Abstract
Hepatitis delta virus (HDV) uses a host-encoded RNA-editing activity to express its two essential proteins from the same coding sequence. Adenosine deaminase that acts on RNA (ADAR)1 and ADAR2 are enzymes that catalyze such reactions, and each, when overexpressed, are capable of editing HDV RNA in vivo. However, the enzyme responsible for editing HDV RNA during replication has not been determined. Mammalian cells express two forms of ADAR1, a large form (ADAR1-L) that mainly localizes to the cytoplasm and a small form (ADAR1-S) that resides in the nucleus. Recently, we found that the specific activity of ADAR1-L within cells is much higher than that of ADAR1-S but only when the substrate can be edited in the cytoplasm. Here we observed that although both ADAR1-S and ADAR1-L were expressed throughout HDV replication, no ADAR2 could be observed at any time. Using expression vectors that individually overexpress either form of ADAR1, we found that ADAR1-S could stimulate editing during replication more efficiently. We next reduced ADAR1 levels during HDV replication. After transfection of an ADAR1-L-specific small interfering RNA (siRNA), we observed a significant loss of that protein and its associated cytoplasmic editing activity while the level of ADAR1-S remained unchanged. Transfection of this siRNA, however, did not reduce editing during HDV replication. In contrast, transfection of an siRNA that targets both forms of ADAR1 greatly reduced the expression of both proteins and potently inhibited editing during replication. We conclude that ADAR1-S edits HDV RNA during replication and that editing occurs in the nucleus.
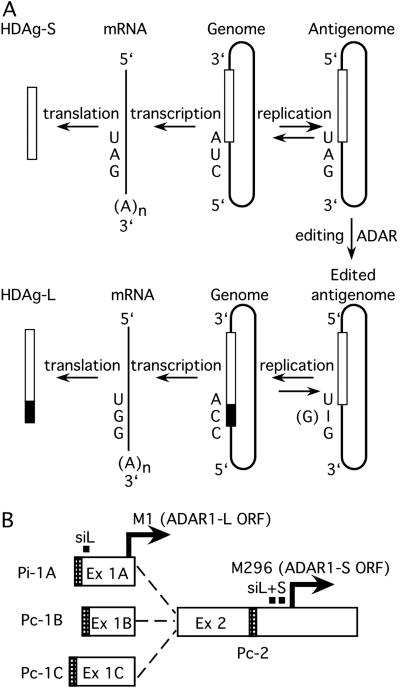
Hepatitis delta virus (HDV) is a subviral pathogen that relies on its helper, hepatitis B virus, to provide envelope proteins needed for virion assembly (1). The HDV genome is a 1,679-nt, single-stranded, negative sense circular RNA that is 70% self-complementary and is thought to fold into an unbranched rod-like structure (2). HDV uses a double rolling-circle replication mechanism, and that process creates the antigenome, a circular replication intermediate that is complementary to the genome (3). Throughout HDV infection, the virus expresses the small delta antigen (HDAg-S). This protein is required for replication and is translated from a subantigenomic message that lacks the characteristic unbranched rod-like structure of the complete antigenome (4). Later during infection, some antigenomes are edited (5) at a specific position, referred to as the amber/W site, such that the UAG that encodes the amber codon of the HDAg-S is converted to a tryptophan codon (UIG, where I = inosine) (see Fig. 1A). The cellular transcriptional machinery recognizes inosine as guanosine, and therefore after replication and transcription, the resulting mRNA has an extended ORF that encodes the large delta antigen (HDAg-L). The HDAg-L inhibits replication and is required for virion assembly (6, 7). Hence, RNA editing is an essential process in HDV replication as it modulates the switch from the early to the late phase and enables the virus to express two essential and functionally distinct proteins from the same coding sequence.
Fig 1.
(A) RNA editing of the antigenome during HDV replication enables the virus to express two proteins from one coding sequence. The genome and antigenome are represented as rods, and the open boxes within the rods represent the sequences corresponding to the ORF of the HDAg-S. The black region of the box is the additional 19 amino acids of the HDAg-L. (B) Transcripts and promoter organization of hADAR1 as determined by Samuel and coworkers (19, 20). The diagram shows the three splice variants, with exons (Ex) 1A, 1B, and 1C spliced to exon 2. The promoters are shown as shaded regions. Exon 1A arises from an IFN-inducible promoter (Pi-1A) and contains the first methionine (M1) of the hADAR1-L. Exons 1B and 1C are constitutively expressed from promoters Pc-1B and Pc-1C, respectively, and lack an in-frame AUG. Only transcripts with exon 1A encode ADAR1-L, whereas the others encode ADAR1-S. A fourth transcript arising from a constitutive promoter at exon 2 (Pc-2) also encodes ADAR1-S. The black squares denote the positions of the siRNAs targeted against only ADAR1-L (siL) or both forms of ADAR1 (siL+S). Sequences targeted by siL+S are duplicated naturally.
The HDAg-S and HDAg-L interact with one another and with both genomes and antigenomes to form ribonucleoprotein complexes (8). Although HDV RNA synthesis is thought to occur in the nucleus, Tavanez et al. (9) recently showed that HDV ribonucleoprotein complexes shuttle between the nucleus and cytoplasm. Furthermore, at least some antigenomic RNA is observed in cytoplasmic fractions (10, 11). Hence, editing of HDV antigenomic RNA potentially could occur in either compartment. Virions that contain genomes copied from edited antigenomes are nonproductive, because after infection, only HDAg-L is expressed. Thus, although editing is an essential process, it must be limited such that HDAg-S-expressing genomes can persist and be assembled into virions. In addition to the extent of editing, the timing of editing is also critical, because the production of the replication inhibitor, HDAg-L, too early during replication would cause that process to abort. Although it is known that HDAg-S can inhibit amber/W editing (12), this protein is present throughout HDV replication and apparently is not responsible for the temporal regulation of editing. Currently, there is no understanding of how such temporal regulation might be accomplished. An obvious first step toward understanding the regulation of editing during HDV replication would be the identification of the host enzyme that catalyzes the event.
Two members of the adenosine deaminase that act on RNA (ADAR) family, ADAR1 and ADAR2, catalyze the deamination of adenosine to inosine in perfect and imperfect duplex RNA (13–16). Mammalian cells express two different molecular mass forms of ADAR1, the large, full-length 150-kDa form (ADAR1-L) and a small, 110-kDa form (ADAR1-S) (17). ADAR1-S lacks the first 295 amino acids that are present in the amino terminus of ADAR1-L. ADAR1-S is localized to the nucleus and is expressed constitutively, whereas ADAR1-L is localized mainly to the cytoplasm (18), and its expression can be induced by IFN (17). The ADAR1-L message is synthesized from an IFN-inducible promoter containing the first exon (exon 1A) with the initiator AUG (see Fig. 1B; ref. 19). ADAR1-S messages, however, can be expressed from three constitutive promoters (20). Transcripts encoding ADAR1-S are translated from an initiator AUG in the second exon of the message. This AUG corresponds to the second methionine of ADAR1-L at position 296. In most cells, ADAR1-L is expressed at a much lower level than is ADAR1-S. However, we recently observed that, even at very low levels of expression, ADAR1-L was remarkably efficient at editing transcripts in the cytoplasm, and under these conditions its specific activity was 80-fold higher than that of ADAR1-S (S.K.W., S. Sato, and D.W.L., unpublished data).
Substrates that can be edited by mammalian ADARs include pre-mRNAs for glutamate (21–23) and serotonin (24) receptors and the rat ADAR2 pre-mRNA (25). Xenopus ADAR specifically and efficiently edits the HDV amber/W site in vitro (26). In addition, when overexpressed by transient transfection, both human ADAR1 and ADAR2 are able to edit this site in vivo with comparable efficiency (27, 28). However, the identity of the activity endogenously expressed in cells that is responsible for editing has been established for only a few sites within these substrates where ADAR2 was found to be responsible (29).
Here we used RNA interference (RNAi) as a tool to identify the enzyme responsible for editing replicating-HDV RNA in tissue culture. The RNAi pathway induces sequence-specific inhibition of gene expression in response to double-stranded RNA of the same sequence (30). A protein known as dicer first cleaves the double-stranded RNA into small duplexes of roughly 21 nt in length (31). These 21-nt small interfering (si)RNAs then are bound by a complex of proteins known as the RNA-induced silencing complex (RISC) (32). The RISC is thought to anneal the antisense strand of the siRNA to messages, and when a perfect match is encountered, the message is degraded. Recently, Elbashir et al. (33) showed that introduction of siRNAs into mammalian cells can specifically decrease the level of expression of the targeted transcript without eliciting nonspecific responses that are induced by longer RNA duplexes in a sequence-independent manner (e.g., IFN-regulated pathways). Using RNAi as well as other approaches, we found that ADAR1-S edits HDV antigenomic RNA in the nucleus during replication. Endogenously expressed ADAR1 was shown definitively to be responsible for the editing event.
Methods
Transfections, Western Analyses, and Quantitation.
Huh7 and human embryonic kidney (HEK)293 cells were cultured in DMEM (Cellgro) supplemented with 10% FBS. DNAs were transiently transfected by either a modified calcium-phosphate precipitation method (34) or by Lipofectamine 2000 (Invitrogen).
Total cellular proteins were analyzed by immunoblotting with anti-ADAR1, anti-ADAR2, anti-delta antigen polyclonal antisera, anti-hemagglutinin (HA) monoclonal antibody (27, 28), or anti-β-actin monoclonal antibody (clone AC-15, Sigma). Quantitation of Western analyses was performed by using the Molecular Dynamics Storm PhosphorImager and IMAGEQUANT software.
Detection of ADARs During HDV Replication.
Huh7 cells were cotransfected with pDL456 (34), a cDNA that expresses replication-competent antigenomic HDV RNA, a puromycin resistance plasmid (pSKW041), and a GFP-expressing plasmid (pEGFP-c1, CLONTECH) and were selected for puromycin resistance. Controls with and without pSKW041were included to determine the efficiency of the puromycin enrichment of transfected cells. GFP expression of the control with pSKW041 indicated that >90% of the surviving cells were transfected. Very few cells (<5%) survived under puromycin selection in the control that lacked pSKW041. Protein and RNA samples were taken 4, 5, 6, 7, 8, 10, and 12 days posttransfection. The protein samples were immunoblotted with anti-ADAR1, anti-ADAR2, or anti-delta antigen polyclonal antisera, and the RNA samples were probed with an RNA probe of antigenomic sequence so as to detect the genomic strand. pSKW041 was derived from pBabe-Puro (35) by subcloning the 0.7-kb HindIII–ClaI fragment of pBabe-Puro into an expression vector, pSS43 (28), that contains a cytomegalovirus promoter and an HDV polyadenylation signal.
Overexpression of HA-Tagged ADARs During HDV Replication.
HEK293 cells were cotransfected with pDL538 (34), which expresses replication-competent HDV genomic RNA, and vectors that express either HA-tagged ADAR1-L (pDL707; S.K.W., S. Sato, and D.W.L., unpublished data) or HA-tagged ADAR1-S (pDL701; S.K.W., S. Sato, and D.W.L., unpublished data).
RNAi Using siRNA.
The siRNAs used (synthesized by Dharmacon Research) were: siADAR2 top strand, 5′-UACAUGAGUGAUCGUGGCCUU-3′; bottom strand, 5′-GGCCACGAUCACUCAUGUAUU-3′; siADAR1-L+S top strand, 5′-CCAGCACAGCGGAGUGGUAUU-3′; bottom strand, 5′-UACCACUCCGCUGUGCUGGUU-3′; siADAR1-L top strand: 5′ GACCCGCGGAGUUUCCCGUUU-3′; and bottom strand, 5′-ACGGGAAACUCCGCGGGUCUG-3′.
The top and bottom strands of the RNA were annealed at a concentration of 80 μM in 10 mM Tris (pH7.7)/1 mM EDTA/100 mM NaCl by heating to 90°C for 1 min and then cooling in a thermocycler at a rate of 0.1°C/s until 22°C was reached. HEK293 cells were seeded into six-well dishes to ≈50% confluence. The cells were transfected initially with 2.5 μl per well of the annealed RNA mixture by using Oligofectin (Invitrogen), and 1 day later this transfection was repeated. One day after the second transfection, cDNAs expressing either a nonreplicating editing reporter, pSS106 (28), or replicating-HDV antigenomic RNA, pDL456 (34), were cotransfected with more siRNA by using Lipofectamine 2000 (Invitrogen). pSS106 is a nonreplicating-HDV mutant derived from the wild-type HDV amber/W reporter and was observed previously to be highly edited by the endogenous editing activity of HEK293 cells (28). Cells transfected with pSS106 were lysed for Western analyses 2 days posttransfection. The cells that were transfected with replicating-HDV RNA were retransfected with siRNAs every 4 days, and samples were harvested for Western analyses 4, 8, and 12 days posttransfection.
Results
ADAR1 but Not ADAR2 Was Expressed During HDV Replication.
We first examined whether either ADAR1 or ADAR2 is expressed and/or induced in human hepatoma cells that harbor replicating HDV. HDV replication can be initiated in tissue culture by transfection of a cDNA that expresses greater than unit-length HDV RNA. To examine the ADAR levels specifically in cells expressing replicating-HDV RNA, untransfected cells were eliminated by puromycin selection. To monitor the levels of expression of the endogenous ADAR1 or ADAR2, total cellular protein then was immunoblotted with either anti-ADAR1 or anti-ADAR2 polyclonal antisera and then detected by using radiolabeled protein A (28). Because the level of HDAg-L expression during replication reflects the extent of editing, the level of that protein was monitored by Western analysis using anti-delta antigen polyclonal serum, whereas the extent of replication was monitored by Northern analysis.
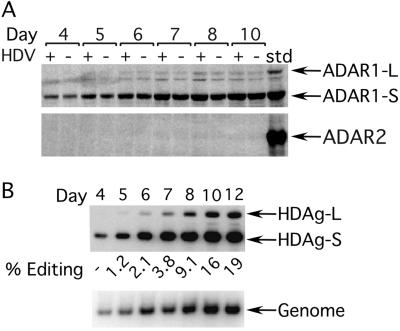
Both forms of ADAR1 were expressed (Fig. 2A Upper) throughout the HDV replication time course. Similar levels of the two forms were observed in the control transfection performed in parallel in which no HDV cDNA vector was included (Fig. 2A, compare HDV+ and HDV− lanes). In contrast to ADAR1, no ADAR2 was detected (Fig. 2A Lower) in the cells harboring HDV, although efficient editing had occurred (Fig. 2B Upper). Previously we determined that the ADAR2 antiserum can detect lower levels of protein than can the ADAR1 antiserum (28). Hence, we concluded that during HDV replication, Huh7 cells expressed much more ADAR1 than ADAR2. Furthermore, we note that based on comparison with cells that did not harbor HDV, replication did not cause any significant induction in the expression of either form of ADAR1 in these cells.
Fig 2.
Expression of endogenous ADAR1 and ADAR2 during HDV replication in Huh7 cells. (A) Western analyses were performed on total cell lysates expressing replicating-HDV RNA (HDV+) or control vector (HDV−) 4–10 days posttransfection and immunoblotted with either anti-ADAR1 (Upper) or anti-ADAR2 (Lower) polyclonal antisera. std, standard. (B) Efficient editing was occurring during the course of replication. (Upper) A Western analysis performed with anti-delta antigen polyclonal antiserum showing the edited (HDAg-L) and unedited (HDAg-S) products. (Lower) A Northern blot probed to detect the HDV genome.
Compared with ADAR1-L, ADAR1-S Was More Efficient at Editing Replicating-HDV RNA.
Recently, we observed that ADAR1-L was much more active than ADAR1-S at editing a nonreplicating mRNA reporter that contains the HDV amber/W site (S.K.W., S. Sato, and D.W.L., unpublished data). With that substrate, the difference in specific activity of the two forms was ≈80-fold, and much of the difference could be attributed to editing that occurred in the cytoplasm with ADAR1-L. When the activities of the two forms of ADAR1 were assayed with a substrate that could be edited only in the nucleus, ADAR1-S was found to be 3-fold more active than ADAR1-L (S.K.W., S. Sato, and D.W.L., unpublished data).
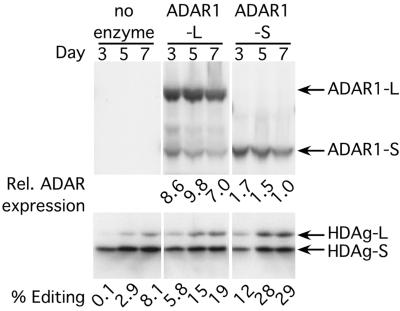
It is not known whether editing occurs in the nucleus or the cytoplasm during HDV replication. We next addressed this issue by comparing the ability of each form of ADAR1 to edit replicating-HDV antigenomic RNA. Vectors expressing HA-tagged ADAR1-L or ADAR1-S were cotransfected with the cDNA that initiates HDV replication, and the levels of ADAR expression and HDAg-L expression were monitored by Western analysis. Although ADAR1-L was expressed to at least a 5-fold higher level than ADAR1-S (Fig. 3 Upper), it was less efficient at stimulating editing during HDV replication at all time points (Fig. 3 Lower). Because ADAR1-S is more efficient than ADAR1-L at editing substrates in the nucleus but much less efficient at editing substrates in the cytoplasm, we concluded that during replication, HDV antigenomic RNA is edited in the nucleus.
Fig 3.
Overexpression of HA-tagged ADAR1-L and ADAR1-S during HDV replication. Western analyses were performed on total cell lysates expressing replicating-HDV RNA and either HA-tagged ADAR1-L or ADAR1-S. Posttransfection (3, 5, and 7 days), the samples were blotted with either anti-HA monoclonal antibody (Upper) to monitor protein expression or anti-delta antigen polyclonal antiserum (Lower) to monitor editing. The relative expression of the HA-tagged ADARs was obtained by comparison with a HA-tagged ADAR1 standard. An equal aliquot of this standard was loaded on all of the anti-HA Westerns, and the signal obtained for this standard was given a value of 10 (27).
siRNA Targeted to ADAR1 Lowered Both the Exogenous and Endogenous Expression of That Protein.
We next used RNAi to test the hypothesis that ADAR1-S is the enzyme expressed by cells that is responsible for editing replicating-HDV RNA. Toward that end, we first evaluated the activity of an siRNA targeted to all ADAR1 transcripts (ADAR1-L+S) as well as that of an siRNA targeted to ADAR2 transcripts. The appropriate siRNA was cotransfected with a HA-tagged ADAR expression vector and with a vector that expresses a nonreplicating-HDV amber/W-editing reporter (28). The reporter was used to monitor editing activity. It expresses an mRNA that encodes the HDAg-S and includes two-thirds of the antigenomic rod-like structure. After editing of this reporter in either the nucleus or the cytoplasm, the resulting transcript expresses the HDAg-L.
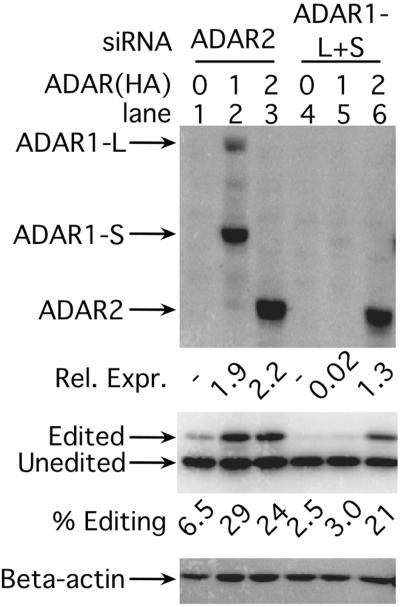
As shown in Fig. 4, the siRNA directed against ADAR2 was not able to reduce expression of that protein (compare lanes 3 and 6). In this and all subsequent experiments, this nonfunctional siRNA serves to control for the nonspecific and sequence-independent effects that might result from the introduction of 21-nt duplex RNA into cells. In contrast, the siRNA targeted to both forms of ADAR1, ADAR1-L+S, was very functional and lowered ADAR1 expression nearly 100-fold (Fig. 4, compare lanes 2 and 5). Furthermore, the siRNA was very specific and did not alter ADAR2 expression significantly (Fig. 4, compare lanes 3 and 6).
Fig 4.
Expression of transfected ADAR1(HA) but not ADAR2(HA) is inhibited by siADAR1-L+S. The top anti-HA Western analysis shows expression of transfected pDL700 (carboxyl-terminally HA-tagged ADAR1) (lanes 2 and 5, ref. 27), pMS040 (ADAR2) (lanes 3 and 6, ref. 27), or empty vector, pDL668 (lanes 1 and 4). (Middle) The anti-delta antigen Western analysis monitored the corresponding editing of the cotransfected HDV amber/W-editing reporter, pSS74 (28). (Bottom) The anti-β-actin Western blot is a control for total protein loading.
We next tested whether ADAR1-L+S siRNA was also effective at inhibiting the endogenous expression of both forms of ADAR1. We also tested whether we could reduce the endogenous expression ADAR1-L specifically by using an siRNA targeted to a sequence within exon 1A of the ADAR1-L message. In the previous experiment, untransfected cells were not a concern. Because the HA-tagged ADAR1 expression vector was co-delivered with the siRNA, any cell not transfected with the siRNA would also not be transfected with the HA-tagged ADAR1 expression vector and therefore would not express a HA-immunoreactive signal. However, in experiments in which the endogenous message is targeted, transfection efficiency is a concern. If only half the cells are transfected but all are harvested, then the siRNA could cause no more than a 2-fold reduction in the level of the targeted protein. To maximize transfection efficiency, multiple sequential transfections were performed. In the final transfection, the siRNA in question was cotransfected with the vector that expresses an editing reporter such that editing activity could be monitored. To monitor transfection variability, the experiment was performed in duplicate.
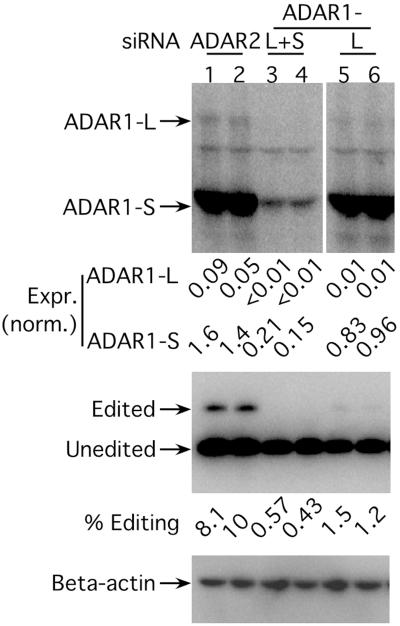
As shown in Fig. 5, ADAR1-L+S siRNA was effective at lowering the endogenous expression of both ADAR1-L and ADAR1-S, and ≈8-fold reductions were observed (compare lanes 1 and 2 with lanes 3 and 4). It is important to remember that this fold reduction is a minimum estimate because of the possible presence of untransfected cells. Nevertheless, given the fold reduction observed, we concluded that the transfection efficiency must have been ≈90%. In the presence of ADAR1-L+S siRNA, almost no editing of the reporter was observed. From this we concluded that ADAR1 is responsible for virtually all the activity expressed by HEK293 cells capable of editing the reporter. Introduction of the ADAR1-L siRNA caused a similar 7-fold reduction in the ADAR1-L level without significantly affecting the level of ADAR1-S (Fig. 5, compare lanes 1 and 2 with lanes 5 and 6). It is interesting to note that the introduction of this siRNA caused a nearly 7-fold reduction in the editing of the reporter. Thus ADAR1-L is responsible for most of the editing that occurs with the reporter despite the fact that it accounts for only 5% of the total ADAR1 signal in untreated cells. This is entirely consistent with our recent observation that, with respect to the reporter, the specific activity of ADAR1-L is nearly 80-fold higher than that of ADAR1-S (S.K.W., S. Sato, and D.W.L., unpublished data).
Fig 5.
siRNAs targeted against ADAR1 decreased ADAR1 endogenous expression and activity. (Top) An anti-ADAR1 Western analysis to detect endogenous ADAR1 expression when siRNAs targeted against either ADAR2 or both forms of ADAR1 (siADAR1-L+S) or ADAR1-L alone (siADAR1-L) were introduced into HEK293 cells. Each siRNA was tested in duplicate. (Middle) An anti-delta antigen (HDAg) Western analysis to monitor the corresponding editing activity assayed with an HDV amber/W reporter, pSS140 (28). (Bottom) An anti-β-actin Western analysis to serve as a total protein loading control. ADAR expression was normalized to β-actin expression.
ADAR1-S Edited HDV RNA During Replication.
Having established the effectiveness of two siRNAs, one that lowered the expression of both forms of ADAR1 and one that specifically targeted ADAR1-L, we next used these reagents to identify the activity expressed by HEK293 cells that edits HDV RNA during replication. The same protocol was used as in the prior experiment except that the vector used to initiate HDV replication was cotransfected with the siRNA in the last transfection. Furthermore, in other experiments, we observed that within 4–6 days after siRNA treatment, ADAR1 protein levels began to increase (data not shown). For this reason, the cells harboring replicating HDV were split 1:3 and retransfected with siRNA 4 and 8 days after transfection of the HDV cDNA.
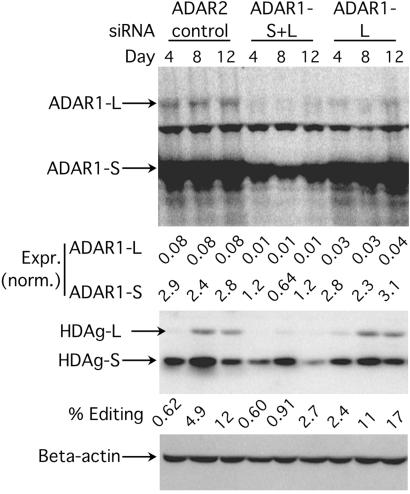
Fig. 6 shows that introduction of the ADAR1-L+S siRNA caused a decrease in the expression of both forms of endogenous ADAR1 (Top) and dramatically decreased editing of replicating HDV at all three time points (Middle) when compared with the ADAR2 siRNA control. In contrast, the ADAR1-L siRNA decreased the expression of ADAR1-L at all time points but did not decrease the editing of replicating HDV. These results established that ADAR1-S edited HDV antigenomic RNA during replication. Because editing of the nonreplicating reporter but not replicating-HDV RNA was inhibited by ADAR1-L siRNA, these results also highlight the differences in the subcellular locations where these two distinct substrates are edited.
Fig 6.
The effect of lowering the endogenous levels of ADAR1 on editing during HDV replication in HEK293 cells. (Top) An anti-ADAR1 Western analysis showing the endogenous ADAR1 expression during HDV replication 4, 8, and 12 days posttransfection. The first three lanes were cotransfected with siADAR2, the next three lanes with siADAR1-L+S, and the last three lanes with siADAR1-L. The cells were retransfected with siRNAs every 4 days during the course of the experiment. (Middle) An anti-delta antigen Western analysis showing the products of HDV editing (HDAg-L and HDAg-S). (Bottom) An anti-β-actin Western analysis for total protein loading control.
Discussion
Here were provided multiple lines of evidence first to suggest and then establish that ADAR1-S edited HDV antigenomic RNA during replication. Although ADAR2 is capable of editing the HDV amber/W site, we found that this protein is not expressed in a liver hepatoma cell line harboring replicating HDV, whereas both forms of ADAR1 were expressed. Recently, we found that ADAR1-L is much more efficient than ADAR1-S at editing a nonreplicating-HDV editing reporter and that such editing by ADAR1-L occurred mainly in the cytoplasm (S.K.W., S. Sato, and D.W.L., unpublished data). In other experiments, we found that ADAR1-S is more efficient than ADAR1-L when editing must occur in the nucleus. Here we found that when expressed exogenously, ADAR1-S was more efficient than ADAR1-L at editing HDV antigenomic RNA during replication, and this enabled us to conclude that editing occurred in the nucleus. Using RNAi, we were able to lower the level of ADAR1-L specifically without significantly affecting the level of ADAR1-S. Such treatment greatly reduced the editing of a nonreplicating-HDV amber/W site reporter that can be edited in the cytoplasm but had no effect on amber/W editing during HDV replication. In contrast, when the levels of both ADAR1-S and ADAR1-L were lowered, amber/W editing during replication was nearly abolished (12% with control siRNA vs. 2.4% with ADAR1-S+L siRNA).
We concluded that in HEK293 cells, ADAR1-S was responsible for at least 80% of all amber/W-editing events. Furthermore, given that siRNA treatment only reduced and did not abolish ADAR1 expression, the actual contribution to amber/W-editing by ADAR1-S in tissue culture is likely to be much higher. We do not know whether the same is also true in the human liver during a natural infection, and this issue cannot be addressed experimentally. However, it should be possible to co-deliver HDV RNA and ADAR1 siRNA in the hydrodynamic mouse model so that the importance of ADAR1-S in that setting could be tested (36, 37).
RNAi is clearly a very powerful method to examine virus–host interactions. One drawback to the method in human cells, however, is the lack of a persistent effect. Because siRNA is not infinitely stable and apparently is not replicated in human cells, we were forced to split cells and retransfect at days 4 and 8 (see Fig. 6). The reduction in the delta antigen signal observed between days 8 and 12 occurred because on day 8, the day-12 sample was split 1:3, and two-thirds of the signal were discarded, whereas the day-8 sample was harvested without splitting. The recent development of cDNA-based systems that express siRNA hairpins (38–40) is likely to obviate this technical difficulty, because transfected cDNA is known to persist in cells for several weeks. In plants, Neurospora, and Caenorhabditis elegans, the RNAi response is very persistent, can be spread to adjacent cells and tissues, and can even be transmitted to offspring (30, 41). These organisms express RNA-dependent RNA polymerases that are thought to amplify siRNA and thereby perpetuate the persistent effect (42, 43). The lack of persistent RNAi in human cells may indicate the absence of an RNAi-specific RNA-dependent RNA polymerase. Consistent with this possibility, BLASTP searches failed to identify a homolog of any known RNAi-specific RNA-dependent RNA polymerase in the human genome.
The replication of certain plant and insect RNA viruses can induce an RNAi response in the host, and this response is fully capable of inhibiting further viral replication. As a retort to the RNAi response, these viruses express a protein that inhibits the function of the RNAi pathway. An example of such an RNAi antagonist is the B2 protein expressed by Flock House virus (44). If HDV had a similar capability to inhibit the RNAi response, then the experiment shown in Fig. 6 would have failed, and no reduction in ADAR1 levels would have been observed. We therefore conclude that HDV replication does not antagonize RNAi and that HDV replication could be sensitive to inhibition by that process. It is possible that siRNA directed against the HDAg-S message would be effective in lowering the expression of the protein. Given that the HDAg-S provides an essential role in replication, RNAi could inhibit or limit HDV replication.
Editing of the amber/W site during replication seems to be a highly regulated process. From days 4 through 7 posttransfection, there was an apparent lag in editing, and very little HDAg-L was expressed even though the level of HDAg-S was increasing steadily during that period (see Fig. 2B). Suddenly, from day 7 to day 8, the proportion of HDAg-L increased from 3.8 to 9.1% and to 16% by day 10. After day 10, the rate of increase in the proportion of HDAg-L began to slow. We have observed this pattern repeatedly, and in experiments of longer duration we observed that a plateau was reached at approximately day 14–16, when the proportion of HDAg-L approached 30% and did not change with further time (data not shown). This apparent regulation of editing is entirely consistent with the HDV life cycle. An initial lag in HDAg-L expression provides a window of time in which replication can occur in the absence of that protein. This is likely to be very important, because it is known that when present at the onset of replication, the HDAg-L inhibits that process in a potent dominant-negative manner (6). The editing plateau observed at very late times posttransfection ensures that the majority of antigenomes remain unedited and hence that the majority of genomes express the HDAg-S. This is also important because in a natural infection, these genomes would be packaged into virions, and only HDAg-S-expressing genomes can complete another cycle of infection.
The identification of ADAR1-S as the enzyme responsible for amber/W site-editing during HDV replication is an important first step in unraveling the mechanism that regulates this process. One could hypothesize that amber/W editing is regulated by modulation of the level of editing enzyme. However, the evidence provided here is inconsistent with such a hypothesis, because no significant change in the amount of ADAR1-S was observed throughout the replication time course (see Fig. 2). It remains possible, however, that the activity rather than the amount of ADAR1-S is modulated during HDV replication. It is also possible that amber/W-editing occurs only on nascent antigenomic RNA as it is being synthesized. If this were true, then editing would be coupled to and regulated by replication. Additional experiments will be needed to test these and alternate models.
Acknowledgments
We thank John Coffin (Tufts University) for helpful discussions. This work was supported by National Institutes of Health Grant R01-AI40472 and the Raymond and Beverly Sackler Research Foundation.
Abbreviations
HDV, hepatitis delta virus
HDAg-S, small delta antigen
HDAg-L, large delta antigen
ADAR, adenosine deaminase that acts on RNA
ADAR1-L, large form of ADAR1
ADAR1-S, small form of ADAR1
RNAi, RNA interference
si, small interfering
HEK, human embryonic kidney
HA, hemagglutinin
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rizzetto M., Hoyer, B., Canese, M. G., Shih, J. W., Purcell, R. H. & Gerin, J. L. (1980) Proc. Natl. Acad. Sci. USA 77, 6124-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuo M. Y., Goldberg, J., Coates, L., Mason, W., Gerin, J. & Taylor, J. (1988) J. Virol. 62, 1855-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazinski D. W. & Taylor, J. M. (1994) Adv. Virus Res. 43, 187-231. [DOI] [PubMed] [Google Scholar]
- 4.Hsieh S. Y., Chao, M., Coates, L. & Taylor, J. (1990) J. Virol. 64, 3192-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casey J. L. & Gerin, J. L. (1995) J. Virol. 69, 7593-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao M., Hsieh, S. Y. & Taylor, J. (1990) J. Virol. 64, 5066-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang F. L., Chen, P. J., Tu, S. J., Wang, C. J. & Chen, D. S. (1991) Proc. Natl. Acad. Sci. USA 88, 8490-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryu W. S., Netter, H. J., Bayer, M. & Taylor, J. (1993) J. Virol. 67, 3281-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tavanez J. P., Cunha, C., Silva, M. C., David, E., Monjardino, J. & Carmo-Fonseca, M. (2002) RNA 8, 637-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gudima S., Chang, J., Moraleda, G., Azvolinsky, A. & Taylor, J. (2002) J. Virol. 76, 3709-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macnaughton T. B. & Lai, M. M. (2002) J. Virol. 76, 3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polson A. G., Ley, H. L., III, Bass, B. L. & Casey, J. L. (1998) Mol. Cell. Biol. 18, 1919-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim U., Wang, Y., Sanford, T., Zeng, Y. & Nishikura, K. (1994) Proc. Natl. Acad. Sci. USA 91, 11457-11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai F., Drakas, R. & Nishikura, K. (1995) J. Biol. Chem. 270, 17098-17105. [DOI] [PubMed] [Google Scholar]
- 15.O'Connell M. A., Krause, S., Higuchi, M., Hsuan, J. J., Totty, N. F., Jenny, A. & Keller, W. (1995) Mol. Cell. Biol. 15, 1389-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melcher T., Maas, S., Herb, A., Sprengel, R., Seeburg, P. H. & Higuchi, M. (1996) Nature 379, 460-464. [DOI] [PubMed] [Google Scholar]
- 17.Patterson J. B. & Samuel, C. E. (1995) Mol. Cell. Biol. 15, 5376-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poulsen H., Nilsson, J., Damgaard, C. K., Egebjerg, J. & Kjems, J. (2001) Mol. Cell. Biol. 21, 7862-7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.George C. X. & Samuel, C. E. (1999) Gene 229, 203-213. [DOI] [PubMed] [Google Scholar]
- 20.Kawakubo K. & Samuel, C. E. (2000) Gene 258, 165-172. [DOI] [PubMed] [Google Scholar]
- 21.Sommer B., Kohler, M., Sprengel, R. & Seeburg, P. H. (1991) Cell 67, 11-19. [DOI] [PubMed] [Google Scholar]
- 22.Higuchi M., Single, F. N., Kohler, M., Sommer, B., Sprengel, R. & Seeburg, P. H. (1993) Cell 75, 1361-1370. [DOI] [PubMed] [Google Scholar]
- 23.Lomeli H., Mosbacher, J., Melcher, T., Hoger, T., Geiger, J. R., Kuner, T., Monyer, H., Higuchi, M., Bach, A. & Seeburg, P. H. (1994) Science 266, 1709-1713. [DOI] [PubMed] [Google Scholar]
- 24.Burns C. M., Chu, H., Rueter, S. M., Hutchinson, L. K., Canton, H., Sanders-Bush, E. & Emeson, R. B. (1997) Nature 387, 303-308. [DOI] [PubMed] [Google Scholar]
- 25.Rueter S. M., Dawson, T. R. & Emeson, R. B. (1999) Nature 399, 75-80. [DOI] [PubMed] [Google Scholar]
- 26.Polson A. G., Bass, B. L. & Casey, J. L. (1996) Nature 380, 454-456. [DOI] [PubMed] [Google Scholar]
- 27.Wong S. K., Sato, S. & Lazinski, D. W. (2001) RNA 7, 846-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato S., Wong, S. K. & Lazinski, D. W. (2001) J. Virol. 75, 8547-8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higuchi M., Maas, S., Single, F. N., Hartner, J., Rozov, A., Burnashev, N., Feldmeyer, D., Sprengel, R. & Seeburg, P. H. (2000) Nature 406, 78-81. [DOI] [PubMed] [Google Scholar]
- 30.Fire A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806-811. [DOI] [PubMed] [Google Scholar]
- 31.Zamore P. D., Tuschl, T., Sharp, P. A. & Bartel, D. P. (2000) Cell 101, 25-33. [DOI] [PubMed] [Google Scholar]
- 32.Hammond S. M., Bernstein, E., Beach, D. & Hannon, G. J. (2000) Nature 404, 293-296. [DOI] [PubMed] [Google Scholar]
- 33.Elbashir S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- 34.Lazinski D. W. & Taylor, J. M. (1994) J. Virol. 68, 2879-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgenstern J. P. & Land, H. (1990) Nucleic Acids Res. 18, 3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang J., Sigal, L. J., Lerro, A. & Taylor, J. (2001) J. Virol. 75, 3469-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCaffrey A. P., Meuse, L., Pham, T. T., Conklin, D. S., Hannon, G. J. & Kay, M. A. (2002) Nature 418, 38-39. [DOI] [PubMed] [Google Scholar]
- 38.Paddison P. J., Caudy, A. A., Bernstein, E., Hannon, G. J. & Conklin, D. S. (2002) Genes Dev. 16, 948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brummelkamp T. R., Bernards, R. & Agami, R. (2002) Science 296, 550-553. [DOI] [PubMed] [Google Scholar]
- 40.Sui G., Soohoo, C., Affarel, B., Gay, F., Shi, Y. & Forrester, W. C. (2002) Proc. Natl. Acad. Sci. USA 99, 5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cogoni C. & Macino, G. (1997) Proc. Natl. Acad. Sci. USA 94, 10233-10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cogoni C. & Macino, G. (1999) Nature 399, 166-169. [DOI] [PubMed] [Google Scholar]
- 43.Sijen T., Fleenor, J., Simmer, F., Thijssen, K. L., Parrish, S., Timmons, L., Plasterk, R. H. & Fire, A. (2001) Cell 107, 465-476. [DOI] [PubMed] [Google Scholar]
- 44.Li H., Li, W. X. & Ding, S. W. (2002) Science 296, 1319-1321. [DOI] [PubMed] [Google Scholar]