Abstract
Photoperiod controls several responses throughout the plant life cycle, like germination, flowering, tuber formation, onset of bud dormancy, leaf abscission, and cambium activity. From these processes, flowering has been most extensively studied, especially in Arabidopsis thaliana. Photoperiod sensing by the function of photoreceptors and the circadian clock appears to regulate flowering time via Arabidopsis CONSTANS (AtCO), a putative transcription factor that accelerates flowering in response to long days. The genetic factors controlling plant photoperiodic responses other than flowering are little known. However, interspecific grafting experiments demonstrated that the flower-inducing (florigen) and tuber- inducing (tuberigen) signals are functionally exchangeable. Here we show that constitutive overexpression in potato of the Arabidopsis flowering-time gene AtCO impairs tuberization under short-day inductive conditions; AtCO overexpressing lines require prolonged exposure to short days to form tubers. Grafting experiments using these lines indicated that AtCO exerts its inhibitory effect on tuber formation by acting in the leaves. We propose that a conserved photoperiodic functional module may be involved in controlling distinct photoperiod-regulated evocation responses in different species. This module would involve the action of CONSTANS in the production of the elusive and long-distance acting florigen-tuberigen signal(s) in the leaves.
As day length increases in the spring, many plants respond by flowering [long day (LD) plants], and as day length shortens in the fall, some plants respond by flowering and others, like potato, by tuberizing [short day (SD) plants] (1–5). Flowering and tuberization are distinct reproduction strategies, both of which involve the sensing of the photoperiod and generation of a signal in the leaves (a process referred to as induction), the subsequent transport of the signal (known as florigen or tuberigen), and the response in a distant organ, the vegetative meristem, or stolon tips (also called evocation) (4). Genetic analyses of flowering in the LD plant Arabidopsis thaliana identified mutations that have been placed in three genetic pathways: an autonomous pathway, a gibberellin pathway, and a photoperiod-dependent pathway (2, 3). However, to date it has not been clarified whether these genetic components are involved in the induction mechanism in the leaf or the floral evocation in the apical meristem, two intimately related processes rarely discriminated. Arabidopsis CONSTANS (AtCO) is one of the best-studied representatives of the photoperiod-dependent pathway. Transcriptional regulation of AtCO is an important determinant of photoperiodic regulation of flowering time. AtCO overexpression in Arabidopsis results in early flowering plants that are almost completely insensitive to day length (6–9). CONSTANS-like (COL) genes have been identified in other plant species and shown to have a role in day length induction of flowering, in both SD and LD plants (10–12).
In some species of potato, like Solanum tuberosum ssp. andigena, tuberization strictly depends on photoperiod and is induced in SD (5). At least two independent pathways controlling tuber formation in potato have been proposed: a photoperiod-dependent pathway and a gibberellin-dependent pathway (13). To date, several strategies have been used to identify genes affecting tuberization (14–16), but none of the genes isolated appear to be directly involved in the photoperiodic control of tuber induction. Despite the obvious differences between flowering and tuberization, there are many similarities between them (5, 13, 17). Interspecific grafting experiments (5, 18) suggested that common factors might control these two distinct photoperiodic evocation responses. We have addressed this possibility by using AtCO as a probe for function by overexpressing it in potato. The resulting lines exhibit a delayed tuberization phenotype, suggestive of a function of CONSTANS in the photoperiodic pathway controlling tuber formation.
Materials and Methods
Plant Material and Growth Conditions.
WT photoperiodic S. tuberosum spp. andigena, anti-phytochrome B (phyB) (15), and potato AtCO (pACo) (this work) plants were used. Plants were vegetatively propagated in vitro, from tubers or through stem cuttings, with propagation methods standardized within a given experiment. After transferring to the soil, plants were grown in the greenhouse (LD), and around the fourth week from potting they were transplanted into 10-cm pots. Plants were fertilized once every 2 weeks. Under these standard conditions, they remained growing for several months without producing tubers unless transferred to SD conditions. Plant height analysis was performed with plants grown in the greenhouse or a growth chamber under LD (16 h light/8 h dark, 22°C) or SD (8 h light/16 h dark, 22°C) conditions (see below). Light intensity was about 200 μmol⋅m−2⋅s−1 and was provided by high-pressure sodium lamps SON-T AGRO 400 (Philips Belgium, Brussels).
Plasmid Generation.
Plasmid SLJ1711, containing 35S:AtCO (8), was ClaI–BamHI cut, and an isolated fragment containing the full-length AtCO cDNA downstream of the constitutive 35S promoter was subcloned into the pBIN19 binary vector to generate pBIN19–35S:AtCO. Transformation of potato plants with the pBIN19–35S:AtCO construct was carried out as described (16). More than 30 independent kanamycin-resistant plants were generated, and these primary transformants were taken for analysis of the levels of expression of AtCO.
Measurement of Stem Length, Tuber Formation, and Flowering.
Stem elongation was measured as the length of the five upper internodes, as they include the whole stem actively growing region. As indicated (13), numbers were given from the apical part of the plant: internode 1 was assigned to the uppermost internode longer than 2 mm, and the leaf located at the base of this internode was referred to as leaf 1. For tuber induction studies (Fig. 1), plants were grown for 6 weeks in the greenhouse (LD, until they had 10–15 leaves), and then transferred to a growth chamber under either LD or SD conditions. After transfer to SD, tuber examination was made with the frequency indicated (Fig. 1d and see Fig. 3). Flowering was recorded when flower buds were first visible, although the produced buds normally aborted and very rarely developed into mature flowers.
Fig 1.
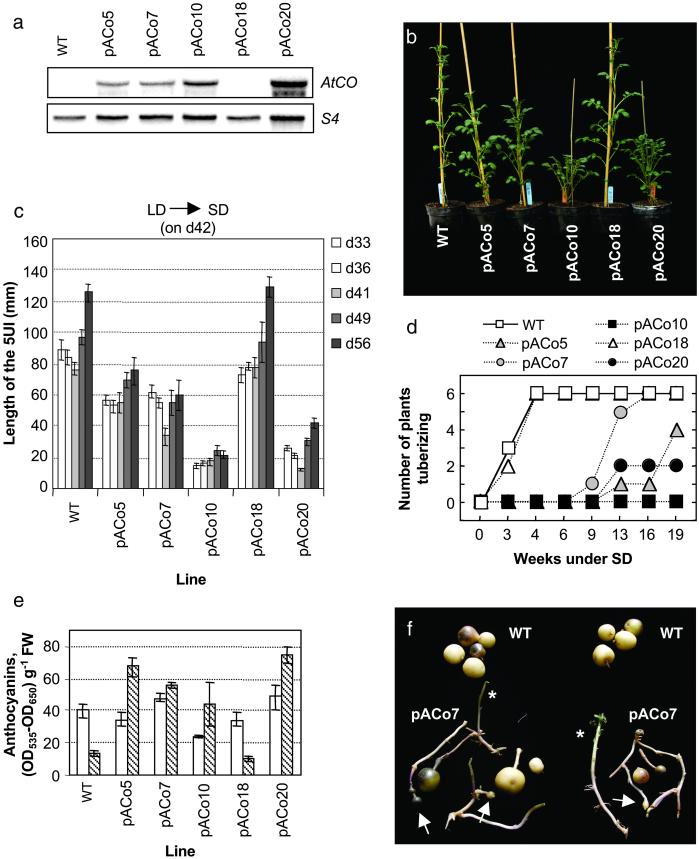
AtCO overexpression in potato. (a) RNA blot analysis of AtCO mRNA abundance in WT and pACo lines. (b) Potato WT and pACo lines grown in the greenhouse for 6 weeks. (c) Stem length of plants shown in b at different times after potting. Values are means of six individuals ± SE. 5UI, five upper internodes. (d) Time course of tuber formation in WT and pACo plants shown in b. The day of transfer to SD (day 42, indicated in c) refers to week 0 under tuber-inducing conditions. Tuberization (six individuals per line) was recorded 3–19 weeks after transfer to SD; note that the values along the abscissa are not evenly spaced. (e) Anthocyanin contents in WT and pACo leaves from plants shown in d the day of transfer to SD conditions (open bar) and 6 weeks later (hatched bar). Values are the means of five samples ± SE. (f) Tubers harvested from WT and pACo7 plants grown for 8 weeks under LD and then for 9–10 weeks under SD. Asterisks indicate the presence in the same plant of tubers and stolons not committed to tuber production. Arrows indicate the resulting morphology of tubers poorly induced to tuberize.
Fig 3.
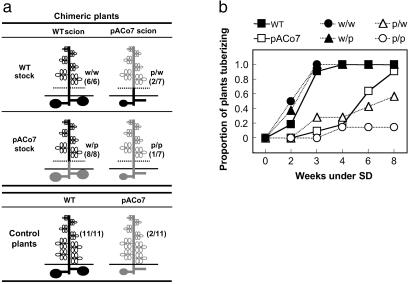
Grafting experiments between WT and pACo7 plants. (a) Diagram of the chimeric plants generated and the control plants used. Dotted lines indicate the graft union. Control plants refers to not manipulated WT (black) and pACo7 (gray) plants. The values in parentheses refer to the number of plants with tubers/total number of plants analyzed, 4 weeks after SD transfer. (b) Time course of tuber formation in chimeric and control plants shown in a. The day of transfer to SD refers to week 0; note that the values along the abscissa are not evenly spaced. w/w, WT onto WT; w/p, WT onto pACo7; p/w, pACo7 onto WT; p/p, pACo7 onto pACo7.
Grafting.
Plants were ≈6–10 weeks old when grafted (19). The transgenic line pACo7 was selected for this experiment because, as the other pACo lines, it exhibited a clear inhibition on potato tuberization (Fig. 1), but the effect on stem elongation was moderate, which made the plants easier to manipulate. The graft union was usually performed at the internode 2–3. After grafting, plants were covered with a transparent plastic bag for 4–6 days and kept in the greenhouse (LD) until successful grafts were observed (within 1 week after grafting). Chimeric plants were grown under LD conditions until the scions contained 8–15 fully developed leaves (3–5 weeks later). Then, the remaining leaves from the stock were removed, and the plants were transferred to inductive SD conditions. Grafting was repeated to obtain at least six chimeric plants per combination and, consequently, sets of chimeric plants generated through the experiment were transferred at three different times to SD conditions (when the scions had enough leaves). To control any effect of plant age, two sets of control plants (WT and pACo7, not grafted) were also transferred to SD conditions together with the first and third group of chimeric plants (see Fig. 3).
Anthocyanin Extraction and Quantification.
Anthocyanins were extracted from leaves and quantified as described (13). Each sample contained a leaflet from leaf 3–5.
Identification of Potato StCOL1 Gene.
StCOL1 (for S. tuberosum COL1) was defined by alignment of the EST sequences EST423924, EST425152, EST425164, EST460331, EST460716, and EST462225 from potato. These sequences were identified by searching in the EST databases (www.ncbi.nlm.nih.gov/BLAST/) with the AtCO query sequence.
Analysis of Gene Expression.
Total RNA was isolated from young leaves (leaves 2–4) as described (20). RNA (20 μg, Fig. 2a; or 30 μg, Figs. 1a and 2 b and c) was separated on 1.2% agarose denaturing formaldehyde gels and transferred onto Hybond N nylon membranes. Hybridization was carried out in Church buffer [125 mM Na2HPO4, pH 7.2/7% (wt/vol) SDS/1 mM EDTA] at 65°C overnight and washed three times for 20 min in 20 mM Na2HPO4 (pH 7.2), 1% (wt/vol) SDS, 1 mM EDTA at 65°C. The S4 cDNA fragment used as a constitutive probe has been described (21). The whole AtCO cDNA was PCR-amplified with two specific oligonucleotides by using the pBIN19–35S:AtCO plasmid as a template, and directly used as a probe. StCOL1 was obtained by PCR, from a cDNA library made from potato leaves and specific primers designed after the available sequence in the EST database. PCR products were subcloned into pGEM-Teasy (Promega) to give pJF273 (StCOL1) and sequenced for identity confirmation. The insert was excised by EcoRI digestion and used as a probe. DNA probes were radioactively labeled by using a random primed DNA kit (Roche Molecular Biochemicals) following the manufacturer's instructions. Images were visualized by using a Molecular Imager FX (Bio-Rad), and band intensities were quantified by using quantity one (Bio-Rad) software. Expression levels were calculated relative to the lowest value of each set of samples after normalization with the S4 control (StCOL1:S4).
Fig 2.
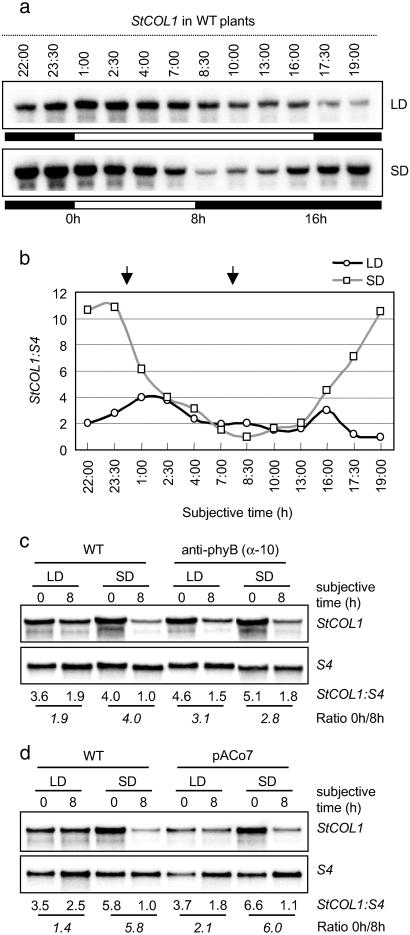
StCOL1 gene expression in WT, anti-phyB, and pACo7 lines. (a) RNA blot analysis of StCOL1 mRNA abundance in WT potato plants grown under LD or SD. Open and filled bars represent light and dark periods, respectively. Time 0h represents subjective dawn. Samples were harvested at different times of the day from plants growing for 6 weeks in greenhouse and differently entrained for 2 weeks to either LD or SD conditions. (b) Quantitative determination of the relative levels of StCOL1 transcript shown in a; reference value is at 19:00 (LD) and 8:30 (SD); note that the values along the abscissa are not evenly spaced. (c and d) StCOL1 expression pattern in WT and anti-phyB plants (α-10 line) (15) (c), and WT and pACo7 plants (d), grown under LD and SD. Samples were harvested at 0h and 8h (as indicated with arrows in b) from plants growing for 6 weeks in greenhouse and differently entrained for 1 week to either LD or SD conditions. Numbers under autoradiograms indicate relative levels of StCOL1. Numbers in italics indicate the ratio of transcript levels between 0h and 8h.
Results and Discussion
AtCO Overexpression Alters Vegetative Development and Impairs Potato Tuberization.
A search in the EST databases allowed us to identify six potato ESTs with a significant degree of homology to the AtCO protein sequence, which partly overlapped, to define one gene that we have designated StCOL1 (see Materials and Methods). The identification of this sequence suggests the existence of a CONSTANS function in potato, but gives no clues as to its role. Likewise, database searches denoted the existence of several COL genes in Arabidopsis, with two of them shown not to be redundant to AtCO (22). Because genetic approaches in potato are difficult to assess (23), we took advantage of the well-established AtCO function and studied the effect of constitutive expression of this protein in potato. This approach has the additional advantage of overcoming the absence of information on the functionality of the endogenous potato COL gene(s). In fact, heterologous overexpression of genes of defined function has proven in several cases to result in phenotypes consistent with those of homologous overexpression and complementary to those of the loss-of-function mutants (24–30). The AtCO cDNA was overexpressed in S. tuberosum ssp. andigena under the control of the constitutive 35S promoter, and the resulting transgenic lines were referred to as pACo plants. mRNA levels of AtCO expression were assessed by Northern analysis, and five lines were selected for further molecular and phenotypic characterization. pACo18 did not express AtCO at detectable levels, like WT plants; pACo5, pACo7, and pACo10 displayed intermediate levels; and pACo20 showed the highest levels of AtCO expression (Fig. 1a).
Under greenhouse conditions (LD), before any SD tuberization studies, a reduction of plant height could be observed in the pACo lines that expressed AtCO at detectable levels (Fig. 1b). In Arabidopsis, overexpression of AtCO also resulted in a reduction of the length of the main flower stem (7), although the mechanisms underlying this reduced stem elongation have not been studied. As shown in Fig. 1c, the five upper internodes of the pACo10 and pACo20 lines were about a fourth as long as those of WT plants, whereas the pACo5 and pACo7 lines showed an intermediate degree of shortening. After transferring the plants to SD for tuberization studies (on day 42 or 6 weeks from potting, Fig. 1c), the length of the five upper internode region increased in all plants, but the relative differences in height among them did not vary (Fig. 1c, compare d49 and d56). All of the lines produced between 13 and 19 nodes during the 4-week period of stem length measurement and displayed a similar root growth (not shown), indicating that the dwarf phenotype of the pACo plants results from a reduction in the internode length rather than from a general decrease in growth. Although the dwarf phenotype was observed only in the pACo lines expressing the transgene, a clear correlation between phenotypic severity and levels of expression of the transgene was not observed (Fig. 1 a and b). This finding suggests that factors other than the overall amount of AtCO transcript detected in the leaves regulate the degree of dwarfing. Taken together, our data indicate that an active AtCO protein is produced in the pACo potato plants and that it affects stem length as reported in Arabidopsis.
After transfer to SD (inductive) conditions, WT and pACo18 plants started to form tubers within 3 weeks (Fig. 1d). None of the other pACo lines produced tubers by week 6, when WT and pACo18 plants begun to show signs of senescence. After 16 weeks under SD inductive conditions, all lines but pACo10 had produced tubers (Fig. 1d). Tubers formed by these lines were typically small and poorly developed (like those shown in Fig. 1f, *), and plant senescence associated with tuber growth and development was delayed. Senescence of the aerial part of the tuberizing plant is associated with a decrease in the leaf anthocyanin levels (13). Accordingly, after 6 weeks under inducing conditions (Fig. 1d), anthocyanin levels decreased only in tuber-bearing WT and pACo18 plants (Fig. 1e). None of the WT or pACo plants produced tubers under noninducing LD conditions, even after >4 months (not shown). These results indicate that overexpression of AtCO inhibits tuberization under SD inductive conditions and, therefore, that AtCO functions as a negative regulator of photoperiod-controlled tuberization in potato, in contrast with the positive control exerted over flowering in Arabidopsis (7, 8). Flowering in potato is not under photoperiodic control (17) and, as expected, flowering time was not found to be altered in the transgenic plants (not shown). Therefore, our results suggest that CONSTANS function has a general influence on photoperiodic-controlled evocation responses, in both Arabidopsis and potato, rather than a specific effect on flowering.
It is worth mentioning that a dwarf phenotype similar to that of pACo transgenic lines has been observed in plants bearing an antisense construct for a gibberellin 20-oxidase gene, involved in gibberellin biosynthesis. The dwarf phenotype in these lines was not associated to an inhibition of tuber formation but instead resulted in early tuberization under SD conditions (21). Even more, the extremely dwarf S. tuberosum ssp. andigena ga1 mutant, exhibiting a highly reduced leaf area and very short internodes, behaves as strongly induced to tuberize and can form tubers even under LD conditions (31). This finding indicates that the dwarf phenotype of the pACo lines is not responsible for the inhibited tuberization observed in these plants and supports our conclusion that this phenotype is a direct consequence of AtCO overexpression.
AtCO Overexpression Does Not Alter Photoperiod Perception.
The pACo plants exhibit a phenotype somehow opposed to that of transgenic anti-phyB potato with reduced levels of the photoreceptor phyB (15). Whereas pACo plants are short and tuberize very late under SD inducing conditions, anti-phyB plants show an elongated phenotype and form tubers under LD noninducing conditions. It appears that the reduction of phyB levels impairs LD photoperiod perception and constitutively switches on the potato responses to SD, resulting in a slender stem and constitutive tuber formation (13). The opposite phenotypes of pACo plants raises the possibility that AtCO overexpression may constitutively switch off the responses to SD (or alternatively switch on the responses to LD) because it impairs SD perception or signaling. To address this possibility we looked for a molecular marker whose expression in the leaves is photoperiod-dependent. We observed that in WT potato leaves, StCOL1 expression fluctuates daily in a photoperiod-dependent manner (Fig. 2 a and b). In SD, mRNA levels have a peak before subjective dawn and a valley at about 8 h after lights are on. In LD-grown plants, however, the difference in the expression level ratio at those time points is attenuated (Fig. 2b). Specifically, the differences in the mRNA level ratio between 0 h and 8 h are much less pronounced in LD (1.9–1.4) than SD (4.0–5.8) (Fig. 2 c and d). This result makes the StCOL1 gene a photoperiod-dependent marker, suitable for our diagnostic purposes. When putting StCOL1 to the test in anti-phyB transgenic plants (α-10 line), a similar daily pattern of StCOL1 expression under both LD (3.1) and SD (2.8) was observed (Fig. 2c), which suggested that photoperiod perception is indeed impaired in these transgenic plants. In pACo7 plants, by contrast, StCOL1 expression follows a similar pattern than in WT (Fig. 2d), which suggested that photoperiod perception is not altered in pACo plants. Together, constitutive expression of AtCO in potato does not appear to alter photoperiod perception in the leaves but it may affect a later step in the photoperiod signaling pathway leading to tuberization. Therefore, our data suggest that AtCO overexpression uncouples photoperiod perception from tuber induction by acting somewhere in the induction or evocation responses.
AtCO Inhibits Tuberization by Acting in the Leaves.
Classical studies on flowering indicated that day-length perception in the leaves induces the production of a graft-transmissible signal that moves to the apex to activate or repress the floral evocation response (1, 32). In Arabidopsis, some of the photoperiodic flowering-time genes, such as GIGANTEA (GI) and AtCO, are expressed both in the leaves (the site of photoperiod perception) and the shoot apices (the site of photoperiod response) (6, 7, 33, 34). This fact, together with the spatial proximity between leaves and shoot apex, has impeded determining which is the specific site of action of these gene products and indeed their specific role in generating, transmitting, or perceiving the flowering signal has not been clarified to the date. Similarly to flowering, the signals that regulate tuber evocation originate outside the stolon tips, in the leaves (5, 19). But, in contrast, the spatial distance between the sites of signal production and evocation in potato offers the possibility to study AtCO site of action. Hence, to determine whether AtCO function is required in the leaves, at the stolon tips, or in both places for inhibition of tuber formation, scions from WT and pACo7 plants were grafted onto WT and pACo7 stocks (see Materials and Methods), and tuber formation under SD inductive conditions was studied in the resulting chimeras (Fig. 3a). When WT control plants were transferred to SD, all of the plants developed normal tubers by week 4 (Fig. 3b), and by week 8 all of the plants were dead (not shown). By contrast, only a few pACo7 plants had formed tubers 4 weeks after transfer to SD (Fig. 3b), and none of them were dead by week 8 (not shown). Moreover, the tubers that eventually formed on pACo7 plants were usually small and attached to alive stolons, suggesting a weak induction of tuberization compared with WT plants (Fig. 1f, arrows). The chimeras WT onto WT and pACo7 onto pACo7 tuberized as the WT and pACo7 plants, respectively (Fig. 3b). When WT scions were grafted onto pACo7 stocks (w/p), the resulting chimera tuberized like WT plants (Fig. 3b), indicating that AtCO expression in the stolons is not sufficient to inhibit tuber formation. On the contrary, when pACo7 scions were grafted onto WT stocks (p/w), the resulting chimeras tuberized as pACo7 control plants (Fig. 3b), indicating that AtCO overexpression in the leaves is sufficient to inhibit tuber formation in the underground stolons. These experiments demonstrate that AtCO functions in the leaves to impair photoperiod-controlled tuberization. This finding does not exclude that AtCO might have additional functions locally in plant development, as indicated by the fact that scions from pACo-grafted plants maintained their semidwarf phenotype (not shown). Taken together, our data indicate that AtCO acts upstream of the generation or transport of the evocation controlling signal(s), that is, the induction response, in the leaves.
A Common Photoperiodic Functional Module Controlling Different Photoperiodic-Regulated Evocation Responses in Distinct Species.
Functional modules have been proposed as fundamental elements of biological organization and regulation, and identifying, defining, and characterizing such modules are key to understanding complex biological systems (35). We claim that overexpression of AtCO, an Arabidopsis gene involved in photoperiod-regulated flowering, alters, and hence reveals, a functional module conserved in potato, which is specifically and primarily involved in controlling photoperiodic evocation responses. A formal possibility is that the identified photoperiodic module involves the action of COL orthologues in potato. Accordingly, other authors have observed that heterologous overexpression of genes of known function, such as phyA, phyB, LFY, SPY, or KN1, among others, resulted in phenotypes consistent with the intrinsic function of the gene in its homologous context. These data also brought evidence on the existence of a conserved regulatory pathway in the new host species, and in some cases the orthologue genes involved in this pathway were subsequently isolated and characterized (11, 24–30).
The photoperiodic module connections (35) should differ among species: whereas in potato the photoperiodic module would interact with a tuber control module, still to be identified, in Arabidopsis it would interact with a flowering control module(s), which is currently relatively well characterized. AtCO encodes a putative transcription factor that in Arabidopsis promotes flowering by acting in the nucleus and by directly activating the expression of specific genes, like AGL20/SOC1 and FT (7, 36). It is possible that the direct control of AGL20/SOC1 expression by AtCO in Arabidopsis mediates connections between the photoperiodic and flowering control modules (36). This finding would imply that AGL20/SOC1 themselves do not belong to the photoperiodic module but to the flowering control one, this hypothesis being in agreement with the photoperiod-responsive flowering phenotype observed in the Arabidopsis agl20/soc1 mutant (36–38). Indeed, we have observed that AtCO overexpression in potato does not affect the expression of an endogenous AGL20 gene (not shown), in agreement with our hypothesis that module connections may differ among species, with the flowering control module and therefore AGL20 in potato not being connected to the photoperiodic module.
The Role of AtCO in Producing the Evocation Controlling Signal(s).
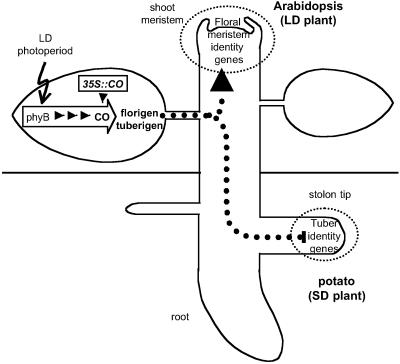
The observation that AtCO likely acts after photoperiod perception (Fig. 2) and that it functions in the leaves to inhibit tuber formation (Fig. 3) suggests that it might be involved, together with phyB (19) in the generation of the evocation controlling signal(s) in the leaves, that is, in the induction response (Fig. 4). These signals, collectively referred to as florigen or tuberigen (see above), include both the inducing and inhibiting signals that ultimately control evocation (4, 5, 17). Therefore, the relative levels of inhibiting and inducing signals would determine whether evocation occurs or not, tuberization or flowering being possible by either an increase in the inducer or a reduction in the inhibitor (17). Grafting of tobacco scions derived from SD, LD, or neutral-day species induced tuberization onto potato stocks when the photoperiodic conditions induced flowering in the corresponding scion (5, 18), indicating that perception of the inducing photoperiod in both LD and SD plants results in the same evocation signal(s) outcome. However, our results suggest that the molecular role of AtCO in this outcome is opposed depending on whether the species is a SD or a LD plant: AtCO overexpression results in flowering in the Arabidopsis LD plant and nontuberization in the potato SD plant. Based on these results, AtCO seems to have intrinsic antagonistic effects in these LD and SD species. Interestingly, AtCO and Hd1, an orthologue of AtCO in the SD plant rice, also display antagonistic activities under LD (10), suggesting that the mechanisms by which these factors may antagonize each other are intrinsic to the protein. More work is needed to ascertain whether this antagonistic function can be generalized to other LD and SD plant species.
Fig 4.
Model of the role of CONSTANS in photoperiodic responses in the whole plant context. AtCO identifies a leaf-acting functional module (empty arrow) involved in photoperiod perception and ulterior florigen/tuberigen signal(s) generation. Overexpression of AtCO (35S:CO) impairs photoperiodic responses. The molecular role of AtCO in the functional module is, however, antagonistic in both types of species: AtCO results in the production of inducing signals in the LD Arabidopsis and inhibitory signals in the SD potatoes.
In summary, we present evidence for the existence of common genetic pathways that control distinct photoperiod regulated processes in different plants and suggest that potato and Arabidopsis have evolved a common and conserved regulatory photoperiodic functional module, defined by AtCO, which acts in the leaves to control flowering and tuberization, two distinct evocation responses. Studying the intrinsic activity of the different AtCO orthologues may help to understand the mechanisms used by SD and LD plants to couple their evocation responses to photoperiod. Defining the function of the primary AtCO target genes in different species, on the other hand, may help to elucidate the molecular identity of the elusive mobile signals controlling flower and tuber formation.
Acknowledgments
We are grateful to G. Coupland for supplying the AtCO cDNA construct; the anonymous referees for their constructive comments; M. Badías, P. Fontanet, and A. Sanz for plant care; A. Sánchez for help with plant pictures; S. D. Jackson for advice on potato grafting; C. Romera for technical help; and M. Boter, J. Casacuberta, J. Casanova, D. Ludevid, M. Rodríguez-Concepción, and P. Suárez-López for helpful comments and suggestions after critical reading of the manuscript. This work was supported by Plan Nacional grants from the Comisión Interministerial de Ciencia y Tecnología (BIO96-0532-C02-02 and BIO99-0961) (to S.P.). J.F.M.-G. thanks the Spanish Ministry of Science and Technology and Institució Catalana de Recerca i Estudis Avançats for funding.
Abbreviations
LD, long day
SD, short day
AtCO, Arabidopsis CONSTANS
COL, CONSTANS-like
pACo, potato AtCO
phyB, phytochrome B
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bernier G., Havelange, A., Houssa, C., Petitjean, A. & Lejeune, P. (1993) Plant Cell 5, 1147-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koornneef M., Alonso-Blanco, C., Peeters, A. J. M. & Soppe, W. (1998) Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 345-370. [DOI] [PubMed] [Google Scholar]
- 3.Piñeiro M. & Coupland, G. (1998) Plant Physiol. 117, 1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas B. (1998) in Biological Rhythms and Photoperiodism in Plants, eds. Lumsden, P. J. & Millar, A. J. (BIOS Scientific, Oxford), pp. 151–165.
- 5.Jackson S. D. (1999) Plant Phys. 119, 1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puterill J., Robson, F., Lee, K., Simon, R. & Coupland, G. (1995) Cell 80, 847-857. [DOI] [PubMed] [Google Scholar]
- 7.Simon R., Igeño, M. I. & Coupland, G. (1996) Nature 384, 59-62. [DOI] [PubMed] [Google Scholar]
- 8.Onouchi H., Igeño, M. I., Périlleux, C., Graves, K. & Coupland, G. (2000) Plant Cell 12, 885-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suárez-López P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F. & Coupland, G. (2001) Nature 410, 1116-1119. [DOI] [PubMed] [Google Scholar]
- 10.Yano M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., Baba, T., Yamamoto, K., Umehara, Y., Nagamura, Y. & Sasaki, T. (2000) Plant Cell 12, 2473-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J., Yu, J., McIntosh, L., Kende, H. & Zeevaart, J. A. D. (2001) Plant Phys. 125, 1821-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samach A. & Gover, A. (2001) Curr. Biol. 11, R651-R654. [DOI] [PubMed] [Google Scholar]
- 13.Martínez-García J. F., García-Martínez, J. L., Bou, J. & Prat, S. (2001) J. Plant Growth Regul. 20, 377-386. [DOI] [PubMed] [Google Scholar]
- 14.van den Berg J. H., Ewing, E. E., Plaisted, R. L., McMurry, S. & Bonierbale, M. W. (1996) Theor. Appl. Genet. 93, 307-316. [DOI] [PubMed] [Google Scholar]
- 15.Jackson S. D., Heyer, A., Dietze, J. & Prat, S. (1996) Plant J. 9, 159-166. [Google Scholar]
- 16.Amador V., Monte, E., García-Martínez, J. L. & Prat, S. (2001) Cell 106, 343-354. [DOI] [PubMed] [Google Scholar]
- 17.Jackson S. & Thomas, B. (1997) Plant Cell Environ. 20, 790-795. [Google Scholar]
- 18.Ewing E. E. & Struik, P. C. (1992) Hortic. Rev. 14, 89-198. [Google Scholar]
- 19.Jackson S. D., James, P., Prat, S. & Thomas, B. (1998) Plant Phys. 117, 29-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodríguez-Concepción M. & Gruissem, W. (1999) Plant Physiol. 119, 41-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carrera E., Bou, J., García-Martínez, J. L. & Prat, S. (2000) Plant J. 22, 247-256. [DOI] [PubMed] [Google Scholar]
- 22.Ledger S., Strayer, C., Ashton, F., Kay, S. A. & Putterill, J. (2001) Plant J. 26, 15-22. [DOI] [PubMed] [Google Scholar]
- 23.Fernie A. R. & Willmitzer, L. (2001) Plant Physiol. 127, 1459-1465. [PMC free article] [PubMed] [Google Scholar]
- 24.Lincoln C., Long, J., Yamaguchi, J., Serikawa, K. & Hake, S. (1994) Plant Cell 6, 1859-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quail P. H., Boylan, M. T., Parks, B. M., Short, T. W., Xu, Y. & Wagner, D. (1995) Science 268, 675-680. [DOI] [PubMed] [Google Scholar]
- 26.Weigel D. & Nilsson, O. (1995) Nature 377, 495-500. [DOI] [PubMed] [Google Scholar]
- 27.Chuck G., Lincoln, C. & Hake, S. (1996) Plant Cell 8, 1277-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitelam G. C., Patel, S. & Devlin, P. F. (1998) Philos. Trans. R. Soc. London B 353, 1445-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izhaki A., Swain, S. M., Tseng, T.-s., Borochov, A., Olszewski, N. E. & Weiss, D. (2001) Plant J. 28, 181-190. [DOI] [PubMed] [Google Scholar]
- 30.Peña L., Martín-Trillo, M., Juárez, J., Pina, J. A., Navarro, L. & Martínez-Zapater, J. M. (2001) Nat. Biotechnol. 19, 263-267. [DOI] [PubMed] [Google Scholar]
- 31.van den Berg J. H., Simko, I., Davis, P. J., Ewing, E. E. & Halinska, A. (1995) J. Plant Physiol. 146, 467-473. [Google Scholar]
- 32.Colasanti J. & Sundaresan, V. (2000) Trends Plant Sci. 25, 236-240. [Google Scholar]
- 33.Fowler S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., Coupland, G. & Putterill, J. (1999) EMBO J. 18, 4679-4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park D. H., Somers, D. E., Kim, Y. S., Choy, Y. H., Lim, H. K., Soh, M. S., Kim, H. J., Kay, S. A. & Nam, H. G. (1999) Science 285, 1579-1582. [DOI] [PubMed] [Google Scholar]
- 35.Hartwell L. H., Hopfield, J. J., Leibler, S. & Murray, A. W. (1999) Nature 402,Suppl., C47-C52. [DOI] [PubMed] [Google Scholar]
- 36.Samach A., Onouchi, A., Gold, S. E., Ditta, G. S., Schwarz-Sommer, Z., Yanofsky, M. F. & Coupland, G. (2000) Science 288, 1613-1616. [DOI] [PubMed] [Google Scholar]
- 37.Borner R., Kampmann, G., Chandler, J., Gleißner, R., Wisman, E., Apel, K. & Melzer, S. A. (2000) Plant J. 24, 591-599. [DOI] [PubMed] [Google Scholar]
- 38.Lee H., Suh, S.-S., Park, E., Cho, E., Ahn, J. H., Kim, S.-G., Lee, J. S., Kwon, Y. M. & Lee, I. (2000) Genes Dev. 14, 2366-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]