Abstract
The developmental program of nodulation is regulated systemically in leguminous host species. A mutant astray (Ljsym77) in Lotus japonicus has lost some sort of its ability to regulate this symtem, and shows enhanced and early nodulation. In the absence of rhizobia, this mutant exhibits characteristics associated with defects in light and gravity responses. These nonsymbiotic phenotypes of astray are very similar to those observed in photomorphogenic Arabidopsis mutant hy5. Based on this evidence, we predicted that astray might contain a mutation in the HY5 homologue of L. japonicus. The homologue, named LjBzf, encodes a basic leucine zipper protein in the C-terminal half that shows the highest level of identity with HY5 of all Arabidopsis proteins. It also encodes legume-characteristic combination of motifs, including a RING-finger motif and an acidic region in the N-terminal half. The astray phenotypes were cosegregated with LjBzf, and the failure to splice the intron was detected. Nonsymbiotic and symbiotic phenotypes of astray were complemented by introduction of CaMV35S:LjBzf. It is noteworthy that although Arabidopsis hy5 showed an enhancement of lateral root initiation, Lotus astray showed an enhancement of nodule initiation but not of lateral root initiation. Legume-characteristic combination of motifs of ASTRAY may play specific roles in the regulation of nodule development.
Legumes exhibit a nitrogen-fixing symbiosis with soil bacteria, rhizobia, only in the combined nitrogen limited condition and take advantage of it by using the assimilated nitrogen. During symbiotic interaction with bacteria, host plant legumes develop a unique organ called “nodule.” On bacterial inoculation, host plant roots are exposed to signal molecules secreted by bacteria, thereby triggering nodule development. Nevertheless, host plants are not only susceptible to the bacterial induction, but also actively regulate the nodule development in both positive and negative manners. Impairment of the gene involved in the negative regulation leads to the enhancement of nodule development in the host plant and to the formation of a large number of nodules compared with the wild type. To better understand the mechanism of negative regulation of nodule development, it is necessary to isolate and characterize the regulatory genes.
A hypernodulating mutant that develops an excessive number of nodules was reported in soybean in 1985 (1, 2). Besides soybean, a number of hypernodulating mutants, i.e., developing a considerably large number of nodules, have been isolated from Pisum, Medicago, and Lotus (3–10), though the number of mutated loci is limited, and none of the corresponding genes have been isolated so far.
astray (Ljsym77) was identified from the model legume plant Lotus japonicus as a root mutant that develops an increased number of nodules compared with the wild type, suggesting that the corresponding gene encodes a negative regulator of nodule development (10). Characterization of the symbiotic phenotype of astray demonstrated the uniqueness of this mutant as an enhanced-nodulating mutant in terms of nodulation property, nodule primordia initiation and sensitivity to nodulation inhibitors (11). Of note is that the astray mutant demonstrated the early nodulation phenotype, which is an unprecedented type of symbiotic characteristic (11). The results indicate that astray is distinct from previously reported hypernodulating mutants, suggesting that the corresponding gene may define a novel locus repressing nodule development.
Interestingly, this mutant displayed altered morphology even in the absence of rhizobia, which was very similar to that observed in photomorphogenic Arabidopsis mutant hy5 (12). Based on the nonsymbiotic characteristics of astray, we predicted that the Lotus HY5 homologue is the corresponding gene of the astray mutant and set out to isolate the homologue. In this paper, we report the cloning and molecular identification of the ASTRAY gene which, to our knowledge, is the first documented negative regulator of nodule development. The results highlight the novelty of this mutant at the molecular level by demonstrating the unique combination of several motifs, including seemingly legume-characteristic ones.
Materials and Methods
Plant Growth Conditions.
Plant germination, growth conditions, and bacterial inoculation were as described (11). Freshly cultured Mesorhizobium loti MAFF 30–3099 was used for inoculation.
Isolation of Lotus HY5 Homologue.
The Lotus HY5 homologue was isolated based on the degenerate PCR method with reference to the sequences of HY5 (12) and its homologous genes, STF1 (L28003) from Glycine max (13) and VFBZIPZF (X97904) from Vicia fava. Degenerate primers (5′-G(C/T)TT(C/G)CTGTTGTGTTCTTCA-3′) and (5′-AATGGTAACTGGGT(A/C)GCA-3′) were designed based on the conserved regions of the N-terminal half of STF1 and VFBZIPZF, and the C-terminal half of STF1, VFBZIPZF, and HY5, respectively. Amplification with root cDNA included 25 cycles of PCR with 30-sec denaturation at 94°C, 1-min primer annealing at 55°C, and 2-min extension/synthesis at 72°C with the degenerate primers described above, realizing a 910-bp product. To obtain the flanking region of this cDNA fragment, bordering sequences of this fragment were used to design primers (5′-GAGCTCAAAGAGCAGACTTTCCA-3′) and (5′-CCCATGATCTAGAACCGTGTCAAC-3′) for inverse PCR with the genomic DNA digested with EcoRI restriction enzyme. Amplification included 25 cycles of PCR with 1-min denaturation at 94°C, 2-min primer annealing at 55°C, and 3-min extension/synthesis at 72°C, realizing a 2.8-kb product. By combining the sequences of degenerate PCR product and inverse PCR product, the entire coding region of LjBzf was determined and then sequenced with primers (5′-TGAGCTGGCTAAGCTTC-3′) and (5′-CTCCAGTTCTAAGTTTCC-3′). Sequencing was performed with the ABI PRISM Ready Reaction kit (Perkin–Elmer) and synthetic oligonucleotides on an Applied Biosystems 310 Analyzer.
Segregation Analysis by Derived Cleaved Amplified Polymorphic Sequence (dCAPS) Technique.
For segregation analysis, the F2 population was generated from a cross of the astray mutant (Gifu B-129 background) and Miyakojima MG-20 (14). Genomic DNA was extracted from leaves of F2 plants by using a DNeasy Plant Mini kit (Qiagen, Valencia, CA). Forty-three F2 plants with the astray phenotype were analyzed individually. Amplification of LjBzf cDNA fragment included 30 cycles of PCR with 30-sec denaturation at 94°C, 30-sec primer annealing at 54°C, and 30-sec extension/synthesis at 72°C with primers (5′-AACTAGTGTTGGATTAGTAGA-3′) and (5′-ATACATTGATTCAAGTAATCG-3′), realizing a 102-bp product. The underlined nucleotide was modified from T to C to incorporate the ClaI site in Gifu sequence. Fragments were digested with ClaI restriction enzyme and then separated on 5% NuSieve GTG agarose gel (Takara Shuzo, Kyoto) for 90 min. Visualization was carried out by using ethidium bromide.
The same conditions were used for nine F2 plants with the astray phenotype generated from a cross of the astray mutant and Gifu B-129, except that primers (5′-AGTCGGAATGAGCTCT-3′) and (5′-TAACCTGTATGCGCAATTAA-3′) were used and that the resulting fragments were digested with PshBI restriction enzyme. The underlined nucleotides were mutated from GC to AT to incorporate the PshBI site.
RT-PCR Analysis.
Total RNA was prepared from mature leaves of wild type and the astray mutant by using an RNeasy Plant Mini kit (Qiagen). RT-PCR was performed by using a OneStep RT-PCR Kit (Qiagen) as per the manufacturer's instructions. Amplification of LjBzf cDNA included 30 min of reverse transcription at 50°C, 15 of min initial PCR activation at 95°C, and 40 cycles of PCR with 30-sec denaturation at 94°C, 30-sec primer annealing at 58°C, and 1-min extension/synthesis at 72°C with primers (5′-CGAACTTGTTCGAGTTAGACACG-3′) and (5′-CTGTCTTAGCATTTGATTCTCATT-3′). PCR products were separated on 3.0% NuSieve GTG agarose gel (Takara Shuzo) for 25 min and visualized with ethidium bromide.
To analyze the LjBzf expression, total RNAs were isolated from various organs. Poly(A)+ RNA was purified by using Oligotex-MAG mRNA Purification Kit (Takara Shuzo), and then the first strand of cDNA was synthesized with Omniscript RT kit (Qiagen). Sample volumes were normalized for equal amplification of DNA fragments with primers specific for LjUbiquitin gene. PCR was then performed under the same condition except for primers specific for the astray cDNA. For semiquantitation of mRNA, we evaluated amplified DNA fragments by a series of numbers of PCR cycle. To distinguish the products amplified from mRNAs from those generated from contaminating genomic DNA, control experiments were done without reverse transcriptase. The primer pairs used were as follows: LjUBI-F (5′-ATGCAGATCTTTTGTGAAGAC-3′) and LjUBI-R (5′-ACCACCACGGAAGACGGAG-3′) for Ubiquitin, and AST-1 (5′-CGAACTTGTTCGAGTTAGACACG-3′) and AST-2 (5′-GCTGAGCTGAAACTCTGTTCC-3′) for ASTRAY.
Generation of Transgenic Plants.
The binary vector pBI121 (CLONTECH) contains the β-glucuronidase (GUS) gene and the NTPII gene, a G418-resistance marker, for plant selection. We digested plasmid pBI121 with BamHI and SacI restriction enzymes, and purified the vector (construct pBI121ΔGUS). Then we designed primers (5′-GAGAAAATGGATCCGCATG-3′) and (5′-CATAAGAGCTCAGTATTTTAG-3′) equipped with BamHI and SacI sites (underlined) and amplified cDNA LjBzf from the cDNA library from roots. The product of PCR amplification was digested with BamHI and SacI restriction enzymes and cloned into BamHI and SacI sites of pBI121ΔGUS. This pRN29 construct was sequenced and then mobilized into Agrobacterium tumefaciens strain C58:pGV2260. Transgenic plants were generated by using this A. tumefaciens strain as described by Thykjaer et al. (15). Phenotypes of the transformants were evaluated in the T2 generation. These plants were shown to carry the 35S:LjBzf construct by PCR-based cosegregation analysis.
Lateral Root Count.
Two-day-old seedlings were transferred onto 0.8% agar plate containing half-strength Gamborg's B5 medium and 1% sucrose. Roots of 4-, 5-, 7-, and 10-day-old seedlings were fixed in solution containing 10% acetic acid and 90% ethanol at room temperature overnight. Fixed samples were rinsed briefly with sterile water and immersed in 90%, 70%, 50%, and 30% ethanol successively for 30 min each at room temperature. After brief washing with sterile water, tissues were cleared in an 8:2:1 dilution of chloral hydrate/glycerol/sterile water. Cleared samples were analyzed by bright-field light microscopy.
Results
Nonsymbiotic Phenotypes of astray.
Besides the symbiotic phenotype (11), astray is notably different from the wild type in terms of appearance even in the absence of rhizobia. The astray mutation caused the elongation of hypocotyl and roots (Table 1). Elongated lateral roots spread out horizontally, indicating the alteration of gravitropic response (Table 1). The mutant name astray is derived from the agravitropic lateral roots that go “astray” against gravity. Furthermore, the mutant exhibited reduced greening in shoots and roots under the light condition (data not shown). The accumulation of anthocyanin in the hypocotyl and the shoot was also reduced in the mutant compared with the wild type (data not shown). These nonsymbiotic phenotypes of astray are very similar to those of photomorphogenic Arabidopsis mutant hy5. The initial phenotypes observed in the hy5 mutant are elongated hypocotyl, enhancement of lateral root elongation and initiation, and reduced greening (12). Unlike the hy5 mutant, however, astray did not show the enhancement of lateral root initiation (Table 2). Interestingly, we have recently discovered that nodule initiation was enhanced in astray in place of lateral root initiation (11). Whereas astray and hy5 are very similar to each other in terms of phenotypic characteristics, the astray mutation seems to be involved in a legume-specific property regarding the regulation of lateral organ development in root. Taken together, we predicted that the Lotus HY5 homologue is a good candidate for the corresponding gene of astray.
Table 1.
Comparison of hypocotyl and root length, and lateral root angles between the wild type and astray
| Parameters | Wild type | astray |
|---|---|---|
| Hypocotyl length, mm | 3.0 ± 0.6 | 5.2 ± 0.9 |
| Main root length, mm | 22.2 ± 6.4 | 60.6 ± 15.9 |
| Lateral root length, mm | 18.8 ± 15.2 | 32.8 ± 16.7 |
| Lateral root angle, ° | 19.9 ± 10.0 | 56.3 ± 17.7 |
Seedlings were grown in the absence of M. loti on agar plates containing half-strength Gamborg's B5 medium and 1% sucrose. Seventeen to 24 plants were measured for each value, and the mean and SD are presented.
Seven-day-old seedlings were analyzed.
The longest lateral root of 12-day-old seedlings was analyzed. Angles between the direction of lateral root growth and gravity were measured.
Table 2.
Number of lateral roots in the wild type and astray
| Genotype | 4 DAG | 5 DAG | 7 DAG | 10 DAG |
|---|---|---|---|---|
| Wild type | 1.20 ± 0.91 | 3.20 ± 1.03 | 3.70 ± 0.70 | 4.50 ± 2.07 |
| astray | 1.80 ± 0.42 | 3.00 ± 1.15 | 4.90 ± 1.10 | 4.50 ± 1.18 |
Ten seedlings were measured for each stage. Lateral root primordia as well as lateral roots in main root were counted, and the mean and SD are presented. DAG, days after germination.
Isolation of Lotus HY5 Homologue, LjBzf.
HY5 encodes the basic leucine zipper (bZIP) transcriptional activator that has been shown to bind with several light-regulated promoters, thereby supporting the idea that HY5 is a positive photomorphogenic regulator promoting the light developmental pattern (16). Based on the premise that the wild-type ASTRAY gene encodes a bZIP protein homologous to HY5, we set out to isolate the corresponding gene. From a database search for proteins homologous to HY5, STF1 of soybean (13) and VFZBIPZF of broad bean were found to have significant homologies to HY5. By degenerate PCR with primers designed with reference to the sequences of HY5, STF1, and VFBZIPZF, we successfully isolated the full-length cDNA of the Lotus HY5 homologue, and named it LjBzf (see Materials and Methods for detailed procedure). The name LjBzf comes from L. japonicus, bZIP, and RING-finger.
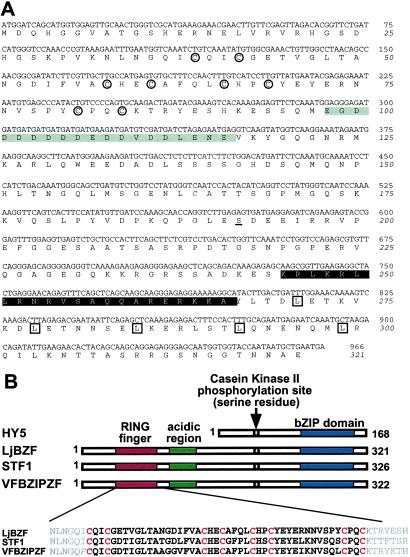
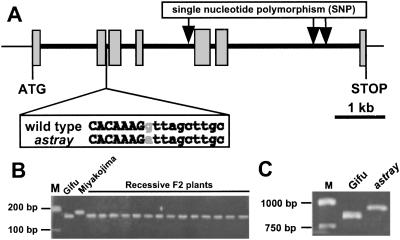
The length of the LjBzf coding region is 966 bp and covers a 5.4-kb genomic region containing seven exons (Figs. 1A and 2A). This gene encodes a 35.7-kDa protein with 321 aa and several domains, such as zinc finger, acidic region, casein kinase II (CKII) phosphorylation site (serine residue) (17) and bZIP (Fig. 2B). The cysteine repeats of the zinc-finger motif, positioned as Cys–X2–Cys–X15–Cys–X2–Cys–X4–Cys–X2–Cys–X11–Cys–X2–Cys, are very similar to those of the RING-finger motif (18). The acidic region shows tandem repeats of aspartic acid and glutamic acid. The bZIP domain has typical leucine repeats preceded by the basic region.
Fig 1.
(A) Sequence of LjBzf cDNA and the predicted protein sequence. Cysteine residues in the RING-finger domain are encircled, and the acidic region is enclosed in a gray box. Serine, the predicted CKII phosphorylation site, is underlined. The basic region and leucine residues in the bZIP domain are indicated in white type and boxed, respectively. (B) Comparison of the primary structures of HY5, LjBZF, STF1, and VFBZIPZF. The RING-finger consensus sequence is highlighted and cysteine repeats are indicated in red type.
Fig 2.
(A) Genomic structure of LjBzf. Exons are enclosed in boxes, and introns are in black lines. LjBzf comprises seven exons and six introns. The nucleotide sequence in the vicinity of the mutation site is presented. Alteration in the astray mutant is indicated in gray type. (B) Segregation analysis of the genomic DNA in the F2 population. M shows size markers. (C) RT-PCR analysis. Primers expected to amplify an 859-bp fragment within the LjBzf ORF were used.
The C-terminal half of LjBZF containing the CKII and bZIP domains is highly conserved between HY5 and LjBZF. Within the bZIP domain, LjBZF shows 81% amino acid identity with HY5. blast search in the Arabidopsis database revealed that the C-terminal half of LjBZF has the highest identity with that of HY5 at an E value of 4e−48. However, the N-terminal half including the RING-finger motif and the acidic region is not shared with HY5 (Fig. 2B). Interestingly, these domains are also present in STF1 and VFBZIPZF, and the astray mutant shows the highest identity with these proteins. Cysteine repeats of the RING-finger motif are completely conserved among the three proteins. Within the RING-finger motif, LjBZF shows 84% and 78% amino acid identities with STF1 and VFBZIPZF, respectively. The acidic regions in these three proteins are, despite the length variation, outnumbered by aspartic acid residues. Another high identity with the N-terminal half of LjBZF was detected near the N-terminal half of RSW (the cellulose synthase subunit) from Arabidopsis thaliana (19). Within the RING-finger motif, LjBZF has 65% identity with RSW. As far as we examined on the gene database, no gene showing overall similarity with LjBzf was detected except legume.
Molecular Genetic Analysis of LjBzf in astray.
To confirm the genetic linkage between astray and LjBzf, we analyzed the segregation of the astray phenotype and LjBzf by using the F2 population derived from a cross of astray (Gifu B-129 background) and Miyakojima MG-20 (14). Nucleotide polymorphism between Gifu B-129 and Miyakojima MG-20 for segregation analysis was searched for by genomic sequencing of LjBzf. There was no polymorphism between Gifu B-129 and Miyakojima MG-20 in any of the exons, but two indels in the sixth intron and one single-nucleotide polymorphism (SNP) in the fourth intron were observed (Fig. 2A). The SNP was used for further analysis.
To circumvent the problem that this SNP did not alter a recognition site for an available restriction enzyme for cleaved amplified polymorphic sequence (CAPS), a modified technique was adopted to detect the SNP. The dCAPS technique is a modified version of CAPS in that it utilizes mismatched PCR primers to create a unique restriction site based on the targeted mutation in one of the alleles (20, 21). Forty-three astray plants from the F2 population were examined, and LjBzf of the Gifu allele was observed, indicating that LjBzf and astray phenotypes are tightly linked (Fig. 2B).
Because the segregation analysis suggested the likelihood of astray being a resultant of LjBzf disruption, the genomic DNA sequences of LjBzf from the wild type and astray were compared. Sequencing the LjBzf in astray revealed no mutation in the coding region, but a G to A transition was observed inside the second intron (Fig. 2A). It appears that this transition resulted in the conversion of the splice donor site from GT to AT, thereby giving rise to an astray mRNA that retained the second intron. This proposition was verified by RT-PCR analysis (Fig. 2C). This mobility shift well corresponds to what would be expected for transcripts that fail to splice out the 83 bp of the second intron. Failure to splice out the 83 bp of the second intron would result in the premature release of the truncated LjBZF protein of 93 aa caused by the presence of a stop codon after five nucleotides of the second intron. It is reasonable to assume that this protein retains no normal function.
To see whether the astray phenotype is related to the presence of this base replacement, the segregation of the base replacement and the astray phenotype was investigated by the dCAPS technique. Nine astray plants from the F2 progeny from a cross between Gifu and astray were investigated and found to exhibit mutated polymorphism (data not shown), consistent with the prediction that a mutation in LjBzf induces the astray phenotype.
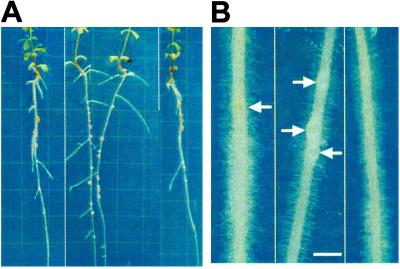
To confirm that astray results from a mutation in LjBzf, we introduced CaMV35S:LjBzf into astray. The phenotypes of the transformants were evaluated after rhizobial inoculation and growth on agar plates in the T2 generation. The transformants showed recovery of both symbiotic and nonsymbiotic phenotypes, including wild-type nodulation property, the same hypocotyl length as that found in the wild type, normal root growth, greening, and anthocyanin accumulation (Fig. 3A and Table 3). With regard to the timing of nodule initiation, primordia in the transformants appeared in almost the same manner as that in the wild type (Fig. 3B).
Fig 3.
(A) Complementation of the astray mutant with 35S:LjBzf. Plants used were the wild type, the astray mutant, and the astray mutant carrying 35S:LjBzf (from left to right). Plants were incubated on agar plates 2 weeks after inoculation. Grids were placed at 1-cm intervals. (B) Comparison of nodule development. Plant roots are the wild type, the astray mutant, and the astray mutant carrying 35S:LjBzf (from left to right). Pictures were taken 5 days after inoculation. Arrows indicate nodule primordia. (Bar = 1 mm.)
Table 3.
Complementation analysis of astray by LjBzf
| Genotype | Nodule no. | Hypocotyl length, mm | Main root length, mm | Nodulation zone, % |
|---|---|---|---|---|
| Wild type | 2.44 ± 0.25 | 1.8 ± 0.1 | 30.2 ± 1.3 | 19.80 ± 2.78 |
| astray | 5.43 ± 0.26 | 3.5 ± 0.1 | 70.1 ± 1.8 | 29.53 ± 2.08 |
| 35S:LjBzf | 1.69 ± 0.30 | 1.9 ± 0.1 | 26.2 ± 1.6 | 18.05 ± 7.66 |
Plants were analyzed 2 weeks after inoculation. Numbers of plants analyzed were 30, 27, and 15 for the wild type, the astray mutant, and astray carrying 35S:LjBzf, respectively. Ratio of nodulation zone was calculated as described in ref. 11. The mean and SD are presented.
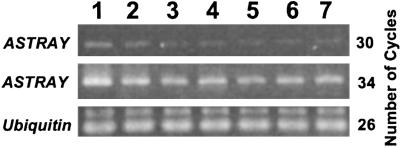
Transcripts of ASTRAY were detected in detected in leaves, cotyledons, stems, hypocotyls, roots, and nodules (Fig. 4). The amount of transcripts was relatively high in the leaves. The expression was not affected in the root by the infection of M. loti (Fig. 4, lanes 5 and 6).
Fig 4.
Accumulation of ASTRAY (LjBzf) transcripts. LjUbiquitin was used as internal control. Lanes 1–7: leaves, cotyledons, stems, hypocotyls, uninfected roots, infected roots, and 18-day nodules, respectively.
Discussion
To fully understand the mechanism by which the ASTRAY gene regulates nodule organogenesis, it is necessary to determine the nature of its encoded product. The isolated gene, designated LjBzf, encoded the CKII phosphorylation site and the bZIP motif, both of which show highest level of identity with those of HY5. In addition, LjBzf encoded the RING-finger motif and the acidic region, both of which are lacking in HY5. Interestingly, these domains were highly conserved in HY5 homologues reported from other leguminous plants, suggesting that they may be legume-characteristic combination of motifs.
Loss of ASTRAY activity caused enhanced and early nodulation, indicating that ASTRAY acts as a negative regulator of nodulation. In Arabidopsis, HY5 interacts with constitutively photomorphogenic protein 1 (COP1) for proteasome-mediated degradation to regulate its abundance negatively (22, 23). Although lateral root initiation is enhanced in the hy5 mutant, it is markedly inhibited in the cop1 mutant (24). Furthermore, transgenic plant overexpressing active HY5 that is mutated to bind DNA constitutively shows a considerable delay in lateral root initiation (25). Interestingly, the pea mutant 1ip shows similar phenotypes to the cop1 mutant and the mutation results from the partial duplication of COP1 (26–28). These findings lead us to speculate that the regulatory system via HY5-COP1 interaction is also applicable to legume plants in terms of nodulation. Because a motif (the core sequence V–P–E–F–G) responsible for the interaction with the COP1 WD-40 repeat (29) is conserved in ASTRAY, interaction with other proteins having the WD-40 repeat may also occur in ASTRAY. In this context, ccs52, recently reported from Medicago truncatula, is also worth noting. The ccs52 product shows a high homology with WD-40 repeat cell regulator protein and appears to be involved in differentiation and endoreduplication (30). It would be interesting to see whether ASTRAY and the ccs52 product inter-relate in nodule development regulation.
Among the features of ASTRAY, of particular interest are the RING-finger domain and the acidic region in the N-terminal half. Both domains are highly conserved among the three species of legumes investigated here. Given that the ASTRAY potentially functions as a transcriptional activator, the N-terminal half may serve as an activation domain that interacts with transcriptional machinery either to enhance expression or to induce specific expression (31, 32). Considering that the N-terminal half of ASTRAY shows high identity with that of RSW from A. thaliana, the cellulose synthase subunit, where the identical region is suggested to be important for protein–protein interaction, the protein–protein interaction via these domains is plausible (19).
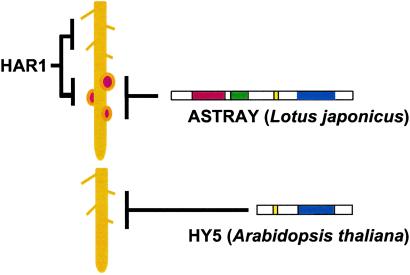
Developmentally, there exist similarities between nodule and lateral root formation. These lateral organs originate endogenously in the division of differentiated cells, namely the nodule is derived from cortical and pericycle cells (33), whereas the lateral root is from pericycle cells. Lateral root initiation is enhanced in the hy5 mutant (12). In this regard, it is of interest whether lateral root development is enhanced in astray as well as nodule development. Curiously, the results indicated that the astray mutation does not affect the frequency and timing of lateral root initiation (Table 2). For cell division to occur, the antiproliferative activities in these stem cells must be relieved. In astray, differentiated cortical cells might easily reenter the cell cycle in response to rhizobial infection, as a similar criterion was suggested in the hy5 mutant. Assuming that ASTRAY regulates cell division in a tissue-specific manner, the legume-characteristic combination of motifs may be related to the specificity. It would also be interesting to determine the structure of the ASTRAY homologue in peanut, which develops nodules from pericycle cells. In contrast to astray, the hypernodulating mutant har1 is characterized by such drastically altered root morphologies as an overall shortening of root length and abundant lateral root formation (9). This observation suggests the existence of a common regulatory element for both root and symbiotic development. In summary, the roles of HY5, HAR1, and ASTRAY are illustrated in Fig. 5. Genetic interaction will be clarified by epistatic analysis of the astray har1 double mutant. Further molecular characterization of the ASTRAY protein as a transcriptional activator will unravel the role of the ASTRAY gene in the development of nodules.
Fig 5.
A plausible role of ASTRAY in comparison with HAR1 and HY5. HY5 is involved in repression of lateral root development. HAR1 represses nodule and lateral root development. ASTRAY acts as a negative regulator of nodule development.
Acknowledgments
We thank T. Aoki at Nihon University for plasmid pBI121, T. Oyama and K. Okada at Kyoto University for A. tumefaciens strain C58, and E. Kobayashi and S. Akao at National Institute of Agrobiological Sciences for horticultural services. This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas 13017206 and Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science and Technology (Japan) (to M.K.).
Abbreviations
DAG, days after germination
CKII, casein kinase II
SNP, single nucleotide polymorphism
dCAPS, derived cleaved amplified polymorphic sequence
bZIP, basic leucine zipper
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the DDBJ database [accession nos. AB092677, ASTRAY (LjBzf) cDNA; AB092678, LjBzf genomic DNA (Gifu); AB092679, LjBzf genomic DNA (Miyakojima)].
See commentary on page 14616.
References
- 1.Carroll B. J., McNeil, D. L. & Gresshoff, P. M. (1985) Plant Physiol. 78, 34-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll B. J., McNeil, D. L. & Gresshoff, P. M. (1985) Proc. Natl. Acad. Sci. USA 82, 4162-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobsen E. & Feenstra, Q. J. (1984) Plant Sci. Lett. 33, 337-344. [Google Scholar]
- 4.Duc G. & Messager, A. (1989) Plant Sci. 60, 207-213. [Google Scholar]
- 5.Sagan M., Morandi, D., Tarenghi, E. & Duc, G. (1995) Plant Sci. 111, 63-71. [Google Scholar]
- 6.Penmetsa R. V. & Cook, D. R. (1997) Science 275, 527-530. [DOI] [PubMed] [Google Scholar]
- 7.Schauser L., Handberg, K., Sandal, N., Stiller, J., Thykjaer, T., Pajuelo, E., Nielsen, A. & Stougaard, J. (1998) Mol. Gen. Genet. 239, 414-423. [DOI] [PubMed] [Google Scholar]
- 8.Szczyglowski K., Shaw, R. S., Wopereis, J., Copeland, S., Hamburger, D., Kasiborski, B., Dazzo, F. B. & de Bruijn, F. J. (1998) Mol. Plant–Microbe Interact. 10, 59-68. [Google Scholar]
- 9.Wopereis J., Pajuelo, E., Dazzo, F. B., Jiang, Q., Gresshoff, P. M., de Bruijn, F. J., Stougaard, J. & Szczyglowski, K. (2000) Plant J. 23, 97-114. [DOI] [PubMed] [Google Scholar]
- 10.Kawaguchi M., Imaizumi-Anraku, H., Koiwa, H., Niwa, S., Ikuta, A., Shono, K. & Akao, S. (2002) Mol. Plant–Microbe Interact. 15, 17-26. [DOI] [PubMed] [Google Scholar]
- 11.Nishimura R., Ohmori, M. & Kawaguchi, M. (2002) Plant Cell Physiol. 43, 853-859. [DOI] [PubMed] [Google Scholar]
- 12.Oyama T., Shimura, Y. & Okada, K. (1997) Genes Dev. 11, 2983-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheong Y. H., Yoo, C. M., Park, J. M., Ryu, G. R., Goekjian, V. H., Nagao, A. T., Key, J. L., Cho, M. J. & Hong, J. C. (1998) Plant J. 15, 199-209. [DOI] [PubMed] [Google Scholar]
- 14.Kawaguchi M., Motomura, T., Imaizumi-Anraku, H., Akao, S. & Kawasaki, S. (2001) Mol. Genet. Genomics 266, 157-166. [DOI] [PubMed] [Google Scholar]
- 15.Thykjaer T., Schauser, L., Danielsen, D., Finneman, J. & Stougaard, J. (1998) in Cell Biology: A Laboratory Handbook, ed. Celis, J. E. (Academic, New York), pp. 518–525.
- 16.Chattopadhyay S., Ang, L. H., Puente, P., Deng, X. W. & Wei, N. (1998) Plant Cell 10, 673-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson R. B. & Kemp, B. E. (1991) Methods Enzymol. 200, 62-81. [DOI] [PubMed] [Google Scholar]
- 18.Takatsuji H. (1998) Cell. Mol. Life Sci. 54, 582-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arioli T., Peng, L., Betzner, A. S., Burn, J., Wittke, W., Herth, W., Camilleri, C., Hofte, H., Plazinski, J., Birch, R., et al. (1998) Science 279, 717-720. [DOI] [PubMed] [Google Scholar]
- 20.Michaels S. D. & Amasino, R. M. (1998) Plant J. 14, 381-385. [DOI] [PubMed] [Google Scholar]
- 21.Neff M. M., Neff, J. D., Chory, J. & Pepper, A. E. (1997) Plant J. 14, 387-392. [DOI] [PubMed] [Google Scholar]
- 22.Torii K. U., McNellis, T. W. & Deng, X. W. (1998) EMBO J. 17, 5577-5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osterlund M. T., Hardtke, C. S., Wei, N. & Deng, X. W. (2000) Nature 405, 462-466. [DOI] [PubMed] [Google Scholar]
- 24.Ang L. H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A. & Deng, X. W. (1998) Mol. Cell 1, 213-222. [DOI] [PubMed] [Google Scholar]
- 25.Hardtke C. S., Gohda, K., Osterlund, M. T., Oyama, T., Okada, K. & Deng, X. W. (2000) EMBO J. 19, 4997-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frances S., White, M. J., Edgerton, M. D., Jones, A. M., Elliottm, R. C. & Thompson, W. F. (1992) Plant Cell 4, 1519-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao L., Wang, C., Zhu, Y., Zhao, J. & Wu, X. (1998) Biochim. Biophys. Acta 1395, 326-328. [DOI] [PubMed] [Google Scholar]
- 28.Sullivan J. A. & Gray, J. C. (2000) Plant Cell 12, 1927-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holm M., Hardtke, C. S., Gaudet, R. & Deng, X. W. (2001) EMBO J. 20, 118-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cebolla A., Vinardell, J. M., Kiss, E., Olah, B., Roudier, F., Kondorosi, A. & Kondorosi, E. (1999) EMBO J. 18, 4476-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ptashne M. (1988) Nature 335, 683-689. [DOI] [PubMed] [Google Scholar]
- 32.Ptashne M. & Gann, A. (1997) Nature 386, 569-577. [DOI] [PubMed] [Google Scholar]
- 33.Timmers A. C. J., Auriac, M.-C., Truchet, A. & Truchet, G. (1999) Development (Cambridge, U.K.) 126, 3617-3628. [DOI] [PubMed] [Google Scholar]