. M., Agard, E., Suh, S. & Czyzyk, L. (1997) Mol. Cell. Biol. 17, 3125-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raghavan S. C., Kirsch, I. R. & Lieber, M. R. (2001) J. Biol. Chem. 276, 29126-29133. [DOI] [PubMed] [Google Scholar]
- 30.Marculescu R., Le, T., Simon, P., Jaeger, U. & Nadel, B. (2002) J. Exp. Med. 195, 85-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamada M., Wasserman, R., Reichard, B. A., Shane, S., Caton, A. J. & Rovera, G. (1991) J. Exp. Med. 173, 395-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zemlin M., Bauer, K., Hummel, M., Pfeiffer, S., Devers, S., Zemlin, C., Stein, H. & Versmold, H. T. (2001) Blood 97, 1511-1513. [DOI] [PubMed] [Google Scholar]
- 33.Breit T. M., Wolvers-Tettero, I. L., Hahlen, K., van Wering, E. R. & van Dongen, J. J. (1991) Leukemia 5, 1076-1086. [PubMed] [Google Scholar]
- 34.Seriu T., Erz, D., Stark, Y. & Bartram, C. R. (1997) Leukemia 11, 759-761. [DOI] [PubMed] [Google Scholar]
- 35.Gu H., Forster, I. & Rajewsky, K. (1990) EMBO J. 9, 2133-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanz I. (1991) J. Immunol. 147, 1720-1729. [PubMed] [Google Scholar]
- 37.Kuppers R. & Dalla-Favera, R. (2001) Oncogene 20, 5580-5594. [DOI] [PubMed] [Google Scholar]
- 38.Reichel M., Gillert, E., Angermuller, S., Hensel, J. P., Heidel, F., Lode, M., Leis, T., Biondi, A., Haas, O. A., Strehl, S., et al. (2001) Oncogene 20, 2900-2907. [DOI] [PubMed] [Google Scholar]
- 39.Xiao Z., Greaves, M. F., Buffler, P. A., Smith, M. T., Segal, M. R., Dicks, B. M., Wiencke, J. K. & Wiemels, J. L. (2001) Leukemia 15, 1906-1913. [DOI] [PubMed] [Google Scholar]
- 40.Atlas M., Head, D., Behm, F., Schmidt, E., Zeleznik-Le, N. H., Roe, B. A., Burian, D. & Domer, P. H. (1998) Leukemia 12, 1895-1902. [DOI] [PubMed] [Google Scholar]
- 41.Wiemels J. L., Alexander, F. E., Cazzaniga, G., Biondi, A., Mayer, S. P. & Greaves, M. (2000) Genes Chromosomes Cancer 29, 219-228. [DOI] [PubMed] [Google Scholar]
- 42.Fuscoe J. C., Zimmerman, L. J., Lippert, M. J., Nicklas, J. A., O'Neill, J. P. & Albertini, R. J. (1991) Cancer Res. 51, 6001-6005. [PubMed] [Google Scholar]
- 43.Breit T. M., Beishuizen, A., Ludwig, W. D., Mol, E. J., Adriaansen, H. J., van Wering, E. R. & van Dongen, J. J. (1993) Leukemia 7, 2004-2011. [PubMed] [Google Scholar]
- 44.Cayuela J. M., Gardie, B. & Sigaux, F. (1997) Blood 90, 3720-3726. [PubMed] [Google Scholar]
- 45.Romanow W. J., Langerak, A. W., Goebel, P., Wolvers-Tettero, I. L., van Dongen, J. J., Feeney, A. J. & Murre, C. (2000) Mol. Cell 5, 343-353. [DOI] [PubMed] [Google Scholar]
- 46.Goebel P., Janney, N., Valenzuela, J. R., Romanow, W. J., Murre, C. & Feeney, A. J. (2001) J. Exp. Med. 194, 645-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marculescu R., Le, T., Bocskor, S., Mitterbauer, G., Chott, A., Mannhalter, C., Jaeger, U. & Nadel, B. (2002) Leukemia 16, 120-126. [DOI] [PubMed] [Google Scholar]
- 48.Nadel B., Marculescu, R., Le, T., Rudnicki, M., Bocskor, S. & Jager, U. (2001) Leuk. Lymphoma 42, 1181-1194. [DOI] [PubMed] [Google Scholar]
- 49.Welzel N., Le, T., Marculescu, R., Mitterbauer, G., Chott, A., Pott, C., Kneba, M., Du, M. Q., Kusec, R., Drach, J., et al. (2001) Cancer Res. 61, 1629-1636. [PubMed] [Google Scholar]
- 50.Hummel M. & Stein, H. (2000) Curr. Opin. Oncol. 12, 395-402. [DOI] [PubMed] [Google Scholar]
- 51.Reichel M., Gillert, E., Nilson, I., Siegler, G., Greil, J., Fey, G. H. & Marschalek, R. (1998) Oncogene 17, 3035-3044. [DOI] [PubMed] [Google Scholar]
- 52.Szczepanski T., Beishuizen, A., Pongers-Willemse, M. J., Hahlen, K., Van Wering, E. R., Wijkhuijs, A. J., Tibbe, G. J., De Bruijn, M. A. & Van Dongen, J. J. (1999) Leukemia 13, 196-205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.