Abstract
Spumaviruses, commonly called foamy viruses (FV), are complex retroviruses that establish lifelong persistent infections without any accompanying pathologies. In tissue culture, cells can be either lytically or latently infected, depending on cell type. Regulation of FV replication is controlled by two promoters: the LTR and a second promoter within the env gene termed the internal promoter (IP). The IP directs expression of the transcriptional activator, Tas, and a second accessory protein, Bet, whose function has been elusive. In this study, we report that expression of exogenous Tas is sufficient to initiate a switch from latent to lytic replication. We also show that treatment with the phorbol ester phorbol 12-myristate 13-acetate (PMA) can lead to an increase in transcription from the IP, and that Bet protein expression abrogates this effect. Finally, we demonstrate that Bet expression severely limits the ability of PMA to activate transcription of latent FV genomes, and that replication of a Bet(-) virus is more easily activated than wild-type FV. Taken together, these data suggest that viral transcription is regulated by a sensitive switch, and that Bet functions as a negative regulator of basal IP activity.
Regulation of retroviral transcription is a complex process that varies widely depending on virus type. Simple retroviruses lack accessory proteins that can modulate transcription. Transcription levels are generally dictated by target cell permissivity and integration site (1). Transcription of complex retroviruses, including the lentiviruses such as HIV, human T cell leukemia virus (HTLV), and the spumaviruses, is also regulated by cell permissivity and integration site. However, the presence of accessory genes in this class of retroviruses provides an additional level of transcriptional regulation. Accessory genes for both HIV type 1 and HTLV type 1 generally promote virus replication. HIV Tat and HTLV Tax are both transcriptional transactivators of their cognate LTRs (2–4). HIV Rev and HTLV Rex both function posttranscriptionally in regulating export of singly spliced and unspliced RNA species (5, 6). None of the accessory proteins from either the lentiviruses or the deltaretroviruses negatively regulate viral replication although, by virtue of exporting unspliced RNA, HTLV Rev indirectly suppresses expression of accessory genes that are encoded on multiply spliced messages (7).
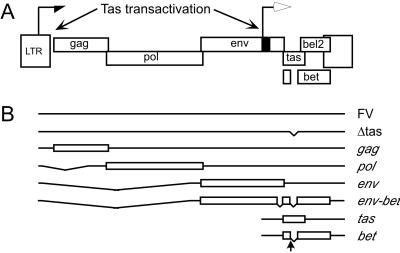
Members of the Spumavirus genus, commonly called foamy viruses (FV), have two promoters: the LTR promoter found in all retroviruses, which directs transcription of gag, pol, and env, while a second promoter unique to FVs, the internal promoter (IP), directs expression of the accessory genes tas and bet (Fig. 1). Like other complex retroviruses, FVs encode a transcriptional transactivator, termed Tas; however, unlike Tat and Tax, FV Tas binds directly to and transactivates both the FV promoters (8–10). Unique among the retroviruses, Bet expression negatively regulates FV replication in latently and persistently infected cells although the mechanism of action is not understood. In contrast to the situation in latently infected cells, nonsense mutations in bet result in a 5- to 10-fold decrease in replication of the prototype FV (PFV), previously called pHFV13 (11), on cell types that support lytic replication (12). In the case of feline FV, Bet is required for efficient virus replication in cells of feline origin (13). These data indicate multiple, complex roles for Bet in FV replication. Using reporter assays, attempts to show that Bet can directly inhibit Tas transactivation of either the LTR or IP have been unsuccessful (ref. 14 and unpublished data). This finding indicates that Bet cannot block transactivation once Tas is synthesized, and that the inhibitory effects of Bet on FV replication are unlikely to be posttranscriptional. Overexpression of FV Bet in target cells can restrict infection by incoming FV through an undefined mechanism (15). Bet is also secreted from infected cells and taken up by uninfected recipient cells (16); however, the function of secreted Bet is unknown. In infected animals, the presence of a tas-deleted, defective, interfering form of FV, termed Δtas, accumulates over time (17). Δtas can express only Bet and does so at elevated levels in the presence of Tas from WT FV (18, 19). Δtas is the predominant proviral form found in latently infected cells that recover from lytic infection. The presence of Δtas proviruses also limits the cytolytic effects of WT FV (19).
Fig 1.
Genome organization of FV. (A) Schematic of the FV genome. LTR promoter (half arrow); IP (black box and open arrow). (B) Schematic of major RNA species expressed. Exons are denoted by open boxes. Location of bet splice donor site is denoted with vertical arrow.
Natural, experimental, and accidental infection with FVs leads to a lifelong persistent infection (20). Most tissues within the host are latently infected, with viral replication detected only in the oral mucosa (21). To better understand FV latency, we have investigated the role of FV Bet in the switch from latent to lytic replication by using an in vitro model of FV latency. In this study, we demonstrate that one function of Bet is to limit expression of the transcriptional transactivator Tas by inhibiting activation of the IP.
Materials and Methods
Cells and Viruses.
Virus titers were determined by using the FAB indicator cell line (12). 293T (ATCC 293tsA1609neo), HT1080 (ATCC CCL-121), and FAB cells were grown in DMEM containing 10% FBS, and antibiotics. Jurkat cells (ATCC TIB-152) were grown in RPMI 1640 medium supplemented with 10% FBS. Jurkat cells were infected by coculture as previously described (22), to produce JurFV cells.
Plasmids.
All FV constructs used in this study were derived from the clone pHFV13 (11), referred to in this document as FV. The vector LTasSD was generated by cloning the region from nucleotide 9201 to nucleotide 10441 encompassing the tas ORF into BclI/HindIII-digested LXSN. The Bet splice donor at nucleotide 9700 was mutated by using oligo-directed mutagenesis (Stratagene). FVBet(-) was generated by first mutating the bet splice donor site in pSub5 (23) by site-directed mutagenesis, then subcloning into the BlpI/SalI-digested pHFV13. The cytomegalovirus (CMV)-driven Bet expression construct C-Bet was generated by PCR cloning the Bet ORF into XbaI-digested pCR3.0 (Invitrogen). The retroviral vector LNCZ, which contains the LacZ gene under control of the CMV immediate early promoter, was obtained from D. Miller (Fred Hutchinson Cancer Research Center, Seattle).
Western Blot.
Western blotting was performed essentially as described (23). Gag was detected by using rabbit anti-Gag serum at 1:2,000 dilution. Tas and Bet were detected by using rabbit anti-Bel1 antiserum (11) at 1:2,000 dilution.
Retroviral Transduction.
Murine leukemia virus (MLV) vectors were pseudotyped with the vesicular stomatitis virus glycoprotein (VSV-G) and concentrated by ultracentrifugation essentially as described (24). Briefly, 3 × 106 293T cells plated the day before were transfected by using 63 μl of Fugene-6 (Roche) and 11 μg of LTasSD or LNCZ, 1 μg of Lgfp, 6 μg of VSV-G, 6 μg of JK3, and 1.5 μg of CMV-tat. After 48 h, filtered supernatants were concentrated by ultracentrifugation. Jurkat or JurFV cells (1 × 106) were transduced in the presence of 2 μg/ml polybrene (Sigma).
Luciferase Reporter Assays.
Luciferase assays were performed as described (23). Briefly, in 24-well plates, 8 × 105 293T or 4 × 105 HT1080 cells per well were cotransfected with 0.31 μg of DNA per well. Transfections were done in triplicate, by using Fugene-6 with 0.1 μg of IPluc, LTRluc, or promoterless pGL3 plasmid, 0.1 μg of effector plasmid (LTasSD or C-Bet), 0.1–0.2 μg of LN filler plasmid, and 0.01 μg of LNCZ, which expresses β-galactosidase (β-gal) from the CMV immediate early promoter, as a cotransfection control. After 48 h, cells were lysed in 250 μl of 1× reporter lysis buffer (Promega), and 20 μl of cleared lysate were assayed by using the luciferase activity system (Promega) and measured by using a 96-well luminometer (Berthold, Nashua, NH). All luciferase values were normalized to cotransfected β-gal.
FV Transfection of 293T Cells.
293T cells (1.1 × 105 cells per well), plated the previous day in 12-well dishes, were transfected by using 5.0 μl of Fugene-6 and 2.05 μg of DNA. Each well was transfected with 0.4 μg of FV and 0.05 μg of the β-gal expression construct LNCZ. The remaining 1.6 μg of DNA contained increasing amounts of C-Bet and LN filler plasmid. Phorbol 12-myristate 13-acetate (PMA; 50 ng/ml) was added at 6–8 h post transfection where appropriate. Duplicate plates were used to determine titers and Western blot analysis.
Results
Expression of Exogenous Tas Reactivates Latent FV in Jurkat Cells.
Infection of cells in vitro by FV can result in lytic replication and cell death, persistent infection, or latent infection (Table 1). We have previously shown that treatment of JurFV cells with PMA resulted in lytic replication (23), suggesting that cellular activation may help overcome any putative inhibitory factors in latently infected cells. Mitogens such as phytohemagglutinin also result in reactivation of latent FV, but to a lesser extent than PMA (unpublished data).
Table 1.
Replication of FV in vitro
| Cell name | Infection | Titer | PMA |
|---|---|---|---|
| BHK-21 | Lytic | 1 × 106 –2 × 106 | 1.0–1.3 |
| HT1080 | Lytic | 1 × 106 –2 × 106 | 0.8–1.0 |
| Jurkat | Latent | 1 × 101 –9 × 101 | 200–2,000 |
| 293T | Latent | 1 × 101 –6 × 101 | 50–2,500 |
Classification of cells infected with FV. Lytic infection characterized by syncytium formation, cytoplasmic vacuolization, and cell death. Latent infection is characterized by lack of any notable cytopathic effects or impairment of normal cell growth and low titers.
Titers were determined as described in Materials and Methods, and are given for cells without PMA treatment.
Fold change in virus titer following PMA treatment.
293T cells were transfected and titer was determined.
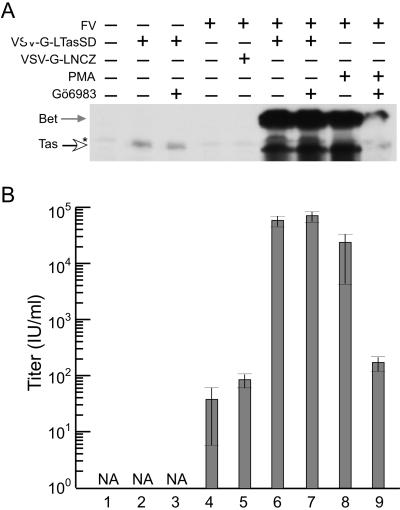
Because there is a significant delay in virus production after PMA treatment (23), we hypothesized that the addition of PMA indirectly results in increased levels of FV Tas, which then becomes available for transactivation of the two FV promoters (Fig. 1A). To test whether additional Tas could reactivate FV replication in latently infected JurFV cells, we used an overexpression strategy using VSV-G pseudotyped MLV particles. MLV virions packaging the MLV LTR-driven Tas expression vector, LTasSD, were pseudotyped with VSV-G envelope and used to transduce uninfected Jurkat cells and JurFV cells. The vector LTasSD contains the tas ORF, with a mutation at the Bet splice donor site (Fig. 1B), so that Bet is not synthesized. This mutated form of Tas (TasSD) is expressed via the MLV LTR promoter. All virus stocks contain ≈10% Lgfp vector that expresses GFP from the MLV LTR. Transduction efficiencies measured by GFP fluorescence were similar for all virus stocks (data not shown). Tas expression in uninfected Jurkat cells is barely detectable by Western blot and is not affected by treatment with the PKC inhibitor Gö6983 (Fig. 2, lanes 2 and 3). When the same amount of virus was used to transduce JurFV cells, virus replication was induced to levels similar to those observed by PMA treatment (Fig. 2, lanes 6 and 8). The control virus carrying the LNCZ vector had no effect on FV replication (Fig. 2, lane 5). Treatment with Gö6983 had no effect after transduction with VSV-G-LTasSD virus, indicating that Tas does not act through PKC (Fig. 2, lane 7). As expected, Gö6983 drastically reduced virus expression and titers after PMA treatment (Fig. 2, lanes 8 and 9). These results indicate that Jurkat cells are not inherently deficient for FV replication and that expression of a small amount of additional Tas is sufficient to initiate a switch from latent to lytic replication.
Fig 2.
Tas expression induces a switch from latent to lytic replication. (A) Western blot showing the level of Tas expression (open arrow) and Bet expression (gray arrow) 48 h after the treatments indicated above the blot. (B) FV titers as assayed on FAB cells 48 h after treatments indicated in A. Lane 1, untransduced, uninfected Jurkat cells. Lane 2, VSV-G-LTasSD-transduced, uninfected cells. Lane 3, same as lane 2, treated with PKC inhibitor Gö6983. Lane 4, FV-infected Jurkat. Lane 5, FV-infected, transduced with VSV-G-LNCZ. Lane 6, FV-infected, LTasSD-transduced. Lane 7, same as lane 6, treated with Gö6983. Lane 8, FV-infected, PMA-treated. Lane 9, same as lane 8, treated with Gö6983. NA, not applicable. *, Nonspecific background.
FV Reactivation from Latency Is More Rapid After Tas Addition than PMA Treatment.
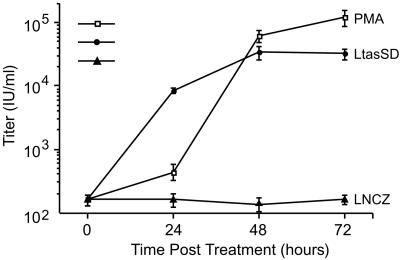
To better understand the mechanism of reactivation from latency in JurFV cells, we compared the kinetics of virus production after PMA treatment and transduction of JurFV cells with VSV-G-LTasSD virus (Fig. 3). At 24-h intervals after either addition of PMA or VSV-G-LTasSD virus, cells and supernatants were harvested and assayed for infectious FV by FAB assay. Significant FV titers were not observed until 48 h after PMA treatment. In contrast, despite the need for virus entry, integration, transcription, and translation of Tas, a large increase in the levels of infectious FV was observed 24 h after addition of VSV-G-LTasSD virus. A similar experiment with more frequent time points showed a profile similar to Fig. 3. In this experiment, a dramatic increase in infectious virus was observed at 36 h for LtasSD and 60 h for PMA. Because protein expression from retroviral vectors usually takes from 6 to 12 h to reach detectable levels, the appearance of replication competent FV by 24–36 h posttransduction indicates that Tas may act directly on the IP. In contrast, PMA is likely to act indirectly by increasing the levels of some putative cellular factor(s) necessary for efficient Tas expression from integrated FV genomes. This model is supported by data showing that a 3-h exposure to the protein synthesis inhibitor emetine, at any point during or after addition of PMA, dramatically reduced IP gene expression (data not shown).
Fig 3.
Kinetics of FV induction after PMA treatment or VSV-G-LTasSD transduction of JurFV cells. □, Titers at indicated times after PMA treatment. •, Titers at indicated times after VSV-G-LTasSD transduction. ▴, Titers at indicated times after transduction with LNCZ control vector. Time 0 reflects the addition of VSV-G pseudotyped virus, or PMA, to cultures.
Mechanism of Reactivation from Latency by PMA.
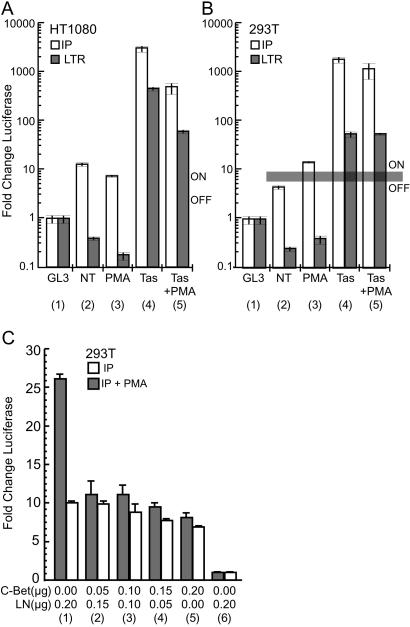
In JurFV cells, we previously noted a small, but reproducible, 2.5-fold increase in IP activity after addition of PMA (23). This finding suggests that the profound effect on virus replication in JurFV cells after PMA treatment may be due to only a small increase in the basal activity of the IP. To further address the effect of PMA on the individual FV promoters in cells that support latent or lytic infection, luciferase reporter assays were performed in 293T (latent) and HT1080 (lytic) cells, which are readily transfected. Cells were cotransfected with LTRluc or IPluc constructs and either a Tas expression construct, LTasSD, or a control expression construct. Triplicate transfections were then treated with PMA, and all cells were assayed for luciferase activity (Fig. 4). Expression of cotransfected CMV-β-gal was used to normalize all luciferase values.
Fig 4.
Promoter activity in 293T and HT1080 cells. Fold change in luciferase activity relative to promoterless control vector GL3. All luciferase values were normalized to cotransfected β-gal. (A and B) Open bars, IP; filled bars, LTR. Column 1, activity of promoterless control vector GL3; column 2, basal activity of promoters; column 3, basal activity of promoters in the presence of PMA; column 4, activity of promoters in the presence of cotransfected Tas; column 5, activity of promoters in the presence of cotransfected Tas and PMA treatment. The horizontal gray line indicates the putative threshold of IP activity required for virus expression in nonpermissive cells. (A) HT1080 cells. (B) 293T cells. NT, no treatment. (C) Measurement of IP promoter activity in the presence of increasing amounts of Bet-expressing plasmid in 293T cells. Open bars, no PMA added. Filled bars, 50 ng/ml PMA added 6 h post transfection. All values normalized to cotransfected β-gal. Columns 1–6, 0.25 μg total DNA transfected. Columns 1–5, 0.05 μg IPluc transfected. Column 6, 0.05 μg control, promoterless GL3 vector transfected. Columns 2–5, increasing amounts of C-Bet.
In the permissive HT1080 cell line, the basal activity of the LTR was significantly lower than the promoterless control (GL3), and PMA treatment further repressed LTR transcription (Fig. 4A, columns 1–3). As expected, addition of Tas, or Tas and PMA, resulted in a large increase in LTR activity (Fig. 4A, columns 5 and 6). Significant basal IP activity and the ability of Tas to transactivate both promoters are expected and are observed; the basal activity of the IP was ≈13-fold above the promoterless control vector and was slightly reduced by the addition of PMA (Fig. 4A, columns 1–3). Addition of Tas, or Tas and PMA, resulted in a dramatic increase in IP activity (Fig. 4A, columns 4 and 5). We have previously reported similar levels of basal IP activity and IP transactivation in the fully permissive BHK-21 cell line (23).
In contrast, in 293T cells, which are not permissive for FV replication, low IP activity or a block to Tas transactivation of the IP would be expected. We found that the basal activity of the LTR was below the promoterless control in 293T cells (Fig. 4B, columns 1 and 2), and PMA treatment had no effect on the basal activity of the LTR (Fig. 4B, column 3). Cotransfection of Tas resulted in increased LTR activity in the absence or presence of PMA (Fig. 4B, columns 4 and 5). The basal activity of the IP was 4- to 5-fold above the promoterless control (Fig. 4B, columns 1 and 2). Similar to the results observed in Jurkat cells (23), PMA treatment enhanced the basal IP activity ≈3-fold to a level about 14 times above GL3 (Fig. 4B, columns 1–3). Cotransfection of Tas resulted in a dramatic increase in IP activity in the presence or absence of PMA (Fig. 4B, columns 4 and 5). The ability of Tas to fully transactivate both promoters indicates that all of the components required for transactivation are present in 293T cells. These data suggest that the lack of sufficient Tas expression is the primary defect limiting FV replication in nonpermissive cells.
Two significant differences in promoter activity were observed that could account for the differences in permissivity between HT1080 and 293T cells. First, compared with the promoterless control vector, the basal activity of the IP was slightly higher in HT1080 cells than 293T cells. Second, PMA treatment stimulated the basal IP activity in 293T cells. Although it is difficult to make direct comparisons of promoter activity between different cell types, the basal IP activity in HT1080 cells and the IP activity in 293T cells after PMA treatment are sufficient to permit FV replication. Despite some basal IP activity, the absence of FV replication in 293T cells indicates that this level of transcription is insufficient to initiate a positive feedback loop at the IP.
The simplest explanation for the lack of FV replication in 293T despite significant basal IP activity is the presence of a saturable Tas inhibitor. If the putative inhibitor is in excess over Tas, no transcription from the LTR can be achieved. The level of Tas expression needed to overcome the inhibitory effects of such an inhibitor is represented by a horizontal gray line in Fig. 4. The only known inhibitor of FV Tas is the promyelocytic leukemia protein (PML; ref. 25); however, PML does not appear to play a role in FV latency (unpublished data). In HT1080 cells, the basal activity of the IP results in expression of sufficient Tas to overcome any Tas inhibitor present, which in turn permits initialization of the positive feedback loop. In contrast, the basal IP activity in 293T cells is insufficient to overcome any inhibitory effects, and the positive feedback loop cannot commence. However, in the presence of PMA, the basal activity of the IP is able to overcome the inhibitory effects, and the positive feedback loop is initialized.
Bet Expression Prevents the Switch to Lytic Replication.
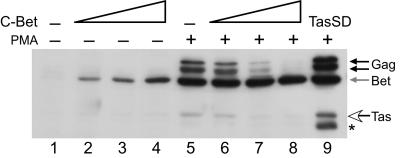
We next asked whether treatment of FV-transfected cells with PMA could stimulate virus expression in a manner similar to that observed in latently infected JurFV cells. Because Jurkat cells are difficult to transfect, we used readily transfectable 293T cells. We had previously determined that 293T cells transfected with WT FV vector and treated with 50 ng/ml PMA behaved the same as JurFV cells, showing increased titers (Table 1) and significant increases in Gag and Bet protein synthesis (Fig. 5, lanes 1 and 5). Cotransfection of the control vector LN had no effect on the ability of PMA to stimulate FV expression (Fig. 5, lane 1). Surprisingly, cotransfection of C-Bet resulted in a dramatic, dose-dependent decrease in FV protein expression after PMA treatment (Fig. 5, lanes 6–8). In lanes 6–8, the level expression of Bet remains relatively constant because, as pC- Bet expression increases, there is a concomitant decrease in Bet expression from the IP. Transfection of cells with LTasSD and treatment with PMA showed a synergistic effect on protein expression (Fig. 5, lane 9). These data indicate that Bet expression potently inhibits the PMA-induced switch from latent to lytic replication.
Fig 5.
Inhibition of FV activation in PMA-treated 293T cells by Bet. Western blot analysis of FV proteins in cotransfected 293T cells. See Materials and Methods for amounts of each vector used. Lane 1, LNCZ control vector. Lanes 2–4, increasing amounts of C-Bet vector. Lane 5, LNCZ control vector plus PMA. Lanes 6–8, increasing amounts of C-Bet vector plus PMA treatment. Lane 9, LTasSD vector plus PMA. Gag protein, black arrows. Bet protein, gray arrows. Tas protein, open arrows. *, Breakdown products observed only with Gag antiserum.
Both bet and tas are transcribed from overlapping reading frames (Fig. 1); therefore, the production of one mRNA precludes expression of the other. Because infected cells produce large amounts of Bet and minimal amounts of Tas, we wished to determine whether a virus that can transcribe only tas mRNA is able to replicate to higher levels in 293T cells, is more readily activated by PMA, or both, compared with WT FV. To address these possibilities, we cloned tas from LTasSD into the WT FV vector, resulting in FVBet(-). Similar to WT FV, 293T cells transfected with FVBet(-) vector did not express any detectable levels of FV proteins in the absence of PMA, and cotransfection of C-Bet before PMA treatment resulted in a dramatic, dose dependent decrease in FV protein expression (data not shown). These data indicate that bet is not required for the establishment of latency and suggest that Bet functions to restrict de novo synthesis of Tas.
Bet Inhibits the Effects of PMA on IP Transcription.
Fig. 5 demonstrated that cotransfection of Bet limits the ability of PMA to induce virus replication in 293T cells. Next, we asked whether Bet acts at the level of IP transcription. 293T cells were cotransfected with 0.5 μg IPluc and increasing amounts of C-Bet, and treated with PMA; then, luciferase values were measured. All luciferase values were normalized to cotransfected LNCZ. The values presented in Fig. 4C are the fold change in luciferase values compared with promoterless control pGL3 after normalization to cotransfected β-gal. In the absence of Bet, treatment with PMA resulted in a 2.6-fold increase in basal IP activity (Fig. 4C, column 1). In the absence of PMA, increasing amounts of cotransfected C-Bet slightly diminished the basal activity of the IP (Fig. 4C, columns 2–5). In contrast, the 2.6-fold increase in IP activity observed in the presence of PMA is dramatically reduced by cotransfection of increasing amounts of C-Bet (Fig. 4C, columns 2–5). These data indicate that Bet has only minimal effects on the basal activity of the IP, but has profound effects on the up-regulation of the IP after PMA treatment.
Activation of FV and FVBet(-) Virus After PMA Treatment.
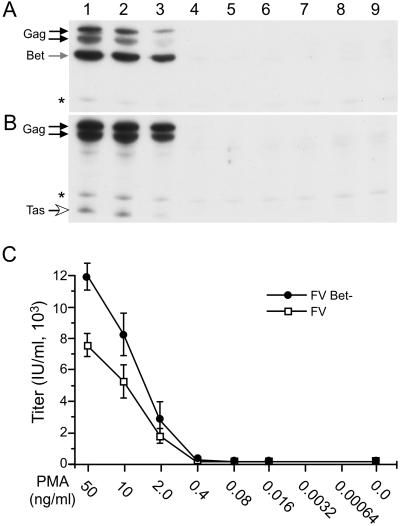
As evidenced by transfection of 293T cells with FVBet(-), a lack of Bet expression has no effect on FV replication in the absence of PMA stimulation in nonpermissive cells (data not shown). We therefore examined whether a virus lacking Bet showed differences in reactivation after PMA treatment. FV and FVBet(-) vectors were transfected into 293T cells, which were treated with serial dilutions of PMA. After normalization to cotransfected β-gal, virus replication was monitored by Western blot and infectious titer (Fig. 6). FV protein expression was observed at PMA concentrations of 50, 10, and 2 ng/ml, but was not detectable at levels of 0.4 ng/ml or lower (Fig. 6A). FVBet(-) expression was also observed only at 50, 10, and 2 ng/ml; however, Gag expression was higher when compared with WT FV at each concentration (Fig. 6B). When normalized for transfection efficiency, at 50, 10, and 2 ng/ml PMA, FVBet(-) titers were significantly higher than those observed with wt FV (Fig. 6C). These data indicate that the ability to make Bet reduces virus levels after activation with PMA.
Fig 6.
PMA Activation of FV and FVBet(-) in 293T. Black arrows, Gag. Gray arrow, Bet. Open arrow, Tas. (A) FV protein expression in 293T cells transfected with FV vector and treated with amounts of PMA indicated below. (B) Same as A, except FVBet(-) vector transfected. (C) Titers from duplicate transfections as in A and B for FV (□) and FVBet(-) (•). *, Nonspecific band.
Discussion
FV latency involves limiting production of the transactivating protein, Tas, while promoting expression of a putative negative regulator, Bet. Because both Tas and Bet are produced from overlapping reading frames, the production of bet messenger RNA precludes tas expression (11, 26). Infection of many cell types in vitro results in production of large amounts of Bet protein, but only limited amounts of Tas protein. Thus, even under optimal conditions for replication, FVs severely limit production of their positive regulator and favor production of the putative negative regulator, Bet.
In the current study, we examined reactivation of FV from latently infected cells. PMA, which activates the PKC pathway, greatly enhanced virus replication. Expression of exogenous Tas protein also stimulates reactivation from latency and does so with rapid kinetics, consistent with Tas acting directly on the IP. Conversely, overexpression of Bet protein significantly limits reactivation from latency. PMA activation increases the basal activity of the IP in latently infected cells, but not in cells that support lytic infection. The ability of Bet to inhibit reactivation occurs by abrogating the increase in IP activity observed in the presence of PMA. Thus, the initiation of latency does not require Bet; however, Bet expression does inhibit virus replication after activation with PMA.
The simplest explanation for the differences in Tas and Bet expression is mRNA export. Fig. 1 shows that all bet transcripts are spliced and thus likely to be efficiently exported to the cytoplasm. In contrast, tas messages do not contain any introns, but do contain a splice donor/acceptor pair, used to generate env-bet mRNA (16, 27), in the 5′ untranslated region. The importance of this env-bet intron in tas expression is not known. Another possibility is that, in addition to down-regulating transcription from the IP, Bet alters tas at the posttranscriptional level. Although there is evidence that FV does not require a posttranscriptional regulator for expression of FV Gag (28), there is no information as to whether Bet regulates expression of tas posttranscriptionally. One posttranscriptional mechanism we have ruled out is the destabilization of Tas by Bet. Pulse–chase analysis of PMA-treated JurFV cells indicates half-lifes of 8.5 and 8.2 h for Tas and Bet, respectively (data not shown). A nearly identical half-life was observed for Tas in the absence of Bet (data not shown). Perhaps, Bet expression alters levels of tas mRNA through mechanisms such as preferentially splicing IP transcripts into the bet form of the message, altering the subcellular localization of tas mRNA, or decreasing the stability of tas mRNA.
The regulation of FV transcription is a complex balance of positive and negative regulatory elements. Reactivation from latency tilts the balance in favor of the positive regulators. Because FV transcription involves the presence of a potent positive feedback loop that generates the transcriptional activator Tas, FV replication can be viewed as an on/off switch (Fig. 4, horizontal gray line). In a given cell line, if sufficient Tas is produced to overcome any inhibitory effects, the switch is turned on; if insufficient Tas is produced, the switch is off. The factors that regulate this switch are not well defined, but some predictions can be made based on cell permissivity. In cells that support latent infection, the basal activity of the IP does not generate sufficient levels of Tas to turn the switch on. However, treatment with PMA or mitogens increases basal transcription from the IP, and sufficient Tas is produced to turn the switch on. Bet expression helps attenuate any positive signals that might elevate Tas levels past the critical threshold required for lytic replication. In contrast, in fully permissive cells the basal activity of the IP is always near or above the level necessary to turn the switch on. The ability of Bet to prevent up-regulation of basal IP activity, but not to prevent transactivation of either promoter by Tas, helps explain how Bet overexpression prevents de novo FV infection of permissive cells, as has been demonstrated by others (15, 19). In both nonpermissive (Fig. 4C), and permissive HT1080 cells (data not shown), Bet overexpression results in a modest decrease in basal IP activity. In permissive cells, Bet overexpression after infection would have minimal effects on virus replication because Bet cannot block Tas transactivation of the IP. Furthermore, in permissive cells, large quantities of Bet are normally produced, indicating that Bet does not inhibit replication after Tas is synthesized. In contrast, overexpression of Bet before infection would dampen basal IP activity and Tas production, preventing transactivation of the IP and virus replication.
The precise mechanism by which phorbol ester activates the FV IP in vitro may provide insight into the mechanism of FV reactivation in vivo. FV infection in vivo is characterized by the presence of viral DNA in most tissues (17, 21), in the absence of detectable levels of viral RNA or protein expression (17, 21, 29, 30). However, reactivation of latent FV frequently occurs on coculture of infected tissues, peripheral blood, or throat swabbings with susceptible cell lines (17, 29–33). The contribution of the immune response to maintenance of FV latency is unknown; however, infected animals generate robust antibody responses primarily against Bet and Gag.
The signals that mediate reactivation of FV in vivo are not understood. In JurFV cells, we have observed increases in FV expression after crosslinking of the T cell receptor (TCR) with anti-CD3 and anti-CD28 antibodies (data not shown). Perhaps productive engagement of the TCR on latently infected T cells provides small bursts of infectious virus. The presence of ΔTas proviruses and uptake of secreted Bet by neighboring cells could limit virus spread. Alternatively, reactivation of latent FV in vivo could be stochastic. Within a given tissue, transgenic mice containing the FV IP region express high levels of FV RNA in some cells, but none in neighboring cells (34). Such a finding is unusual for a transgene under control of a constitutive promoter, but fits nicely with a self-regulated switch like the FV IP. One might expect slight variations in expression of the positive and negative regulators of the FV IP. Thus, within a single tissue, a fraction of the cells might a express the right combination of factors, permitting the switch to lytic replication as Tas levels exceed the threshold and transactivate the IP.
Acknowledgments
We are grateful to Denise Galloway for critical reading of the manuscript. We thank Dr. Martin Lochelt (German Cancer Research Center, Heidelberg) for the Bel1 antiserum and Dusty Miller (Fred Hutchinson Cancer Research Center) for LNCZ. This work was supported by National Cancer Institute Grant CA81297 (to M.L.L.). C.D.M. was funded by National Cancer Institute Training Grant T32 CA80416 and by a Helen Riaboff Whiteley fellowship.
Abbreviations
FV, foamy virus
IP, internal promoter
PMA, phorbol 12-myristate 13-acetate
JurFV cells, FV-infected Jurkat cells
β-gal, β-galactosidase
MLV, murine leukemia virus
VSV-G, vesicular stomatitis virus glycoprotein
CMV, cytomegalovirus
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Goff S. P. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott-Raven, Philadelphia), Vol. 2, pp. 1871–2140. [Google Scholar]
- 2.Shimotohno K., Golde, D. W., Miwa, M., Sugimura, T. & Chen, I. S. (1984) Proc. Natl. Acad. Sci. USA 81, 1079-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sodroski J., Trus, M., Perkins, D., Patarca, R., Wong-Staal, F., Gelmann, E., Gallo, R. & Haseltine, W. A. (1984) Proc. Natl. Acad. Sci. USA 81, 4617-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felber B. K., Paskalis, H., Kleinman-Ewing, C., Wong-Staal, F. & Pavlakis, G. N. (1985) Science 229, 675-679. [DOI] [PubMed] [Google Scholar]
- 5.Felber B. K., Hadzopoulou-Cladaras, M., Cladaras, C., Copeland, T. & Pavlakis, G. N. (1989) Proc. Natl. Acad. Sci. USA 86, 1495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sodroski J., Goh, W. C., Rosen, C., Dayton, A., Terwilliger, E. & Haseltine, W. (1986) Nature 321, 412-417. [DOI] [PubMed] [Google Scholar]
- 7.Hidaka M., Inoue, J., Yoshida, M. & Seiki, M. (1988) EMBO J. 7, 519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erlwein O. & Rethwilm, A. (1993) Virology 196, 256-268. [DOI] [PubMed] [Google Scholar]
- 9.Lochelt M., Muranyi, W. & Flugel, R. M. (1993) Proc. Natl. Acad. Sci. USA 90, 7317-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He F., Blair, W. S., Fukushima, J. & Cullen, B. R. (1996) J. Virol. 70, 3902-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lochelt M., Zentgraf, H. & Flugel, R. M. (1991) Virology 184, 43-54. [DOI] [PubMed] [Google Scholar]
- 12.Yu S. F. & Linial, M. L. (1993) J. Virol. 67, 6618-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alke A., Schwantes, A., Kido, K., Flotenmeyer, M., Flugel, R. M. & Lochelt, M. (2001) Virology 287, 310-320. [DOI] [PubMed] [Google Scholar]
- 14.Keller A., Partin, K. M., Lochelt, M., Bannert, H., Flugel, R. M. & Cullen, B. R. (1991) J. Virol. 65, 2589-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bock M., Heinkelein, M., Lindemann, D. & Rethwilm, A. (1998) Virology 250, 194-204. [DOI] [PubMed] [Google Scholar]
- 16.Giron M. L., de The, H. & Saib, A. (1998) J. Virol. 72, 4906-4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saib A., Neves, M., Giron, M. L., Guillemin, M. C., Valla, J., Peries, J. & Canivet, M. (1997) Virology 228, 263-268. [DOI] [PubMed] [Google Scholar]
- 18.Saib A., Peries, J. & de The, H. (1993) EMBO J. 12, 4439-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saib A., Koken, M. H., van der Spek, P., Peries, J. & de The, H. (1995) J. Virol. 69, 5261-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meiering C. D. & Linial, M. L. (2001) Clin. Microbiol. Rev. 14, 165-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falcone V., Leupold, J., Clotten, J., Urbanyi, E., Herchenroder, O., Spatz, W., Volk, B., Bohm, N., Toniolo, A., Neumann-Haefelin, D. & Schweizer, M. (1999) Virology 257, 7-14. [DOI] [PubMed] [Google Scholar]
- 22.Meiering C. D., Comstock, K. E. & Linial, M. L. (2000) J. Virol. 74, 1718-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meiering C. D., Rubio, C., May, C. & Linial, M. L. (2001) J. Virol. 75, 6574-6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartz S. R. & Vodicka, M. A. (1997) Methods 12, 337-342. [DOI] [PubMed] [Google Scholar]
- 25.Regad T., Saib, A., Lallemand-Breitenbach, V., Pandolfi, P. P., de The, H. & Chelbi-Alix, M. K. (2001) EMBO J. 20, 3495-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muranyi W. & Flugel, R. M. (1991) J. Virol. 65, 727-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindemann D. & Rethwilm, A. (1998) J. Virol. 72, 4088-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee A. H., Lee, H. Y. & Sung, Y. C. (1994) Virology 204, 409-413. [DOI] [PubMed] [Google Scholar]
- 29.Brown P., Moreau-Dubois, M. C. & Gajdusek, D. C. (1982) Arch. Virol. 71, 229-234. [DOI] [PubMed] [Google Scholar]
- 30.Santillana-Hayat M., Rozain, F., Bittoun, P., Chopin-Robert, C., Lasneret, J., Peries, J. & Canivet, M. (1993) Res. Virol. 144, 389-396. [DOI] [PubMed] [Google Scholar]
- 31.Swack N. S. & Hsiung, G. D. (1975) Infect. Immun. 12, 470-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hooks J. J. & Detrick-Hooks, B. (1979) J. Gen. Virol. 44, 383-390. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt M., Niewiesk, S., Heeney, J., Aguzzi, A. & Rethwilm, A. (1997) J. Gen. Virol. 78, 1929-1933. [DOI] [PubMed] [Google Scholar]
- 34.Aguzzi A., Bothe, K., Anhauser, I., Horak, I., Rethwilm, A. & Wagner, E. F. (1992) New Biol. 4, 225-237. [PubMed] [Google Scholar]