Abstract
Homologues of the Musashi family of RNA-binding proteins are evolutionarily conserved across species. In mammals, two members of this family, Musashi1 (Msi1) and Musashi2 (Msi2), are strongly coexpressed in neural precursor cells, including CNS stem cells. To address the in vivo roles of msi in neural development, we generated mice with a targeted disruption of the gene encoding Msi1. Homozygous newborn mice frequently developed obstructive hydrocephalus with aberrant proliferation of ependymal cells in a restricted area surrounding the Sylvius aqueduct. These observations indicate a vital role for msi1 in the normal development of this subpopulation of ependymal cells, which has been speculated to be a source of postnatal CNS stem cells. On the other hand, histological examination and an in vitro neurosphere assay showed that neither the embryonic CNS development nor the self-renewal activity of CNS stem cells in embryonic forebrains appeared to be affected by the disruption of msi1, but the diversity of the cell types produced by the stem cells was moderately reduced by the msi1 deficiency. Therefore, we performed antisense ablation experiments to target both msi1 and msi2 in embryonic neural precursor cells. Administration of the antisense peptide-nucleotides, which were designed to specifically down-regulate msi2 expression, to msi1−/− CNS stem cell cultures drastically suppressed the formation of neurospheres in a dose-dependent manner. Antisense-treated msi1−/− CNS stem cells showed a reduced proliferative activity. These data suggest that msi1 and msi2 are cooperatively involved in the proliferation and maintenance of CNS stem cell populations.
During mammalian CNS development, neurons and glial cells are thought to be generated from common neural precursor cells (CNS stem cells) located in the periventricular area (1). The molecular basis for the maintenance of this cell population is, however, largely unknown. The recent discovery of neural RNA-binding proteins raises the possibility that the development of neural cells from their precursors may be controlled by posttranscriptional gene regulation, including mRNA stabilization, splicing, or translational control. Musashi1 (Msi1) and Musashi2 (Msi2) are RNA-binding proteins that are characterized by two RNP-type RNA recognition motifs (RRMs) and show remarkable similarity to one another, both in their primary structures and their RNA-binding specificities in vitro (2–4). These two molecules seem to define the Msi family of RNA-binding proteins, which is evolutionarily conserved across different species. In mammals, Msi1 and Msi2 expression is developmentally regulated. Our previous studies revealed that Msi1 and Msi2 are coexpressed predominantly in proliferating embryonic pluripotent neural precursors (2, 4–6), as well as in cell populations that are believed to be the source of postnatal and adult CNS stem cells (3, 6). In the cerebral cortex, the expression of Msi1 and Msi2 is rapidly down-regulated in newly generated postmitotic neurons (2), with the exception of some GABAergic interneurons that continue to express Msi2 exclusively (4). Although the molecular functions of the Msi family members remain obscure, their expression profiles suggest that they may play similar roles in the development and maintenance of CNS stem cells through posttranscriptional gene regulation (6).
In the present study, we used targeted disruption of the msi1 gene in mice and antisense ablation of Msi2 to address the roles of these proteins during development. The results suggested that the Msi family genes have critical functions in restricted cell populations, including CNS stem cells.
Materials and Methods
Gene Targeting.
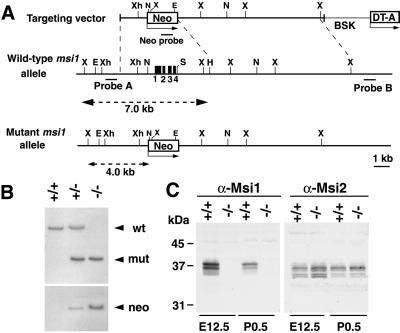
Genomic DNA fragments of msi1 gene (1.5-kb NotI–SalI fragment for the left arm, and 9.4-kb HindIII–SalI fragment for the right arm) were isolated from 129/Sv genomic DNA library and inserted into the targeting vector, which contained a 1.8-kb G418 resistance (neo) cassette and the diphtheria toxin A-fragment (DT-A) cassette. This targeting construct was designed to delete 4.2 kb of genomic DNA, including a 267-bp exonic sequence spanning the translational initiation codon and the first RRM, and to replace them with the neo cassette in the same transcriptional orientation as msi1. Correctly targeted ES cells, chimeric males, and the progeny of heterozygous intercrosses were genotyped by Southern blot or PCR analysis, using WT and mutant allele specific primers (primer sequences available on request). After seven to eight generations of backcrossing heterozygous mutants and C57BL/6 females, the F8 or F9 progeny of heterozygous intercrosses was used for histological and in vitro culture analyses. Histological analyses were performed as described (2, 4).
Antisense Peptide Nucleic Acids (asPNA).
The asPNAs were custom synthesized, purified, and analyzed by PE Biosystems. The sequences of the msi2 asPNA corresponded to the translation initiation codon (PNA1, 5′-CTCCATAGCGGAGCC-3′-Lys) or the coding region (PNA2, 5′-ACCTAATACTTTATCT-3′-Lys). These two asPNAs had similar effects on neurosphere formation, and PNA1 was used throughout the experiments in this report. A scrambled PNA (5′-AACTCTTATATCTCTA-3′-Lys), which did not match significantly any other known RNA or DNA sequence, was used for the control.
Neurosphere Culture.
The basic culture medium, containing 20 ng/ml epidermal growth factor (Sigma) and 10 ng/ml FGF2 (R & D Systems), and the procedures for the neurosphere formation and differentiation assays were as described (7). Briefly, cells from the anterior halves of the E14.5 telencephalon were used for primary sphere formation (1 × 105 cells per ml). The indicated amount (0–10 μM) of asPNA was added to the culture medium when the primary spheres were dissociated and replated for secondary sphere formation (500 cells per 200 μl per well, 96-well plate). The numbers of secondary spheres were counted 4 days later (4 div). Cells used for semiquantitative RT-PCR and immunocytochemistry for Msi2 or nestin (Rat401) were harvested after 24 h (1 div) of treatment with the asPNA. To assess cell proliferation within the asPNA-treated spheres, 2 μM BrdU (5-bromodeoxyuridine, Sigma) was administrated to the cultures at 2 div After 24 h of incubation, spheres were trypsinized, dissociated, and plated onto polyL-lysine-coated coverslips for 3 h, then processed for immunocytochemistry using anti-BrdU antibody (Sigma).
Semiquantitative RT-PCR.
Cells incubated with or without asPNA (10 μM) for 24 h were collected (1.5 × 105 cells). Semiquantitative RT-PCR was carried out using the primer sets for msi2 (5′-GTCTGCGAACACAGTAGTGGAA-3′ and 5′-GTAGCCTCTGCCATAGGTTGC-3′; 340 bp), AUF1 (5′-ACTGCACTCTGAAGTTAGATCCTA-3′ and 5′-TGTAGCTATTTTGATGTCCACCTC-3′; 534 bp), hnRNPA1 (5′-ATGGCTAGTGCTTCATCCAGTCA-3′ and 5′-CTGTGCTTGGCTGAGTTCACAAA-3′; 508 bp), hnRNPC1/C2 (5′-AGTGGATTCAATTCGAAGAGTGGA-3′ and 5′-GATGACCTGGTGTTACTTTCATCT-3′; 513 bp), NKT (5′-TAAGCTGAGTATAGAGGTGCT-3′ and 5′-CATTCACTAGGATACAGATGC-3′; 332 bp), and g3pdh (5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′; 452 bp).
Results
msi1−/− Mice Develop Obstructive Hydrocephalus with Ependymal Abnormalities.
Targeted disruption of the msi1 locus in ES cells was performed by replacing four exons, including the translation initiation codon, with a neo-resistance gene cassette (Fig. 1 A and B). Interbreeding of the heterozygous mutant (msi1+/−) mice yielded homozygous mutant (msi1−/−) pups with the expected Mendelian ratio, indicating that Msi1 is not essential for embryonic viability. The absence of Msi1 expression in embryonic and neonatal brains in homozygous animals was confirmed by immunoblot analysis (Fig. 1C) using an anti-Msi1 monoclonal antibody.
Fig 1.
Targeting strategy, germ-line transmission, and expression analysis of the msi1 gene. (A) Organization of the targeting vector, the msi1 gene, and the allele resulting from homologous recombination. Four exons (black boxes) of the msi1 allele containing the initiation codon were replaced with a Neo cassette. A 0.3-kb Sau3AI fragment (probe A) was used to screen for recombinant alleles, and the sizes of the recombinant and WT fragments after XbaI digestion are shown (broken lines). X, XbaI; E, EcoRV; Xh, XhoI; Not, NotI; S, SalI; H, HindIII; BSK, plasmid vector. (B) Germ-line transmission was confirmed by Southern blot analysis of XbaI- or EcoRV-digested tail DNA from a litter of F6 mice using probe A or the Neo probe, respectively. (C) Immunoblot analysis of brain lysates from E12.5 and P0.5 mice using anti-Msi1 or anti-Msi2 antibodies. Genotypes are indicated.
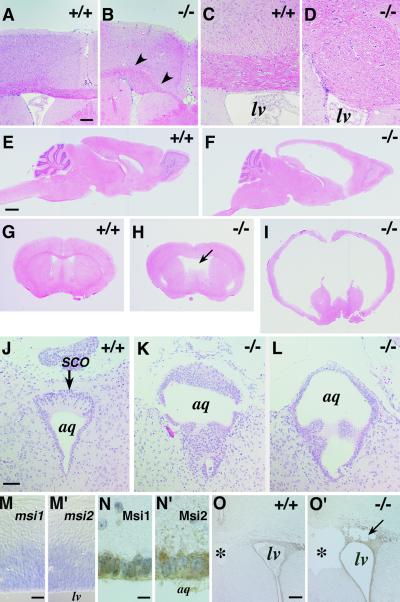
Histological analyses showed that msi1−/− embryos (E10–E17) exhibited normal cortical development. Neuroepithelial cell proliferation in the ventricular zone (VZ) was not perturbed, as determined by light microscopy and BrdU incorporation analysis (not shown). Newborn msi1−/− pups were normal in size and appearance for the first postnatal week. However, by 1–2 postnatal weeks, 70–80% of the msi1−/− animals developed hydrocephalus with progressive dilation of the lateral ventricles and an enlarged, domed cranium (Fig. 2 E–I). In addition, a small population of msi1−/− mice (5–10%) exhibited Probst's bundle (8), an entangled axon tract in the corpus callosum (CC), because of errors in the projection of commissural fibers across the midline in the forebrain (Fig. 2 A–D). This callosal agenesis was observed at E17, when normal CC axons have arrived at the midsagittal plane (not shown). Agenesis or hypoplasia of the CC with Probst bundles is frequently found in hereditary hydrocephalus in humans (X-linked hydrocephalus) (9) and in several targeted and spontaneous mouse mutants, e.g., hyh (10) and E2F-5 (11), suggesting that complex multigenic developmental pathways are disrupted in the msi1−/− mice.
Fig 2.
Development of hydrocephalus in postnatal msi1−/− mice with ependymal abnormalities and the concurrent expression of Msi1 and Msi2 in the developing CNS and aqueduct. Genotypes are indicated. (A–I) Hematoxylin & eosin-stained adult brain. Agenesis of the CC observed in an adult msi1−/− mouse (B, arrowheads) compared with the normal CC in a WT littermate (A). (C and D) A higher magnification view of the normal CC and the Probst's bundle, respectively. (E–I) Hydrocephalus of the adult msi1−/− mice. (E and F) Sagittal sections. (G–I) Coronal sections at the level of the anterior commissure. (F and I) A severe hydrocephalic mutant showing massive ventricular dilation. (H) Moderate dilation of the lateral ventricles accompanied by cavitation of the septum pellucidum and hypoplasia of the septum. (J–L) Coronal sections through the aqueduct of the P7 brain, showing abnormal accumulation and polyposis of ependymal cells surrounding the Sylvius aqueduct and SCO. (M and M′) mRNA in situ hybridization analysis of msi1 and msi2 in the WT E12.5 telencephalon showing their expression in the VZ. (N and N′) Immunohistochemical detection of Msi1 and Msi2 in ependymal cells surrounding the aqueduct of WT P3 mice. Immunoreactivities were visualized by DAB (brown), and the nuclei were counterstained with hematoxylin. (O and O′) Expression of Msi2 in the SVZ and ependymal cells lining the lateral ventricle of a hydrocephalic msi1−/− (O′) and a WT littermate (O) at P3. The msi1−/− tissue shows partial destruction of the SVZ (arrow). lv, lateral ventricle; aq, Sylvius aqueduct; *, septum pellucidum. (Scale bars: A and B, 150 μm; C, D, and J–L, 50 μm; E–I, 1 mm; M, 30 μm; N, 10 μm; O, 100 μm.)
Pathologic examination of the msi1−/− adult brains revealed dilation of the lateral ventricles and the third ventricle, thinning of the cerebral cortices, hypoplasia of the septa, disruption of the white and gray matters accompanied by intracerebral hemorrhage, edema, and necrosis of the periventricular parenchyma. The fourth ventricle was not dilated in msi1−/− mice. The olfactory bulb, cerebellum, thalamus, medulla, and hippocampus were not primarily affected, although they appeared to be compressed by the elevated intracranial pressure (Fig. 2 F and I). The dilation of the lateral ventricles was frequently accompanied by cavitation in the subventricular zone (SVZ) (Fig. 2O′). The areas surrounding the anterior horns of the lateral ventricles were occasionally denuded of ependymal cells or had a disrupted ependymal lining (Fig. 2O′). Analyses of P0 forebrain sections by BrdU labeling, TUNEL staining, and immunohistochemistry using cell-type-specific markers failed to demonstrate significant changes in the number of apoptotic cells, differentiated neurons, or proliferating precursor cells in the msi1−/− SVZ before the onset of ventricular dilation (not shown). The msi1−/− mice with severe hydrocephalus demonstrated ataxic gate and dehydration, and they eventually died within 1–2 months after birth. Moderate cases exhibited the dilated lateral ventricles and cavitation of the septum pellucidum (Fig. 2H) but survived into adulthood.
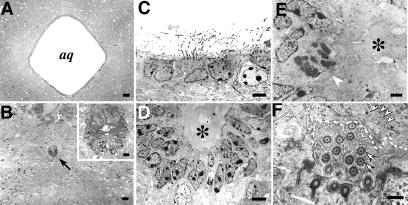
In the msi1−/− mice, abnormal proliferation and polyposis were observed in ependymal cells surrounding the Sylvius aqueduct, as well as in the subcommissural organ (SCO), an ependymal gland located at the entrance of the cerebral aqueduct that secretes glycoproteins into the cerebrospinal fluid (CSF) (12) (Fig. 2 K and L). These glycoproteins are known to aggregate in the CSF to form a fibrous structure (Reissner's fiber) along the aqueduct (12). Notably, Msi1 is expressed in the ependymal cells of both the SCO and the cerebral aqueduct (Fig. 2N). Electron microscopic analysis showed an abnormal stratification of the ependymal cells of the msi1−/− aqueduct that was not evident in heterozygous or WT littermates. Detailed histological analysis demonstrated that the aqueduct of Msi1 null mice was enwrapped by a thick epithelium composed of two or three layers of ependymal cells, in contrast to the well organized single ependymal cell layer in the aqueduct of WT or heterozygous littermates (Fig. 3D). Although the majority of these msi1−/− cells appeared to be terminally differentiated ependymal cells, characterized by cilia (Fig. 3 D and F), some appeared to proliferate heterotopically without nuclear heteromorphism (Fig. 3E), which is consistent with the ependymal polyposis in the cerebral aqueduct and SCO. The lumen of the msi1−/− aqueduct was completely filled with amorphous materials abundant in electron-dense glycogen granules, and the microvilli of ependymal cells were irregularly arranged, as if they were compressed by the packed materials (Fig. 3F).
Fig 3.
Light microscopic and ultrastructural analyses of the msi1−/− ependyma at P14. (A and B) Light micrographs of the msi1−/− brain showing stenosis of the Sylvius aqueduct (B, arrow) and of the WT brain (A). Inset in B shows a low-power view of the ependymal cells in the aqueduct. (C) An electron micrograph of the WT aqueduct showing a single layer of ependymal cells and well organized microvilli protruding into the aqueductal lumen. (D) The aqueduct of an msi1−/− mouse, surrounded by the stratified ependymal epithelium composed of two or three cell layers. (E) A high-power view of msi1−/− ependymal cells, showing heterotopic mitotic figures without nuclear heteromorphism (arrowhead). Cytoplasmic organelles in these cells appear intact. The lumen of the msi1−/− aqueduct is completely filled with amorphous materials (asterisks in D and E). (F) A magnified view of msi1−/− ependymal evaginations and cilia. Cilia, characterized by central and peripheral tubules (arrows), are apposed to and are disarranged by solid materials (dashed line) that are abundant in the electron-dense particles of glycogen granules (arrowheads). (Scale bars: A and B, 40 μm; B Inset, 8 μm; C and D, 5 μm; E, 2 μm; F, 0.5 μm.)
Hydrocephalus could result from impaired CSF absorption from the subarachnoid space or from overproduction of CSF by the choroid plexus; however, histological examination failed to demonstrate abnormalities in these areas (not shown). Taking these observations together, it is reasonable to attribute the hydrocephalus of msi1−/− mice to the obstruction of SCF flow in the aqueduct.
Characterization of CNS Stem Cells Lacking Msi1.
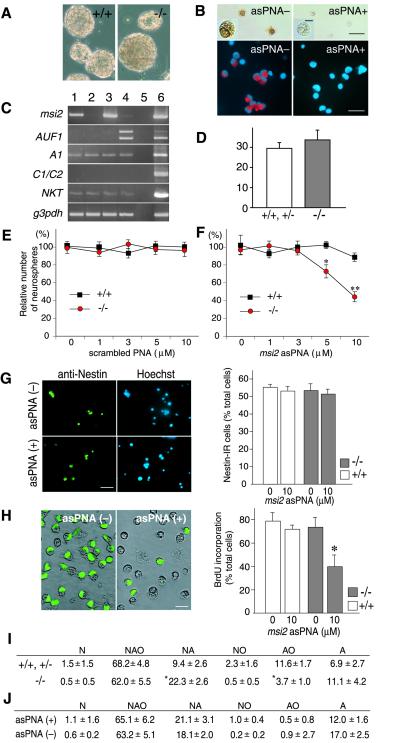
We next examined the role of Msi1 in the CNS stem cell population using a neurosphere assay, which is a selective culture system for CNS stem cells (7, 13). In the presence of mitogens such as epidermal growth factor and fibroblast growth factor 2, each CNS stem cell proliferates to form a floating multicellular structure, the neurosphere. Thus, CNS stem cells can be defined as sphere-forming cells (SFCs) (13). The number and size of the formed neurospheres are believed to reflect the number and proliferative activity of SFCs in the dissociated cell population, respectively (neurosphere formation assay). The multipotency of SFCs, that is, their ability to generate different neural cell types, can be assessed by a neurosphere differentiation assay; each neurosphere can be induced to differentiate into TuJ1+ neurons (N), GFAP+ astrocytes (A), and/or O4+ oligodendrocytes (O), after being plated onto substrates in a culture medium containing serum. Consistent with the histological analysis of the msi1−/− embryos, the number and size of the neurospheres derived from the msi1−/− embryos were quite similar to those derived from WT littermate embryos (Fig. 4 A and D). When the msi1−/− neurospheres were dissociated into single cells, a subset of the cells reformed neurospheres repeatedly, for as many passages as dissociated cells from the WT neurospheres did (not shown). These observations confirmed that the characteristics of the msi1−/− stem cells per se were unchanged; disruption of the msi1 gene alone did not affect the number or self-renewal activity of CNS stem cells.
Fig 4.
Loss of msi1 and msi2 activities decreased the efficiency of neurosphere formation through the reduced proliferation of SFCs. (A) Neurospheres derived from the E14.5 telencephalons of msi1−/− or WT littermates (after three passages). (Scale bar, 100 μm.) (B) Suppression of Msi2 immunoreactivity on the asPNA-treated SFCs. Dissociated cells from the WT (Upper) or msi1−/− (Lower) primary neurospheres were incubated with or without asPNA (10 μM) for 24 h and immunostained with anti-Msi2 antibody. Immunoreactivities to Msi2 were visualized by a DAB reaction (brown, Upper) or Alaxa-568 (red, Lower). Insets are magnified views of representative cells, showing the repression of Msi2 protein expression without pyknotic changes. [Scale bar, 5 μm (Inset) or 25 μm (Upper and Lower).] (C) Semiquantitative RT-PCR analysis of msi2 mRNA, the msi-related genes encoding RNA-binding protein (AUF1, hnRNP A1, and hnRNP C1/C2), NKT mRNA, and an internal control mRNA (g3pdh). The msi1−/− or WT SFCs were incubated with or without asPNA for 24 h, then subjected to the RT-PCR. Lane 1, WT cells; lane 2, WT cells + asPNA; lane 3, msi1−/− cells; lane 4, msi1−/− cells + asPNA; lane 5, no RT control; lane 6, E14.5 WT cerebral cortex. (D) The number of neurospheres per field (×4) formed by the dissociated cells from msi1−/− (−/−, n = 3) or littermates (+/+, +/−, n = 3). (E and F) The number of neurospheres derived from the telencephalons of msi1−/− and their WT littermates in the presence of the scrambled PNA (E) or in the presence of the asPNA (F). The data were presented as the mean ± SEM (WT, n = ≈3–6; msi1−/−, n = ≈3–7). *P < 0.005, **P < 0.0001, in comparison with the WT control (unpaired t test). (G) The number of nestin+ cells after the exposure to asPNA for 24 h. Dissociated cells of the primary neurospheres derived from the WT and msi1−/− telencephalons were incubated with or without asPNA (10 μM) and then immunopositive cells for anti-nestin were counted (WT, n = 6; msi1−/−, n = 6). Photomicrographs showed the msi1−/− cells that were positive for nestin (green). (Scale bar, 25 μm.) (H) Decreased number of BrdU+ cells within the msi1−/− neurospheres exposed to asPNA. During the formation of neurospheres in the presence (+) or absence (−) of asPNA (10 μM), BrdU was administrated at 2 div. After the additional cultivation for 24 h, each sphere was dissociated and immunostained with anti-BrdU (WT, n = 6; msi1−/−, n = 6). Photomicrographs represented the BrdU-labeled msi1−/− cells (green). The data were presented as the mean ± SEM. *, P < 0.001 in comparison with the other conditions (two-tailed Student's t test). (Scale bar, 10 μm.) (I) Differentiation assay of neurospheres that derived from the msi1−/− (−/−) and their littermate cells (+/+, +/−). The differentiation capacity of each primary neurosphere was determined based on the cell types contained in each clone. N, neurons; A, astrocytes; O, oligodendrocytes. The majority (84.5%) of neurospheres from msi1−/− mice generated both neurons and glia (NAO+NA+NO). The clone types were analyzed for 98 spheres from WT or heterozygous mice (n = 5) and 234 spheres from msi1−/− mice (n = 10) and presented as the mean ± SEM. *, P < 0.0001, in comparison with WT and heterozygous controls (Student's t test). (J) Differentiation assay of neurospheres that derived from the msi1−/− cells treated with or without asPNA. The clone types were analyzed for 67 spheres from asPNA (+) and 141 spheres from asPNA (−).
A clonogenic differentiation assay using cell-type-specific markers revealed that the majority of SFCs from msi1−/− mice were multipotent and could give rise to all three types of differentiated cells (NAO) (Fig. 4I). Nonetheless, we noticed that the msi1−/− SFCs showed a slightly limited repertoire for differentiation: the number of clones containing oligodendrocytes (AO+NO+NAO) tended to be slightly smaller in the msi1−/− neurospheres (66.2%) than in the WT neurospheres (82.1%). This reduction could not be restored by supplementing the differentiation medium with retinoic acid or T3, agents that potentiate the differentiation and proliferation of oligodendrocyte precursors (not shown). Thus, a lack of msi1 activity reduces the multipotency of the CNS stem cells.
The Msi Family Functions in the Proliferation of Neural Progenitor Cells Including CNS Stem Cells.
Taking into account the high expression levels of msi1 during embryonic development (2), it was somewhat surprising that msi1−/− mice survived embryonic development with just minor structural abnormalities in restricted populations of ependymal cells. This finding indicates that msi1 is not essential for the development of most tissues and body structures. In addition, a small fraction of msi1−/− mice (5–10%) survived into adulthood without detectable morphological abnormalities. These observations raised the possibility that the loss of msi1 functions might be compensated for, at least in part, by one or more other genes. One such candidate was msi2, another member of the Msi family (4). In the CNS, the expression of Msi2 is developmentally regulated and overlaps with that of Msi1 in neural precursor cells, astrocytes, and ependymal cells (Fig. 2 M–O) (4). The Msi2 expression level was up-regulated 1.4–2.0-fold in the msi1−/− embryonic brains (E12.5), compared with WT brains, as determined by Western (Fig. 1C) and Northern (not shown) blotting. These results support the hypothesis that msi2 compensates for the msi1 deficiency during the CNS development of msi1−/− mice.
To test directly for the cooperative involvement of Msi family proteins in the function of CNS stem cells, we tried making a functional double knockout of msi1 and msi2. To this end, we added antisense compounds specific to the msi2 gene to the medium of a CNS stem cell culture prepared from msi1−/− embryos or WT littermates and assessed the resulting number of neurospheres. Antisense oligonucleotides covering the translation initiation region or the coding region of msi2 (a 16-mer) was synthesized as peptide nucleic acids (asPNA). PNA is a new type of DNA analogue that has an artificial homomorphous peptide backbone with much higher sequence specificity for target genes, a higher resistance to proteases and nucleases, and lower cytotoxicity (14) than other forms of antisense nucleotides, such as the phosphorothioate oligos. Administration of asPNA to cultures derived from the WT or msi1−/− embryonic forebrain led to a specific and marked reduction in msi2 expression at both the mRNA and protein levels, as demonstrated by semiquantitative RT-PCR analysis and immunocytochemical detection with the anti-Msi2 antibody (Fig. 4 B and C). With the concentrations of the asPNA used in this series of experiments, rapid cellular changes such as nuclear condensation were not observed in most of the individual dissociated cells, confirming that there was no or very little cytotoxicity associated with the asPNA. Homology search with this asPNA sequence against the GenBank/EST or the Celera Discovery System and Celera Genomics' associated databases indicated that any other mRNAs did not contain a completely matched sequence to the designed sequence of the msi2 asPNA. We performed the control RT-PCR analysis of the organic cation transporter NKT (GenBank accession no. U52842), which was expressed in the neurospheres and shared a sequence similarity to asPNA with a two-base mismatch. As shown in Fig. 4C, the asPNA treatment had no effect on the level of NKT mRNA expression in the msi1−/− or littermate cultures, confirming the target specificity of asPNA. In the neurosphere formation assay, the presence of the asPNA caused a drastic reduction in the number of neurospheres formed in the msi1−/− cultures in a dose-dependent manner (Fig. 4F). In contrast, the greatest concentration of asPNA used in our study did not decrease the number of neurospheres formed in the WT cultures, indicating that the suppression of Msi2 alone did not affect the neurosphere-forming ability and viability of CNS stem cells. Administration of the nonspecific scrambled PNA did not affect the number of neurospheres formed in both the WT and msi1−/− cultures (Fig. 4E), excluding the possibility that the msi1−/− CNS stem cells are more susceptible to PNA-mediated cytotoxicity than WT cells. We, therefore, concluded that the reduction of neurosphere formation mediated by asPNA is a specific effect and that a cooperative action of both Msi1 and Msi2 is essential for the proliferation and/or maintenance of embryonic CNS stem cells.
We sought to establish the characteristics of msi1−/− CNS stem cells that exposed to the asPNA. Although there are no unambiguous markers for CNS stem cells, we analyzed the number of cells that express nestin, a marker for neural progenitor cells including CNS stem cells (1). Dissociated cells from the primary neurospheres were incubated with the asPNA during the first 24 h before the majority of the SFCs started cell division. Immunocytochemical analysis indicated that nestin+ cells were ≈55% of total cells in WT and msi1−/− cells, and the number of nestin+ cells were unchanged with or without asPNA treatment (Fig. 4G), suggesting that the asPNA treatment did not influence the nestin expression and that there were the equivalent number of progenitor cells at the beginning of their sphere formation. Next, we determined the effect of asPNA on the proliferation of neural progenitor cells including CNS stem cells. BrdU was administrated into the sphere cultures that have pretreated with asPNA for 2 div. Twenty-four hours later (3 div), the individual forming neurospheres, which were in a phase of the exponential proliferation of SFCs, were harvested, and the BrdU-labeled cells were counted. Without the asPNA treatment, the labeling index for BrdU was unchanged in msi1−/− neurospheres relative to the WT (≈75% of total cells); however, the marked reduction of BrdU-labeled cells was evident in the asPNA-treated msi1−/− neurospheres (≈40% of total cells) (Fig. 4H). Neurosphere differentiation assay revealed that the asPNA treatment did not lead a significant change in the differentiation ability in the msi1−/− SFCs (Fig. 4J). Taken together, the reduction of neurosphere formation in msi1/msi2 functional double knockout cells was primarily attributable to the inhibition of the SFC proliferation, rather than due to an aberrant cell-fate choice. Because the short exposure of the asPNA (1 div) seemed to be unaffected for the number of nestin+ cells, msi1 and msi2 may not be involved directly in a survival of the neural progenitor cells including CNS stem cells.
Among the numerous RNA-binding proteins, Msi2 and Msi1 form a unique subgroup in the hnRNP A/B class that contains two copies of RRMs. Within this class, the Msi family has a sequence similarity to AUF1 (hnRNP D) and hnRNP A1, albeit to a lesser degree (4). To assess whether the expression levels of these msi-related genes altered in the functional double knockout cells, we performed a semiquantitative RT-PCR analysis. A significant up-regulation of the mRNA for AUF1 was detected in the asPNA-treated msi1−/− cultures without affecting the expression levels of hnRNP A1 and hnRNP C1/C2 (Fig. 4C).
Discussion
Here, we showed that disruption of the msi1 gene causes hydrocephalus. The abnormal proliferation and/or differentiation of ependymal cells lining the aqueduct may be the primary cause of the obstruction of the CSF passage, which results in the perinatal onset of hydrocephalus in msi1−/− mice. However, we found no significant difference in the total number of ependymal cells lining the whole area of the aqueduct in perinatal msi1−/− pups, as determined by counting the cells in serial sections (not shown). In addition, the BrdU pulse-labeling experiments at P3 and P10 brains showed that there was no difference in the total number of proliferating cells along the aqueductal ependyma between msi1−/− pups and their WT littermates (not shown), in contradiction to the stratification and polyposis formation in the postnatal SCO. These morphologically abnormal ependymal cells were rarely labeled with BrdU at least by 4 h of BrdU labeling at P3 (not shown). They may proliferate slower than other normal ependymal populations during the aqueductal development. Collectively, Msi1 is likely to be essential for the differentiation or proper functions of the highly restricted cell populations in the ependyma lining the aqueduct. Conceivably, these cells may have a higher susceptibility to the deficiency of the msi1 gene than do the ependymal cells located in other regions of the ventricular system. Nevertheless, considering the coexpression of Msi1 and Msi2 in most ependymal populations (Fig. 2N), it remains unclear why the defects in msi1−/− mice are restricted to a certain population of ependymal cells. Msi1 may have a specific function that is required for the development of aqueductal ependymal cells.
The CNS stem cells continue to proliferate in an undifferentiated state (self-renewal proliferation), and also can give rise to the lineage-restricted progenitors, i.e., the neuronal or glial progenitors. The antisense ablation experiments of the msi2 gene indicated the vital functions cooperated with the msi genes during the formation of the neurosphere. Functionally double knockout neurosphere cells showed the reduction of proliferation. This finding suggests that two msi genes are involved in the self-renewal proliferation of CNS stem cells. Alternatively, such phenomena could have been caused by enhanced differentiation of CNS stem cells to lineage-restricted progenitors. However, the differentiation potential of double knockout cells was not significantly changed from that of msi1−/− cells, suggesting that the msi2 gene is not essential for the lineage restriction of CNS stem cells.
Recent in vitro and in vivo studies have provided intriguing evidence regarding a source of CNS stem cells in the postnatal CNS. Johansson et al. (15) showed that CNS stem cells could originate from ependymal cells in the adult brain and spinal cord, whereas Doetsch et al. (16) provided compelling evidence that SVZ astrocytes are precursors for neurogenesis and could possibly serve as stem cells. Although it has been controversial which type of cells serves as the source of postnatal CNS stem cells in vivo, the disorganized proliferation or premature differentiation of msi1−/− ependymal cells may reflect an impairment of the proliferation or maintenance of CNS stem cells residing in the aqueductal ependyma. However, this idea regarding the ependymal proliferation caused by the loss of msi1 seemed to be inconsistent with the proposed function of msi genes that positively regulate the proliferation of CNS stem cells. It is possible that there is a unique function of the msi1 gene that keeps the aqueductal ependymal cells quiescent that the msi2 gene cannot compensate for. Alternatively, the msi2 gene may be functionally “overexpressed” in the absence of msi1 in the restricted aqueductal ependymal cell population. Because the Msi family proteins bind to a similar RNA sequence, excessive binding of Msi2 protein to the target mRNAs normally occupied by the Msi1 protein could account for the ependymal proliferation through the misregulation of target mRNAs. Such ependymal proliferation might be implicated in the genesis of ependymal tumors, ependymomas. In fact, of the various brain tumors, the expression levels of the Msi family proteins are markedly elevated in ependymoma cells (unpublished results). Msi1 expression also correlates well with the proliferative activity and malignancy of human gliomas (6), suggesting the involvement of deregulated Msi1 expression in tumorigenesis. Appropriately and tightly regulated expression levels of the msi family genes may be crucial not only for the proper development of neural precursors but also to prevent the clonal expansion of proliferative tumors.
Msi1 and Msi2 show a marked sequence similarity, especially within their RRM domains (≈90%), which are responsible for specific binding with target RNA molecules. Our previous in vitro RNA-binding assay (4) revealed that Msi1 and Msi2 have a very similar RNA-binding specificity characterized by uridine-rich sequences. The cooperative and redundant functions of msi2 and msi1 in CNS stem cells further support the idea that Msi1 and Msi2 interact with common target RNAs in vivo. In addition, our present RT-PCR analysis indicated the up-regulation of AUF1 mRNA in the asPNA-treated msi1−/− cultures. AUF1 is a key regulatory factor of gene expression that is known to bind uridine-rich elements of 3′-UTRs and regulate the stability of many protooncogene and cytokine mRNAs (17). It is unclear whether the Msi proteins and AUF1 share the common downstream target mRNAs. Therefore, we could not exclude a possibility that the loss of two msi gene activities could have been compensated by the enhanced expression of AUF1 in the neurospheres derived from the msi2 asPNA-treated msi1−/− cells (Fig. 4F). The molecular mechanism underlying the functions of the Msi family should be elucidated by identifying their target mRNAs. One of the candidates is the mRNA of mammalian numb (m-numb), which encodes a membrane-associated antagonist of Notch signaling (18, 19). The Notch signal is required for the self-renewing activity of mammalian CNS stem cells (7, 20, 21). Our recent study (22) indicated that Msi1 binds to the uridine-rich sequence in the 3′-UTR region of m-numb mRNA in vitro and can repress the expression of m-numb at the level of translation in NIH 3T3 cells. These results, combined with the observation that overexpressed Msi1 activates Notch signaling (22), indicate that the Msi family may contribute to the self-renewing activity of CNS stem cells by modulating the m-numb-Notch signaling cascade. However, our immunohistochemical study with an anti-m-Numb antibody failed to demonstrate a significant increase in the m-Numb protein in the neurospheres or brain of msi1−/− mice (not shown). Considering that the up-regulated msi2 acts as a redundant gene in the msi1−/− CNS, loss-of-function experiments with both the msi2 and msi1 genes may unequivocally reveal the function of the Msi proteins in mammalian CNS development. We have generated a homozygous mutant harboring a disrupted msi2 allele, and the phenotypic analyses of double-knockout mice with an msi2/msi1 genotype are currently in progress in our laboratory.
Acknowledgments
We are grateful to Drs. B. Barres, H. Kamiguchi, T. Shimazaki, and H. J. Okano for helpful discussions about the manuscript. This work was supported by grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology and from the Japan Science and Technology Corporation.
Abbreviations
Msi1, Musashi1
Msi2, Musashi2
VZ, ventricular zone
SVZ, subventricular zone
SCO, subcommissural organ
SFC, sphere-forming cell
RRM, RNA recognition motifs
asPNA, antisense peptide nucleic acids
CC, corpus callosum
CSF, cerebrospinal fluid
BrdU, 5-bromodeoxyuridine
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Temple S. & Alvarez-Buylla, A. (1999) Curr. Opin. Neurobiol. 9, 135-141. [DOI] [PubMed] [Google Scholar]
- 2.Sakakibara S., Imai, T., Hamaguchi, K., Okabe, M., Aruga, J., Nakajima, K., Yasutomi, D., Nagata, T., Kurihara, Y., Uesugi, S., et al. (1996) Dev. Biol. 176, 230-242. [DOI] [PubMed] [Google Scholar]
- 3.Sakakibara S. & Okano, H. (1997) J. Neurosci. 17, 8300-8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakakibara S., Nakamura, Y., Satoh, H. & Okano, H. (2001) J. Neurosci. 21, 8091-8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keyoung H. M., Roy, N. S., Benraiss, A., Louissaint, A., Jr., Suzuki, A., Hashimoto, K., Rashbaum, W. K., Okano, H. & Goldman, S. A. (2001) Nat. Biotechnol. 19, 843-850. [DOI] [PubMed] [Google Scholar]
- 6.Okano H., Imai, T. & Okabe, M. (2002) J. Cell Sci. 115, 1355-1359. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura Y., Sakakibara, S., Miyata, T., Ogawa, M., Shimazaki, T., Weiss, S., Kageyama, R. & Okano, H. (2000) J. Neurosci. 20, 283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozaki H. S. & Wahlsten, D. (1993) J. Comp. Neurol. 336, 595-604. [DOI] [PubMed] [Google Scholar]
- 9.Kenwrick S., Watkins, A. & De Angelis, E. (2000) Hum. Mol. Genet. 9, 879-886. [DOI] [PubMed] [Google Scholar]
- 10.Pérez-Fígares J. M., Jiménez, A. J., Pérez-Martín, M., Fernández-Llebrez, P., Cifuentes, M., Riera, P., Rodríguez, S. & Rodríguez, E. M. (1998) J. Neuropathol. Exp. Neurol. 57, 188-202. [DOI] [PubMed] [Google Scholar]
- 11.Lindeman G. J., Dagnino, L., Gaubatz, S., Xu, Y., Bronson, R. T., Warren, H. B. & Livingston, D. M. (1998) Genes Dev. 12, 1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodríguez E. M., Oksche, A., Hein, S. & Yulis, C. R. (1992) Int. Rev. Cytol. 135, 39-121. [DOI] [PubMed] [Google Scholar]
- 13.Reynolds B. A. & Weiss, S. (1996) Dev. Biol. 175, 1-13. [DOI] [PubMed] [Google Scholar]
- 14.Aldrian-Herrada G., Desarménien, M. G., Orcel, H., Boissin-Agasse, L., Méry, J., Brugidou, J. & Rabié, A. (1998) Nucleic Acids Res. 26, 4910-4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson C. B., Momma, S., Clarke, D. L., Risling, M., Lendahl, U. & Frisén, J. (1999) Cell 96, 25-34. [DOI] [PubMed] [Google Scholar]
- 16.Doetsch F., Caillé, I., Lim, D. A., García-Verdugo, J. M. & Alvarez-Buylla, A. (1999) Cell 97, 703-716. [DOI] [PubMed] [Google Scholar]
- 17.Wilson G. M. & Brewer, G. (1999) Prog. Nucleic Acid Res. Mol. Biol. 62, 257-291. [DOI] [PubMed] [Google Scholar]
- 18.Zhong W., Feder, J. N., Jiang, M. M., Jan, L. Y. & Jan, Y. N. (1996) Neuron 17, 43-53. [DOI] [PubMed] [Google Scholar]
- 19.Frise E., Knoblich, J. A., Younger-Shepherd, S., Jan, L. Y. & Jan, Y. N. (1996) Proc. Natl. Acad. Sci. USA 93, 11925-11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaiano N. & Fishell, G. (2000) Soc. Neurosci. Abstr. 26, 1347. [Google Scholar]
- 21.Hitoshi S., Alexson, T., Tropepe, V., Donoviel, D., Elia, A. J., Nye, J. S., Conlon, R. A., Mak, T. W., Bernstein, A. & van der Kooy, D. (2002) Genes Dev. 16, 846-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imai T., Tokunaga, A., Yoshida, T., Hashimoto, M., Mikoshiba, K., Weinmaster, G., Nakafuku, M. & Okano, H. (2001) Mol. Cell. Biol. 21, 3888-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]