Abstract
In the first stage of engineering a herpes simplex virus (HSV)-1 that specifically targets human malignant glioma cells, we constructed a recombinant virus designated R5111 in which we have ablated the binding sites for sulfated proteoglycans in glycoproteins B and C, replaced the amino-terminal 148 aa in glycoprotein C by IL-13 flanked at its amino terminus with a signal peptide, and inserted a second copy of IL-13 after the amino acid 24 of glycoprotein D. In the process, the binding site for HveA, a viral entry receptor, was disrupted. We have also transformed a cell line (J1.1) lacking HSV-1 receptors to express IL13Rα2 receptor (J13R cells). We report the following: the R5111 recombinant virus replicates as well as wild-type virus in a variety of cell lines including cell lines derived from brain tumors. R5111 failed to replicate in the parent J1.1 cell line but multiplied to titers similar to those obtained in other cell lines in the J13R cell line. On the basis of the evidence that R5111 can use the IL13Rα2 receptor for entry, we conclude that HSV-1 can use receptors other than HveA or nectins, provided it can bind to them. The domains of gD that interact with HveA and nectin receptors are independent of each other. Lastly, the fusogenic activities of the glycoproteins in the viral envelope are not dependent on a set of unique interactions between glycoprotein D and its receptor. The construction of R5111 opens the way for construction of viruses totally dependent on selected receptors for entry or imaging of targeted cells.
Malignant gliomas are devastating human brain tumors. The average life span after diagnosis is <1 year, and few patients have been reported to survive 5 years (1). Furthermore, none of the conventional anticancer therapies has been successful in significantly prolonging the lifespan of patients with this disease. In recent years, there have been numerous attempts to use genetically engineered herpes simplex viruses (HSV) as oncolytic agents to treat malignant gliomas (2–6). Because wild-type viruses are highly virulent, the viruses used in preclinical evaluations and lately in phase 1 studies have been extensively debilitated. Whereas several deletion mutants have been tested, the mutants that reached clinical trails lacked either the γ134.5 gene encoding the infected cell protein no. 34.5 (ICP34.5; refs. 2–4) or both Δγ134.5 and the UL39 gene encoding the large subunit of ribonucleotide reductase (5).
The properties of these viruses reflect the strengths and weaknesses of the genetically engineered HSV mutants tested to date. The strength of these mutant viruses is that they have a significantly reduced capacity to replicate in normal, nondividing cells in vivo. Viral ribonucleotide reductase is an essential gene for viral replication in resting cells, and hence the UL39 mutant virus is highly debilitated in the normal environment of the central nervous system (7). The major function of ICP34.5 is to preclude the shutoff of protein synthesis caused by activation of protein kinase R in infected cells. Once activated, this enzyme phosphorylates the α subunit of the translation initiation factor 2 (eIF2α) resulting in the total shutoff of the translation machinery (8–10). Mutants lacking the γ134.5 genes are highly attenuated because they are readily blocked by the IFN pathway. Indeed, γ134.5 mutants are nearly as virulent as wild-type viruses in mice lacking IFN receptor (11). Mutants deleted in both γ134.5 and UL39 are not significantly more attenuated than those lacking the γ134.5 genes, but they provide an added insurance that second-site mutations that compensate for the absence of both genes are not likely to arise.
The weakness of the viruses used to date is that they replicate poorly even in dividing cells; in experimental animal systems, they do not persist long enough to destroy many more tumor cells beyond those infected at the time of administration of the virus. To achieve maximum killing of tumor cells, high doses of virus are thus required. Because these viruses grow poorly even in dividing cells, production of virus pools large enough to enable inocula of >109 plaque-forming units (pfu) becomes a major obstacle.
One solution to the problem is to genetically engineer viruses capable of multiplying solely in tumor cells. In principle, there are two possible ways to target viruses to specific cells. The first approach is to place an essential ORF under a promoter active only in the tumor cells. The disadvantage of the system is that although these viruses could multiply only in cells in which the promoter is active, they can attach, infect, and potentially damage or destroy cells in which the promoter is not active.
The second alternative described in this report is to modify the surface of the virus particle so as to target the virus to a specific receptor present solely or predominantly on the surface of the tumor cell. Relevant to this report are therefore both the process by which HSV enters cells and the properties of the cell-surface receptor to which the genetically engineered virus was targeted.
HSV enters susceptible cells in two steps. In the first, glycoproteins B and C (gB and gC) projecting from the viral envelope attach to heparan sulfate proteoglycans on cell surfaces. The gB and gC sequences interacting with heparan sulfate have been mapped (12). Following the initial attachment, viral glycoprotein D (gD) interacts with one of several receptors. Of these, two are particularly important for entry (13). One, designated HveA, is a member of the family of receptors for tumor necrosis proteins (14). A second, designated HveC, is a member of nectins, a family of proteins that are structurally related to the Ig superfamily and that serve as intercellular connectors (13, 15). Once gD interacts with its receptor, it recruits glycoproteins B, H, and L to effect the fusion of the envelope with the plasma membrane (13, 15). In addition, recent studies have lent significance to an old observation that gD interacts with the cation-independent mannose 6 phosphate receptor. Thus, virus particles taken up by endocytosis are degraded by lysosomal enzymes, but the cells succumb as a consequence of the degradation of its DNA by lysosomal DNase. gD blocks cell death through its interaction with the mannose 6 phosphate receptor (16). gD thus interacts with HveA, nectins, mannose 6 phosphate receptor, and at least one complex of viral glycoproteins involved in the fusion of the cell with the plasma membrane.
The target for entry of the virus into malignant glioma cells is the IL13Rα2 receptor reported to be present in malignant gliomas (17, 18). Unlike the more prevalent IL13αR1 receptor, IL13Rα2 receptor has a shorter cytoplasmic domain and does not interact with IL-4, of which IL-13 is a close relative (19, 20). In the studies reported here, we mutagenized gB and gC to preclude their interaction with heparan sulfate. We have also inserted IL-13 into gD at amino acid 24 to disrupt the binding site of HveA. We report that the IL-13–gD chimeric virus can use IL13Rα2 for entry into cells carrying only that receptor.
Materials and Methods
Cells and Viruses.
SK-N-SH, HEp-2, Vero, and U87 cells were obtained from the American Type Culture Collection and maintained in DMEM supplemented with 10% FBS. J1.1, a derivative of BHK, and thymidine kinase minus cells that lack both HveA and nectin 1 receptors was a kind of gift from G. Campadelli-Fiume (University of Bologna, Italy).
Antibodies.
Monoclonal antibodies against gD (clone H170), gC, and hemagglutinin (HA)-specific polyclonal antiserum described elsewhere were purchased from the Goodwin Institute, Plantation, FL (16, 21, 22). Polyclonal antibodies against IL-13 were purchased from Santa Cruz Biotechnology.
Construction of the R5111 Recombinant Virus.
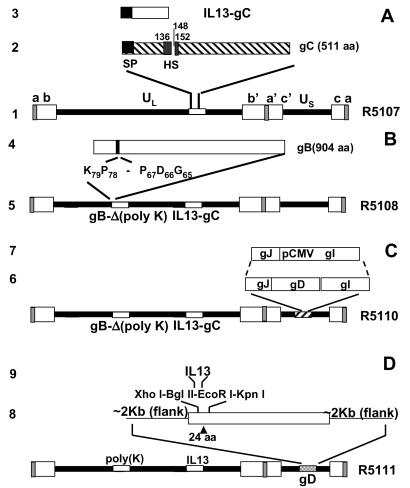
The recombinant virus R5111 was constructed in several steps depicted in Fig. 1.
Fig 1.
Schematic representation of the HSV-1 (F) genome and gene manipulations in glycoprotein C (gC), glycoprotein B (gB), and glycoprotein D (gD). Line 1, sequence arrangement of HSV-1 genome. The rectangular boxes represent the inverted repeat sequences ab and b′a′ flanking the unique long (UL) sequence and inverted repeat c′a′ and ca flanking the unique short (US) sequence. Line 2, sequence arrangement of the domains of the glycoprotein C, signal peptide (SP) domain, and heparan sulfate (HS)-binding domain of gC are highlighted. Line 3, human IL-13 with signal peptide that replaced the N-terminal segment of 148 aa of gC. Line 4, sequence arrangement of the polylysine domain of gB. Line 5, schematic representation of recombinant HSV-1(F) genome, in which the N-terminal domain of gC was replaced with IL-13, and the polylysine domain (from codon 68 to codon 77) of gB was deleted. Line 6, sequence arrangement of glycoprotein J (gJ), glycoprotein D (gD), and glycoprotein I (gI) in US. Line 7, replacement of gD with the immediately early promoter of cytomegalovirus to enable the expression of gI. Line 8, schematic representation of recombinant HSV-1(F) genome, in which the N-terminal domain of gC was replaced with IL-13, the polylysine domain of gB was deleted, and IL-13 was inserted after amino acid 24 of gD. Line 9, a polylinker XhoI-BglII-EcoRI-KpnI was inserted after amino acid 24 of gD, and then IL-13 was inserted into the XhoI and KpnI sites of gD.
Substitution of amino-terminal domain of gC with IL-13 fused to the signal sequence of gC.
Fig. 1A, lines 1–3, schematically depicts the engineering by the primer elongation method of a cDNA consisting of IL-13 coding sequence fused at its amino terminus to its signal sequence. The complete cDNA of IL-13 with the N-terminal signal peptide was engineered by PCR primer elongation method. The primers were as follows: pIL13F1, CATTGCTCTCACTTGCCTTGGCGGCTTTGCCTCCCCAGGCCCTGTGCCTCCCTCTACAGC; pIL13F2, GCAGCTAGCCTCATGGCGCTTTTGTTGACCACGGTCATTGCTCTCACTTGCCTTGGCGGC; and pIL13REcoRI, GAGCTCGGATCCTGAATTCAACCGTCCCTC. First-round PCR used pIL13F1 and pIL13REcoRI as primers and pRB5830 as template. The PCR mixture was then diluted 10-fold, and 1 μl of the diluted reaction mixture was used as template for the second round of PCR amplifications with pIL13F2 and pIL13REcoRI as the primer set. The PCR product was gel purified, digested with NheI/EcoRI, and ligated into pBluescript II KS(+) at XbaI/EcoRI sites to generate pRB5832. To construct the transfer plasmid pRB5835, a 4.8-kbp HindIII/SacI fragment containing the HSV-1 gC was released from cosmid pBC1007 and inserted into pBluescript II KS(+) to generate pRB5833. pRB5833 was cleaved with NheI and EcoRI, and the N-terminal 148 residues of gC were replaced with the gC signal–IL-13 chimeric sequence (pRB5834). The insert in pRB5834 was released by XhoI/SacI digestion and ligated into pKO5Y at the same sites to generate pRB5835.
The recombinant virus R5107 (Fig. 1A, line 1) carrying the IL-13–gC chimera was generated with the aid of BAC-HSV system. RR1-competent cells that harbored bacterial artificial chromosome (BAC)-HSV bacmids were transformed with the transfer plasmid pRB5835 by electroporation. After incubation for 1 h at 30°C in LB broth, the transformed bacteria were plated on prewarmed zeocine plus chloramphenicol (20 μg/ml each) plates and incubated overnight at 43°C for integration. The next day, six colonies were picked and diluted in 1 ml of LB, respectively; 5 μl of the diluted bacteria were then plated on chloramphenicol/10% sucrose (Suc) plates and incubated at 30°C overnight. To further confirm the loss of the replacement vector, 24 chloramphenicol/Suc-resistant colonies (four colonies from each plate) were restreaked in duplicate on chloramphenicol LB and zeocine LB plates, respectively. The Sucr/Cmr/Zeor colonies were further screened by PCR (95°C, 4 min; then 35 cycles of 94°C, 1 min; 60°C, 1 min; 72°C, 1 min). The primers were pgC-F, GACACGGGCTACCCTCACTATCGAGGGC (from nt 96158 to 96185 in HSV-1 strain 17), and pgC-R, GGTGATGTTCGTCAGGACCTCCTCTAGGTC (from nt 96859 to 96830). The DNA fragment amplified from PCR positive clones (Fig. 2B) was sequenced to further confirm the integration of IL-13 in the correct ORF of gC. To verify the viability of the recombinant (R5107), the recombinant BAC-HSV DNA was prepared as described elsewhere (23) and transfected into rabbit skin cells by Lipofectamine reagent (Life Technologies, Grand Island, NY). The resultant virus R5107 was stored at −80°C.
Fig 2.
Amino acid sequence alignment of IL-13–gC, IL-13–gD junction sequence and the HS-binding domain of gB. (A) The amino-terminal sequence of IL-13–gC chimeric protein. The sequences upstream and downstream of the HS-binding site portion are shown. IL-13 (27) was inserted between two restriction enzyme sites that are underlined. (B) The domain of the gB ORF from which the polylysine [poly(K)] sequence was deleted. The underlined sequences (codons 68–77) were not present in gB amplified from R5107. (C) The amino-terminal sequence of IL-13–gD. The first underlined sequence identifies the gD signal peptide. IL-13 (bracketed by arrows) was inserted between residues 24 and 25 (underlined) of gD, between the XhoI and KpnI restriction enzyme sites.
Deletion of the polylysine track from gB.
To make a transfer plasmid for the deletion of gB heparan sulfate binding domain (polylysine), a 4.76-kbp BstEII fragment (from nt 53164 to 57923) containing the UL27 (gB) ORF released from cosmid BC1014 was blunt-ended and cloned into pBluescript II KS (+) at the SmaI site to generate pRB5846 (Fig. 1B). To construct pRB5847, from which the 10-aa polylysine domain of gB was deleted, two fragments flanking the polylysine domain were amplified by PCR from pRB5846. The primer sets were pgB1BamHI, GTTCTTCTTCGGTTTCGGATCCCCCG; pgB2BspEI, CGGCATTTCCGGAATAACGCCCACTC; pgB3BamHI, CAGAAAACCGGATCCCCCAAAGCCGCC; and pgB4BsiWI, GCCAACACAAACTCGTCGTACGGGTAC. PCR-amplified fragments were then cut with BspEI/BamHI or BsiWI/BamHI and ligated into pRB5846, in which the 1.2-kbp BsiWI/BspEI fragment had been removed. To generate the transfer plasmid pRB5848, the 4.76-kbp insert in pRB5847was released by XbaI/EcoRV digestion and ligated into pKO5Y at the sites of XbaI/ScaI. Recombinant HSV-1 virus R5108 is based on R5107 with the additional deletion of the gB heparan sulfate binding domain. It was made by the same procedure as BAC-R5107, except that the transfer plasmid pRB5848 was used instead of BAC-HSV wild type and pRB5835. The sequence of the mutant gB was verified by sequencing of the entire ORF.
Deletion of gD.
The objective was to replace the coding sequence of gD with the human cytomegalovirus immediate early promoter to enable the expression of glycoprotein I (Fig. 1C, lines 6 and 7). A 0.65-kbp fragment containing the promoter was released from pRB5836 by ClaI digestion and inserted into pgD−, a plasmid obtained from G. Campadelli-Fiume. This plasmid, containing the flanking sequences of gD but lacking the gD ORF, had been cut with ClaI to generate pRB5849. pRB5849 was then cut with NotI/PmeI and ligated into pKO5Y at NotI/ScaI to generate the transfer plasmid pRB5850. Recombinant HSV-1 virus R5110 is based on R5108 with the additional deletion of the gD. It was made by the same procedure as BAC-R5107 except that transfer plasmid pRB5850 was used instead of BAC-HSV wild type and pRB5835. The recombinant BAC-HSV DNA was prepared as described elsewhere (23). The mutant virus was designated as R5110.
Construction of the R5111 mutant carrying the IL-13–gD chimeric gene.
Plasmid pRB123 carries a 6584 bp BamHI J fragment containing gD and flanking sequences in the BamHI site of pBR322 (Fig. 1D). To construct the IL-13–gD chimeric plasmid, pRB123 was digested with AflII and HpaI to release two fragments, 7.6 and 3.2 kb, respectively. The 3.2-kb fragment was further digested with FspI to release a 2.5-kb and a 0.7-kb fragment, which contains the amino-terminal 661 bp of gD ORF. A polylinker sequence containing the restriction sites XhoI-BglII-EcoRI-KpnI was inserted into the 0.7-kb fragment downstream of the 24th codon of gD by two PCRs with forward primer 5′-CAGTTATCCTTAAGGTCTCTTTTGTGTGGTG-3′, reverse primer 5′-CCGGAATTCCGGAGATCTTCCCTCGAGGACCGGAAGGTCTTTGCCGCGAAAG-3′, and another forward primer 5′-CCGGAATTCCGGGGTACCCTGGACCAGCTGACCGACCCTCCGG-3′ and reverse primer 5′-CGGGGGGATGCGCAGCGGGAGGGCGTACTTAC-3, respectively. After digestion of the two PCR products by EcoRI, they were ligated and amplified by PCR again to obtain the desired DNA fragment containing the polylinker insertion. IL-13 was amplified by PCR with forward primer 5′-CCGCTCGAGATGGCGCTTTTGTTGACCACGG-3′ and reverse primer 5′-GGGGTACCGTTGAACCGTCCCTCGCGAAA-3′ and then inserted into the XhoI and KpnI site of the 0.7-kb fragment. This new fragment with IL-13 insertion was then ligated with 2.5 and 7.6 kb together to generate the IL-13–gD chimeric transfer plasmid, pRB13–24.
R5111 was generated by cotransfection of transfer plasmid pRB13–24 and the R5110 viral DNA into U87 glioma cells. The progeny of the transfection was plated at a high dilution on Vero and HEp-2 cell cultures so as to yield individual, well spaced plaques. From each of the infected cell cultures, six single plaques were picked, frozen-thawed, sonicated, and then replated for preparation of virus stocks and viral DNA for sequencing on fresh cultures of Vero or HEp-2 cells, depending on the origin of the plaque.
Viral DNA Extraction.
The infected cells were removed from each of the 25-cm2 flasks exposed to the individual plaque isolates, rinsed, and resuspended in 500 μl of Lyse-O-Lot (150 mM NaCl/10 mM Tris/1.5 mM MgCl2) in the presence of 0.1% of Nonidet P-40. The nuclei were removed by low-speed centrifugation. SDS to 0.2%, EDTA to 5 mM, and β-ME to 50 mM were added to the supernatant fluid, which was then extracted twice with phenol/chloroform. Viral DNA was finally precipitated by ethanol and resuspended, and the IL-13 ORF and IL-13–gD chimeric reading frame were amplified by PCR with two sets of primers. The first set designed to detect IL-13 consisted of 5′-CCGCTCGAGATGGCGCTTTTGTTGACCACGG-3′ and 5′-GGGGTACCGTTGAACCGTCCCTCGCGAAA-3′, which included IL-13 ORF. The second set designed to detect the IL-13–gD junction consisted of 5′-CCGCTCGAGATGGCGCTTTTGTTGACCACGG-3′ and 5′-AACTGCAGGTTGTTCGGGGTGGCCGGGGG-3′. All 12 IL-13–gD PCR products were sequenced to determine whether the gD sequence contained deletions or substitutions.
Construction of J13R, a Cell Line Stably Expressing IL13Rα2 Receptor.
IL13α2 ORF was tagged with an HA tag at its carboxyl terminus by PCR with forward primer 5′-AAGATTTGGGCTAGCATGGCTTTCGTTTGC-3′ and reverse primer 5′-TCCCTCGAAGCTTCAAGCATAATCTGGCACATCATATGTATCACAGAAAAA-3′. NheI and HindIII restriction endonucleases were used to create compatible ends. The DNA fragment was then inserted into pcDNA 3.1 (zeocine) vector (Invitrogen) to generate transfer plasmid pRB13-R2. All of the constructs were sequenced to ensure fidelity. J1.1 cells stably transfected with pRB 13-R2 by using Lipofectamine kit (GIBCO/BRL) were selected on the basis of their resistance to zeocine (Invitrogen). Zeocine-resistant clones were amplified and screened for IL13Rα2 expression by immunoblotting with anti-HA polyclonal antibody. Thus, parental and transformed cells solubilized in SDS, where each was electrophoretically separated in a denaturing gel (50 μg/lane), transferred to a nitrocellulose sheet, and probed with antibody against HA (Santa Cruz Biotechnology). The protein bands were visualized by an enhanced chemiluminescent detection (ECL) system (Pierce) according to the instructions of the manufacturer. One (J13R-2) was selected for testing the ability of R5111 to use the IL13Rα2 receptor.
Immunoblotting Electrophoretically Separated Proteins.
The indicated cells were mock-infected or exposed to 10 pfu of recombinant or wild-type HSV-1(F) per cell. The cells were harvested 24 h after infection, disrupted in SDS disruption buffer, boiled, cleared by centrifugation, and electrophoretically separated on a 10% denaturing polyacrylamide gel. After being transferred onto a nitrocellulose membrane, the isolated proteins were reacted with antibodies as indicated in results according to procedures described elsewhere (21).
Results
Verification of the Structure of R5111.
The construction of the R51s11 virus is depicted in Fig. 1 and detailed in Materials and Methods. The objective of the design was to replace the HveA binding site with the IL-13 ligand to enable the recombinant virus to bind IL13α2 receptor on cell surfaces and to delete the sequences reported to bind to heparan sulfate. The verification of the structure of R5111 was done as follows.
(i) The replacement of amino-terminal domain of gC with IL-13 and the disruption of the heparan sulfate binding site was initially verified by sequencing gC from recombinant R5107 (Fig. 2A).
(ii) The deletion of codons 68–77 of gB was verified by sequencing of the gB ORF amplified by PCR from recombinant R5108 (Fig. 2B).
(iii) The presence of chimeric IL-13–gD in R5111 was verified first by PCR illustrated in Fig. 3 and by sequencing of the entire ORF amplified by PCR in Fig. 2. The R5111 recombinant was initially isolated from transfected U87 cells and grown in parallel in Vero and HEP-2 cells, respectively. To determine whether the virus grown in Vero or HEp-2 cells differed with respect to amino acid sequence, we picked from Vero or HEp-2 cultures containing well separated plaques (six plaques each). In this series of verification experiments, we used two sets of primers to confirm the presence of IL-13 insert in gD and to verify the presence of a junction between IL-13 and gD. In a second round of verifications, the 12 clones of gD were sequenced to determine whether the isolates obtained from the viruses passaged in Vero or in HEp-2 cells differed in amino acid sequence. No differences were found. Furthermore, except for the inserted IL-13 sequence, no differences were found between the sequence of HSV-1(F) gD and those of the cloned IL-13–gD chimeric genes (Fig. 2C).
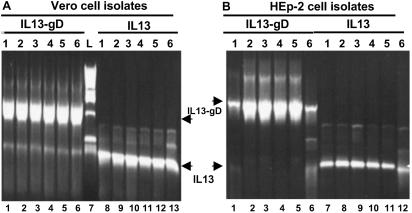
Fig 3.
Verification of R5111 viral DNA by PCR. Photographs of electrophoretically separated PCR products amplified directly from the plaques picked from Vero (A) and HEp-2 cells (B). Viral DNA was extracted as described in Materials and Methods and subjected to PCR with IL-13 primers from IL-13 ORF and IL-13–gD primers, which bracketed IL-13 and the gD ectodomain.
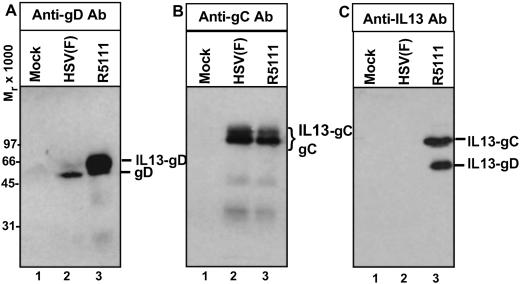
(iv) In denaturing polyacrylamide gels, IL-13 migrated as a protein with an apparent Mr of 15,000–17,000. In the recombinant R5111, IL-13 replaced 148 aa of gC. Fig. 4B shows an immunoblot of electrophoretically separated proteins from a lysate of R5111 mutant-infected cells reacted with an antibody to gC. As illustrated in that figure, the antibody reacted with bands present in lysates of HSV-1(F) and R5111 lysates with a similar electrophoretic mobility. The antibody to IL-13 reacted with a band of similar mobility in R5111 lysates (Fig. 4C, lane 3) but not in lysates of HSV-1(F) (Fig. 4C, lane 2). The IL-13–gD fusion protein in R5111 mutant virus was verified by reacting the cell lysates with gD and IL-13 antibody. Comparison of wild-type gD and the chimeric IL-13–gD chimeric protein (Fig. 4A, lane 3), showed that, as expected, IL-13–gD migrated more slowly than the wild-type gD (Fig. 4A, lane 2). The faster migrating band of gD did not react with the antibody to IL-13 (Fig. 4C, lane 2).
Fig 4.
Photograph of electrophoretically separated proteins from lysates of cells infected with R5111 reacted with antibody to gC, gD, or IL-13. HEp-2 grown in 25-cm2 flasks were exposed to 10 pfu of HSV-1 or R5111 per cell. The cells were harvested 24 h after infection, solubilized, subjected to electrophoresis in 10% denaturing polyacrylamide gels, electrically transferred onto a nitrocellulose sheet, and reacted with a monoclonal antibody against gD (A), gC (B), or IL-13 (C), respectively. The protein bands corresponding to the gC, IL-13–gC fusion protein, gD, and IL-13–gD fusion protein are indicated. IL-13–gC is expected to be the same size as the native gC.
Transfection and Selection of the J13R Cell Line.
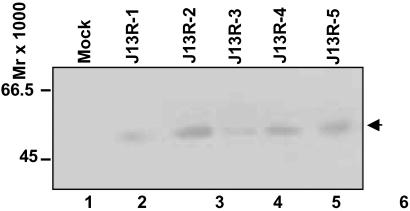
To test whether R5111 was able to use IL13Rα2 protein as a receptor for entry, it was necessary to construct a cell line expressing this protein in the absence of other HSV-1 entry receptors. The selection of the J1.1 cell line was published elsewhere (26). In essence, this cell line lacks the receptors necessary for the entry of virus into cells, and the cell line is not susceptible to infection by wild-type virus. The construction of a plasmid encoding an IL13Rα2 protein fused at its carboxyl terminus with an HA tag, transfection of J1.1 cells with the plasmid encoding the tagged IL13Rα2 protein, and selection of the cell line expressing the protein is detailed in Materials and Methods. To test for the production of IL13Rα2 protein, five clones of the selected cells were harvested, solubilized, subjected to electrophoresis in denaturing polyacrylamide gels, and tested for the expression of the protein. As shown in Fig. 5, all clones expressed a protein band reactive with the anti-HA antibody. The apparent size of the protein was consistent with the reported size of IL13Rα2. Of 5 J13R-positive clones, J13R-2 (Fig. 5, lane 3) was selected for further studies.
Fig 5.
HA-tagged IL13Rα2 expression from the individual clones of stable transfectants of the J1.1 cell line. The individual clones were amplified as detailed in Materials and Methods. The cells were harvested from 25-cm2 flasks, solubilized, and subjected to electrophoresis in 12% denaturing polyacrylamide gels, electrically transferred onto a nitrocellulose sheet, and reacted with a polyclonal antibody to HA tag.
Growth Properties of R5111 Recombinant Virus.
Replicate cultures of SK-N-SH, HEp-2, Vero, U87, J1.1, and J13R were exposed to 0.01 pfu of R5111 virus per cell. After 24 h of incubation, the cells were harvested and titered on Vero cells. The results shown in Table 1 were as follows: R5111 replicated to within a 10-fold range in HEp-2, Vero, U87, and J13R. The titer obtained from J1.1 cells was ≈105-fold lower than that obtained from all other cell lines. To test whether the J13R cell line acquired a receptor for HSV-1(F), J1.1 and J13R cells were also exposed to the wild-type virus. The results, also shown in Table 1, indicate that the cells remain resistant to the wild-type virus. Earlier studies have shown that HEp-2 cells express the nectin but not the HveA (ref. 26; G.Z. and B.R., unpublished studies). The results are consistent with the objectives of the experimental design to replace the HveA receptor with IL-13 insert to serve as a ligand for the IL13α2 protein. The results indicate that R5111 can use the IL13Rα2 as a receptor for entry in a cell line lacking all other HSV-1 receptors.
Table 1.
Replication of R5111 recombinant in various cell lines
| Virus | Cell line | Yield |
|---|---|---|
| R5111 | Vero | 11 × 107 |
| HEp-2 | 1.2 × 107 | |
| SK-N-SH† | 17 × 107 | |
| U87† | 27 × 107 | |
| J1.1 | 2 × 102 | |
| J13R | 11 × 107 | |
| HSV-1(F) | J1.1 | 6 × 103 |
| J13R | 4 × 103 |
The cells were exposed to 0.01 pfu of R5111 or HSV-1(F) per cell and harvested 24 h after infection. Progeny virus was titered on Vero cells.
† Cell lines derived from human brain tumors.
Discussion
In this report, we describe the construction and some properties of recombinant R5111. In R5111, we have ablated the heparan sulfate binding sites on the surface of viral particle to preclude or at least reduce the attachment of virus to nontargeted cells. Attachment even in the absence of fusogenic activity may lead to endocytosis, degradation of the virus particle, and, potentially, damage to the cell by lysosomal enzymes (16, 24). At the same time, we inserted into gC a copy of IL-13 to enhance binding of virus particles to the IL13Rα2 receptor. The major restructuring of the viral genome consisted of insertion of IL-13 at amino acid 24 of gD. Available data indicate that this step ablates the binding site to HveA receptor (25). Our data indicate that the virus retains the capacity to interact with the nectin receptor. The major finding, however, is that R5111 recombinant was able to infect and replicate in J13R cells but not in the parental, J1.1 cells. The significance of the results presented in this report stems from the following considerations.
(i) J1.1 cells lack both HveA and nectin (HveC) receptors. As noted in the Introduction, these cells are not susceptible to infection by wild-type virus (26). The observation that R5111 replicates in J13R cells to titers similar to those observed in SK-N-SH, U87, and other cell lines indicates the R5111 mutant can use the IL13Rα2 as a receptor. This is the first evidence that gD can be modified to use effectively another cellular protein as a receptor and that entry of HSV into cells can be totally dependent on receptors selected to target the virus to specific cells.
(ii) HveA has been reported to interact with domains located at both amino terminus and at a site closer to the carboxyl terminus of gD (25). Although mutations at the amino terminus ablated the interaction with HveA, this observation did not alleviate the possibility that to enable viral entry, HveA had to interact with other viral or cellular proteins, either one of the two interactive domains of gD. Our results indicate that the need for such interactions is unlikely. The data strongly suggest that interaction of gD with a cellular receptor is essential to bring the envelope in a specific juxtaposition to the plasma membrane and that once this is accomplished, the receptor has no major role in viral entry. Furthermore, in light of the evidence that HEp-2 and Vero cells lack IL-13 receptors (unpublished data), the results presented in this report indicate that the receptor-interacting domains of gD do not overlap and can be ablated independently. Work now in progress is to ablate the ability of gD to interact with nectin without affecting the fusogenic and antiapoptotic functions of gD.
(iii) HSV targeted to specific surface markers can be used to visualize the distribution of tumor cells in tissues. This was not possible as long as the virus attached to cells indiscriminately but replicated only in dividing cells. Ablation of indiscriminate binding to heparan sulfate and opportunity to use robust viruses capable of replicating in both dividing and nondividing cells make it possible to visualize the distribution of tumor cells by at least two methods. The first involves viral thymidine kinase-dependent incorporation of a radioactive precursor. A more attractive approach is to fuse noncritical tegument proteins (e.g., US11) present in nearly 2,000 copies per virus particle to GFP capable of being visualized in vivo.
The studies presented in this report make targeting of HSV for both oncolytic activity and visualization of tumor cells feasible.
Acknowledgments
We thank Dr. G. Campadelli-Fiume for the J1-1 cell line. The studies conducted at the University of Chicago were aided by National Cancer Institute Grants CA78766, CA71933, CA83939, CA87661, and CA88860.
Abbreviations
pfu, plaque-forming units
HSV, herpes simplex virus
HA, hemagglutinin
References
- 1.Davis F. G., Freels, S., Grutsch, J., Barlas, S. & Brem, S. (1998) J. Neurosurg. 88, 1-10. [DOI] [PubMed] [Google Scholar]
- 2.Pyles R. B., Warnick, R. E., Chalk, C. L., Szanti, B. E. & Parysek, L. M. (1997) Hum. Gene Ther. 8, 533-544. [DOI] [PubMed] [Google Scholar]
- 3.Rampling R., Cruickshank, G., Papanastassiou, V., Nicoll, J., Hadley, D., Brennan, D., Petty, R., MacLean, A., Harland, J., McKie, E., et al. (2000) Gene Ther. 7, 859-866. [DOI] [PubMed] [Google Scholar]
- 4.McKie E. A., Brown, S. M., MacLean, A. R. & Graham, D. I. (1998) Neuropathol. Appl. Neurobiol. 24, 367-372. [DOI] [PubMed] [Google Scholar]
- 5.Markert J. M., Medlock, M. D., Rabkin, S. D., Gillespie, G. Y., Todo, T., Hunter, W. D., Palmer, C. A., Feigenbaum, F., Tornatore, C., Tufaro, F. & Martuza, R. L. (2000) Gene Ther. 7, 867-874. [DOI] [PubMed] [Google Scholar]
- 6.Mineta T., Rabkin, S. D., Yazaki, T., Hunter, W. D. & Martuza, R. L. (1995) Nat. Med. 1, 938-943. [DOI] [PubMed] [Google Scholar]
- 7.Simard C., Langlois, I., Styger, D., Vogt, B., Vlcek, C., Chalifour, A., Trudel, M. & Schwyer, M. (1995) Virology 212, 734-740. [DOI] [PubMed] [Google Scholar]
- 8.Chou J., Chen, J. J., Gross, M. & Roizman, B. (1995) Proc. Natl. Acad. Sci. USA 92, 10516-10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He B., Chou, J., Brandimarti, R., Mohr, I., Gluzman, Y. & Roizman, B. (1997) J. Virol. 71, 6049-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cassady K. A., Gross, M. & Roizman, B. (1998) J. Virol. 72, 7005-7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leib D. A., Harrison, T. E., Laslo, K. M., Machalek, M. A., Moorman, N. J. & Virgin, H. A. W. (1999) J. Exp. Med. 189, 663-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laquerre S., Argnani, R., Anderson, D. B., Zucchini, S., Manservigi, R. & Glorioso, J. C. (1998) J. Virol. 72, 6119-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spear P. G., Eisenberg, R. J. & Cohen, G. H. (2000) Virology 275, 1-9. [DOI] [PubMed] [Google Scholar]
- 14.Montgomery R. I., Warner, M. S., Lum, B. J. & Spear, P. G. (1996) Cell 87, 427-436. [DOI] [PubMed] [Google Scholar]
- 15.Campadelli-Fiume G., Cocchi, F., Menotti, L. & Lopez, M. (2000) Rev. Med. Virol. 10, 305-319. [DOI] [PubMed] [Google Scholar]
- 16.Zhou G. & Roizman, B. (2002) J. Virol. 76, 6197-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Debinski W., Gibo, D. M., Hulet, S. W., Connor, J. R. & Gillespie, G. Y. (1999) Cancer Res. 5, 985-990. [PubMed] [Google Scholar]
- 18.Mintz A., Gibo, D. M., Slagle-Webb, B., Christensen, N. D. & Debinski, W. (2002) Neoplasia 4, 388-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debinski W. (1998) Crit. Rev. Oncog. 9, 255-268. [PubMed] [Google Scholar]
- 20.Debinski W. & Gibo, D. M. (2000) Mol. Med. 6, 440-449. [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou G. & Roizman, B. (2001) J. Virol. 75, 6166-6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arsenakis M., Tomasi, L. F., Speziali, V., Roizman, B. & Campadelli-Fiume, G. (1986) J. Virol. 58, 367-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye G. J. & Roizman, B. (2000) Proc. Natl. Acad. Sci. USA 97, 11002-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou G., Galvan, V., Campadelli-Fiume, G. & Roizman, B. (2000) J. Virol. 74, 11782-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carfi A., Willis, S. H., Whitbeck, J. C., Krummenacher, C., Cohen, G. H., Eisenberg, R. J. & Wiley, D. C. (2001) Mol. Cell 8, 169-179. [DOI] [PubMed] [Google Scholar]
- 26.Cocchi F., Menotti, L., Mirandola, P. & Campadelli-Fiume, G. (1998) J. Virol. 72, 9992-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Debinski W. & Thompson, J. P. (1999) Clin. Cancer Res. 5, Suppl. 10, 3143-3147. [PubMed] [Google Scholar]