Abstract
Background
When generating transformed plants, a first step in their characterization is to obtain, for each new line, an estimate of how many copies of the transgene have been integrated in the plant genome because this can deeply influence the level of transgene expression and the ease of stabilizing expression in following generations. This task is normally achieved by Southern analysis, a procedure that requires relatively large amounts of plant material and is both costly and labour-intensive. Moreover, in the presence of rearranged copies the estimates are not correct. New approaches to the problem could be of great help for plant biotechnologists.
Results
By using a quantitative real-time PCR method that requires limited preliminary optimisation steps, we achieved statistically significant estimates of 1, 2 and 3 copies of a transgene in the primary transformants. Furthermore, by estimating the copy number of both the gene of interest and the selectable marker gene, we show that rearrangements of the T-DNA are not the exception, and probably happen more often than usually recognised.
Conclusions
We have developed a rapid and reliable method to estimate the number of integrated copies following genetic transformation. Unlike other similar procedures, this method is not dependent on identical amplification efficiency between the PCR systems used and does not need preliminary information on a calibrator. Its flexibility makes it appropriate in those situations where an accurate optimisation of all reaction components is impossible or impractical. Finally, the quality of the information produced is higher than what can be obtained by Southern blot analysis.
Background
Genetic transformation of plants has become a routine procedure in both basic and applied research. In fact, this technology has been widely exploited to study the physiology of plants (biochemical pathways, resistance to pathogens, reaction to stress), as well as to obtain commercial crops with improved agronomic characters (herbicide tolerance, insect resistance, etc.) and, more recently, to develop new types of plants as bio-reactors (pharmaceuticals, vaccines, nutraceuticals, etc.) [1].
Regardless of the scope of the transformation, when new transgenic plants are obtained an early and essential step is their molecular characterization. The reason for analysing many primary transformants (T0) resides in the mechanism of integration itself: since the new DNA is inserted at random in the plant genome, plants with one to several integrated copies are obtained, and the multiple copies can be found in one or more chromosome locations. Usually plants where one or two integration events have occurred are those with the highest level of expression of the new gene. Low and sometimes unstable expression of transgenes has been related with high copy number and subsequent transgene silencing [2,3]. It is therefore clear that the T0 plantlets have to be analysed as soon as possible, so that only the most interesting ones are taken through the steps of acclimatation in soil, flowering, seed production, etc.
Transgene copy number is usually estimated by Southern analysis, a classic molecular biology method. This procedure provides an indication of the number of integrated copies, but is quite costly in terms of reagents, labour, and time, and it requires a relatively large amount of plant material to start with. Moreover, in the presence of rearranged copies (with loss of restriction sites), the estimates are not correct.
In this report we have used a different strategy to estimate transgene copy number in T0 plants, the quantitative real-time PCR, and compared the results with those of Southern analysis.
Real-time PCR has made it possible to accurately quantify starting amounts of nucleic acid during the PCR reaction without the need for post-PCR analyses. A fluorescent reporter is used to monitor the PCR reaction as it occurs. The reporter can be of a nonspecific nature (such as SYBR Green I) or of a specific nature (such as TaqMan probes, molecular beacons, FRET probes). In a TaqMan assay [4], the probe is labeled at the 5' end with a fluorescent reporter molecule and at the 3 ' end with another fluorescent molecule, which acts as a quencher for the reporter. When the two fluorophores are fixed at opposite ends of the 20–30 nt probe and the reporter fluorophore is excited by an outside light source, the normal fluorescence of the reporter is absorbed by the nearby quencher, and no reporter fluorescence is detected. When Taq polymerase encounters the bound probe during extension from one of the primers, it digests the probe by its 5' exonuclease activity, freeing the reporter from the quencher, and the reporter fluorescence can be detected and measured [5]. The fluorescence of the reporter molecule increases as products accumulate with each successive round of amplification.
With real-time PCR, results can be obtained quickly and can be subjected to statistical analysis. Since quantitative data on distinct sequences of the T-DNA can be obtained, lines with possible rearrangements are much easier to recognize than with Southern analysis.
Results and Discussion
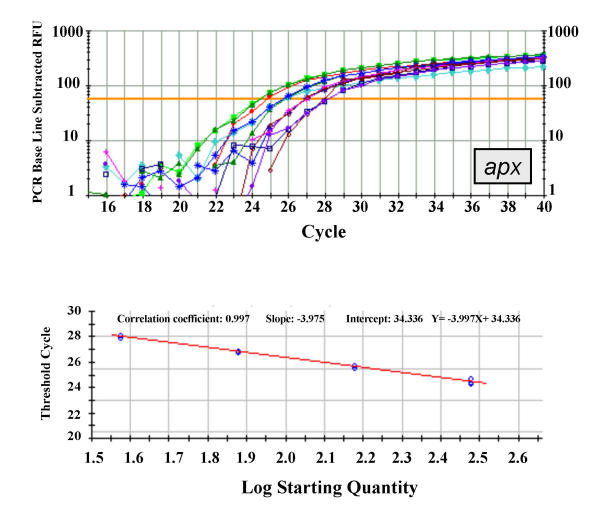
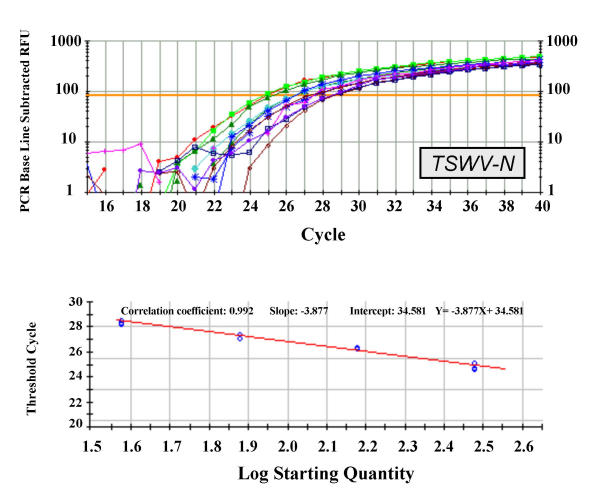
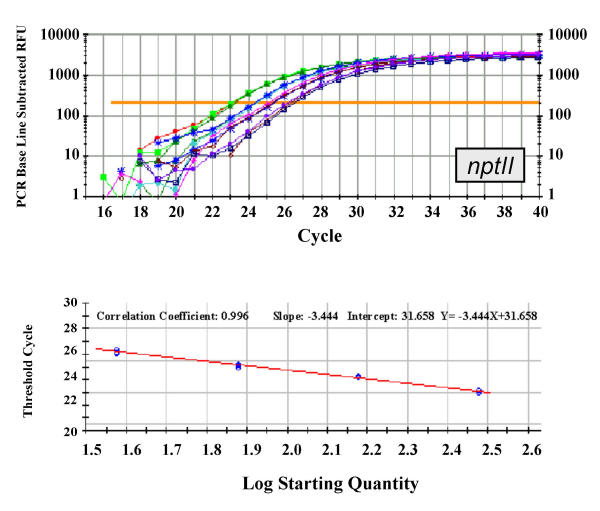
In analysing tomato transgenic plants three genes were considered, one endogenous (tomato ascorbate peroxidase, apx) and two transgenic ones, the nucleocapsid gene of Tomato spotted wilt virus (TSWV-N) and the neomycin phosphoril-transferase II gene (nptII). A total of six experiments (two experiments for each gene) were conducted and standard curves, obtained from serial dilutions of a transgenic line, were produced using the Bio-Rad iCycler software. In all experiments samples were run in triplicate. The correlation coefficients of the standard curves were rather good, being in the range between 0.990 and 0.997. Representative curves for the apx, TSWV-N and nptII are shown in Figures 1, 2 and 3, respectively.
Figure 1.
Real-time PCR amplification and standard curve of apx gene. Upper panel: real-time PCR logarithmic plot resulting from the amplification of four three-fold serial dilutions of a tomato standard DNA (see Methods for details) using the apx-specific primers and probe described in Table 5. Each sample was run in triplicate. Lower panel: Standard curve obtained for the same samples. Correlation coefficient and slope values are indicated. The calculated TC values were plotted versus the log of each starting quantity.
Figure 2.
Real-time PCR amplification and standard curve of TSWV-N transgene. Upper panel: real-time PCR logarithmic plot resulting from the amplification of four three-fold serial dilutions of a tomato standard DNA (see Methods for details) using the TSWV-N-specific primers and probe described in Table 5. Each sample was run in triplicate. Lower panel: Standard curve obtained for the same samples. Correlation coefficient and slope values are indicated. The calculated TC values were plotted versus the log of each starting quantity.
Figure 3.
Real-time PCR amplification and standard curve of nptII transgene. Upper panel: real-time PCR logarithmic plot resulting from the amplification of four three-fold serial dilutions of a tomato standard DNA (see Methods for details) using the nptII-specific primers and probe described in Table 5. Each sample was run in triplicate. Lower panel: Standard curve obtained for the same samples. Correlation coefficient and slope values are indicated. The calculated TC values were plotted versus the log of each starting quantity.
From the standard curves the starting quantities of each gene in each tomato line was determined, again with the iCycler software. The results of the two experiments (six measurements) conducted on each gene were combined and are shown in Table 1 together with their 95% confidence intervals. Most of the data fall inside the upper and lower limits of the standard curves, except one case for apx, and three cases for TSWV-N and nptII. Since the real-time PCR technique is known for producing linear response over a wide range of starting concentrations [6], and these few data are not too distant from the extreme values used to produce the standard curves, we considered them acceptable.
Table 1.
Calculated starting quantities of the three genes for each transgenic tomato line. Starting quantities (SQ), as calculated by the Bio-Rad software, are expressed in arbitrary units (a.u.), with their 95% confidence intervals (δSQ) indicated aside.
| starting quantities (a.u.) | |||
| line | apx | TSWV-N | nptII |
| 111–6 | 238 ± 29 | 54.6 ± 6.4 | 81.9 ± 5.5 |
| 99–1 | 230 ± 39 | 51.5 ± 7.4 | 78.7 ± 8.6 |
| 1–2 c | 328 ± 28 | 75.1 ± 13 | 194 ± 6.3 |
| 110–1 | 241 ± 14 | 114 ± 17 | 173 ± 8.8 |
| 46–1 | 72 ± 6.2 | 25.8 ± 8.2 | 17.5 ± 6.1 |
| 80–1 | 194 ± 15 | 75.3 ± 8.2 | 148 ± 22 |
| 127–1 | 98.8 ± 6.9 | 46.7 ± 10 | 115 ± 10 |
| 133–1 | 140 ± 13 | 100 ± 10 | 122 ± 6.0 |
| 1–1 | 156 ± 28 | 92.9 ± 13 | 99.8 ± 7.3 |
| 30–4 | 112 ± 24 | 86.4 ± 10 | 94.2 ± 6.4 |
| 113–12 | 218 ± 24 | 169 ± 25 | 263 ± 22 |
| 20–2 | 167 ± 11 | 310 ± 67 | 491 ± 42 |
| 197–1 | 277 ± 38 | 524 ± 108 | 741 ± 69 |
| 118–2 | 124 ± 13 | 0 | 191 ± 18 |
| Hom.110–1 | 130 ± 14 | 123 ± 12 | 184 ± 33 |
To estimate the number of transgene copies in each tomato line, the rline values of the ratio between transgenic and endogenous starting quantities were calculated (Table 2), and from these rline values the "virtual calibrator" r1 was calculated, which is the value of such ratio corresponding to one copy of transgene. The virtual calibrator is a weighted combination of all the lines under study, the weight given to each line depending on the accuracy of the determination of rline. This procedure does not require to have a real "calibrator" line, identified with an independent test. As a consequence it allows to perform analysis of copy number even in the absence of previous knowledge on the transgenic lines, provided at least one line among all those analysed contains one copy.
Table 2.
Calculated copy number for TSWV-N and nptII transgenes. For each line and for each gene system, the rline ratio (SQtrans /SQend) is shown together with its 95% confidence interval (δrline) in the left column. The calculated copy number, obtained as the ratio between rline and the virtual calibrator r1 is shown in the right column, together with its 95% confidence interval.
| TSWV-N | nptII | |||
| Line | rline | rline/r1 | rline | rline/r1 |
| 111–6 | 0.23 ± 0.04 | 1.10 ± 0.19 | 0.34 ± 0.05 | 1.10 ± 0.16 |
| 99–1 | 0.22 ± 0.05 | 1.05 ± 0.24 | 0.34 ± 0.07 | 1.10 ± 0.23 |
| 1–2 c | 0.23 ± 0.04 | 1.10 ± 0.19 | 0.59 ± 0.05 | 1.90 ± 0.16 |
| 110–1 | 0.47 ± 0.07 | 2.24 ± 0.33 | 0.72 ± 0.06 | 2.32 ± 0.19 |
| 46–1 | 0.36 ± 0.12 | 1.71 ± 0.57 | 0.24 ± 0.09 | 0.77 ± 0.29 |
| 80–1 | 0.39 ± 0.05 | 1.86 ± 0.24 | 0.76 ± 0.13 | 2.45 ± 0.42 |
| 127–1 | 0.47 ± 0.11 | 2.24 ± 0.52 | 1.16 ± 0.13 | 3.74 ± 0.42 |
| 133–1 | 0.72 ± 0.10 | 3.43 ± 0.48 | 0.87 ± 0.09 | 2.81 ± 0.29 |
| 1–1 | 0.60 ± 0.13 | 2.86 ± 0.62 | 0.64 ± 0.12 | 2.06 ± 0.39 |
| 30–4 | 0.77 ± 0.19 | 3.67 ± 0.90 | 0.84 ± 0.19 | 2.71 ± 0.61 |
| 113–12 | 0.77 ± 0.14 | 3.67 ± 0.67 | 1.20 ± 0.17 | 3.87 ± 0.55 |
| 20–2 | 1.86 ± 0.41 | 8.86 ± 1.95 | 2.94 ± 0.31 | 9.48 ± 1.00 |
| 197–1 | 1.90 ± 0.47 | 9.05 ± 2.24 | 2.68 ± 0.44 | 8.65 ± 1.42 |
| 118–2 | - | - | 1.54 ± 0.22 | 4.97 ± 0.71 |
| Hom.110–1 | 0.95 ± 0.14 | 4.52 ± 0.67 | 1.42 ± 0.30 | 4.58 ± 0.97 |
The copy number for each line was determined as rline/r1 (see Table 2). Of course the copy number determined in this way is a real number: our estimate for the actual, integer copy number is the range of integers that are included in the 95% confidence interval around the ratio rline/r1. In the case of TSWV-N, this range included at least one integer in all lines. For two lines (110–1 and 80–1) in the case of the nptII transgene there is actually no integer included in such interval, so we quote as our estimate of the copy number the integer closest to the rline/r1.
By comparing the results for each line (see Table 3), it appears that in some cases the number of integrated copies of the two transgenes is the same, but in others it is not. This indicates that rearrangements have occurred in the T-DNA during the process of integration in plant chromosomes, and the integrity of the transformation cassette, which included both TSWV-N and nptII genes, was not preserved. This is the case for lines 1–2 c, 46–1, 127–1, 1–1 and 118–2. Line 118–2 appears as an extreme case, where 5 copies of nptII gene have been integrated, but not even one copy of TSWV-N gene. In 5 lines the integration appears to have occurred without loss of one of the transgenes: 111–6, 99–1, 110–1, 80–1 and 133–1. For the remaining four lines, all with estimates of 3 or more copies, no conclusion can be drawn. All this information on rearrangements due to loss of one of the transgenes is usually not available when classical Southern analysis is performed, since normally only the transgene of interest is considered, and not the selectable marker gene.
Table 3.
Real-time PCR estimates of copy number for TSWV-N and nptII transgenes and possible rearrangements. For each line and each gene the estimate of copy number was derived from values in Table 2, as the range of integers that are included in the 95% confidence interval around the ratio rline/r1. The column on the right indicates evidence for rearrangements in the T-DNA.
| Line | TSWV-N | nptII | rearranged |
| 111–6 | 1 | 1 | No |
| 99–1 | 1 | 1 | No |
| 1–2 c | 1 | 2 | Yes |
| 110–1 | 2 | 2 * | No |
| 46–1 | 2 | 1 | Yes |
| 80–1 | 2 | 2 * | No |
| 127–1 | 2 | 4 | Yes |
| 133–1 | 3 | 3 | No |
| 1–1 | 3 | 2 | Yes |
| 30–4 | 3–4 | 3 | ? |
| 113–12 | 3–4 | 4 | ? |
| 20–2 | 7–10 | 9–10 | ? |
| 197–1 | 7–11 | 8–10 | ? |
| 118–2 | 0 | 5 | Yes |
| Hom.110–1 | 2 § | 2 § | No |
* As no integer in the 95% confidence interval was found, the nearest integer to rline/r1 was chosen. §For the homozigous 110–1 line, containing the double amount of transgene, the copy number was calculated as [(rline/r1)/2].
When a line was analysed both as primary transformant, which carries the new DNA only on one allele, and homozygous progeny, carrying it on both alleles, the values obtained for the progeny were, as expected, twice those obtained for the T0. This represents a fast and easy way to distinguish, in the T1 progeny of self-pollinated transformed plants with single insertion site, which plants are homozygous for the transgene from those which are heterozygous. This task is normally accomplished by a time consuming procedure, that requires months, of obtaining and analysing the T2 progeny from a number of T1 plants. This apparently marginal further use of the real-time PCR in transgenic work should not be underestimated, since it can be of great help.
In the case of the TSWV-N gene, data from real-time PCR were compared with data from Southern analysis (Table 4). Initially, the DNAs from the lines were digested with KpnI, a single-cutter in the T-DNA, and analysed with a TSWV-N-specific probe. The results of this first analysis indicated that the two techniques agreed only in 3 cases (111–6, 1–2 c and 118–2). In the attempt to understand the causes of such discrepancies, a deeper Southern analysis was therefore performed, cutting the plant DNAs with other restriction enzymes, again single-cutters, but located in different parts of the T-DNA (Table 4 right column, and unpublished results). The results show that Southern analysis, even if performed extensively on the T-DNA, can produce data of difficult interpretation. However, by comparing the real-time PCR estimates of copy number with those obtained with the extended Southern analysis, most of the differences observed initially can be explained. In fact, in most cases (111–6, 110–1, 46–1, 80–1, 133–1, 30–4, 113–12, 20–2, 197–1, 118–2) the highest value estimated by Southerns is equal or lower to the real-time PCR estimate. There are several reason for a Southern analysis to underestimate the transgene copy number: insertion of more than one T-DNA copies in one locus, deletions of sequences outside the coding parts of genes but containing the restriction site used for the analysis (actually, KpnI is located between the 3'-end of the TSWV-N gene and the left border), generation of DNA fragments of very similar size that are not resolved on the gels, etc.
Table 4.
Comparison between copy number values as estimated by real-time PCR and Southern blotting. The results of estimates of TSWV-N transgene copy number obtained with real-time PCR are compared with those derived from Southern analysis, either limited to one restriction enzyme (KpnI) or extended to several ones.
| Line | real-time PCR | Southern (KpnI) | Southern (extended) |
| 111–6 | 1 | 1 | 1 |
| 99–1 | 1 | 2 | 1–2 |
| 1–2 c | 1 | 1 | 1–2 |
| 110–1 | 2 | 1 | 1–2 |
| 46–1 | 2 | 1 | 1–2 |
| 80–1 | 2 | 1 | 1–2 |
| 127–1 | 2 | 4 | 4–7 |
| 133–1 | 3 | 2 | 2–3 |
| 1–1 | 3 | n.t.* | n.t. |
| 30–4 | 3–4 | 1 | 1–4 |
| 113–12 | 3–4 | 2 | 2–4 |
| 20–2 | 7–10 | 4 | 4–8 |
| 197–1 | 7–11 | 4–6 | 3–6 |
| 118–2 | 0 | 0 | 0 |
| Hom.110–1 | 2 | 1 | 1–2 |
* not tested
For 3 lines (99–1, 1–2 c and 127–1) the real-time PCR estimates were lower than those produced by the Southerns. In those cases deletions or rearrangements probably affected the short 79bp-long sequence recognized by the real-time PCR primers and probe. This sequence does not include recognition sites of the enzymes used in Southern analysis. Alternatively, partial digests may have artifactually increased the estimates obtained by Southern analysis.
As a concluding remark on the comparison between the two methods, it is possible to state that multiple insertions, rearrangements, partial digests and a subjective evaluation of bands on the films are the most important causes that may produce a wrong estimation of copy number by Southern analysis. On the other hand, rearrangements are the only reason for real-time PCR to be unable to detect the presence of a copy of a gene.
Conclusions
The standard curve is the key element for the quantitative assay: since it is based on the standard DNA used, the choice and the preparation of this DNA is extremely important. One of the proposed methods to prepare the standard DNA consists in mixing plant DNA with a plasmid carrying the transgene [7], producing, for example, a solution with one copy of transgene per plant genome. This approach, however, introduces several sources of error that cannot be controlled: previous absolute quantification of both plant and plasmid DNA is necessary, together with precise knowledge of the nuclear genome size of the plant species to be assayed. Unfortunately for most plants only approximate estimates are available. All these problems were by-passed by simply taking the DNA of one transgenic line and using its dilutions to construct the standard curve. As shown above, results of the quantification are then used to build a "virtual calibrator", and finally to estimate the transgene copy number for each line. As an alternative to the standard curve method, relative quantification can also be achieved with the method named "comparative CT" or "delta-deltaCT" [8]. This method has the advantage of not requiring the construction of a standard curve for each experiment, but requires a validation experiment to demonstrate that reaction efficiencies for transgenes (two in our case) and endogenous gene are identical or at least very close [9]. Since reaction efficiencies were good but not identical for the TSWV-N, nptII and apx systems, it would be incorrect to use this method without performing further optimization of the systems, such as testing several combinations of primer concentration, Mg concentration, etc., without any guarantee of finding the right conditions for the three systems. Furthermore, every time a new gene, both transgene or endogenous, is studied the extended optimization must be repeated. The method used in the present work, when compared to other proposed methods [7,9] is more flexible, requiring the least amount of optimization and validation, and, when accompanied by statistical analysis, can be considered an efficient and reliable procedure for estimating the transgene copy number. It can also be used as an indication of the integrity of the DNA transferred in the transgenic lines. In fact, by measuring not only the TSWV-N, but also a second trait (the nptII in our case) present in the T-DNA, we demonstrated that in several lines the transformation cassette must have undergone some kind of modification during the integration process in the plant nuclear DNA. Therefore rearrangements during integration appear to be relatively frequent events, and may never be recognized if only a limited molecular analysis, such a single Southern blot, is performed.
Methods
Transgenic plants and DNA preparation
The transgenic tomato plants used in this work were generated via Agrobacterium tumefaciens-mediated transformation [10] with a binary plasmid containing in its T-DNA an expression cassette for the TSWV-N gene, together with a second cassette for expression of the nptII gene, as selectable marker [11]. The genomic DNA was isolated from the primary transformants (T0) lines, using 1 g of leaf material, with the CTAB method, as described by [12] and not quantified. In the case of line 110–1 homozygous T2 progeny (Hom. 110–1) was also analysed.
Outline of the method
For monitoring the real-time PCR reactions we used the Bio-Rad i-Cycler System, with specific fluorescent oligonucleotide probes (TaqMan probes, PE Biosystems) [4]. This assay exploits the 5' exonuclease activity of Taq polymerase to cleave a labeled hybridization probe during the extension phase of PCR [5]. The fluorescence of the reporter molecule increases as products accumulate with each successive round of amplification. The point at which the fluorescence rises appreciably above the background has been called the threshold cycle (TC), and there is a linear relationship between the log of the starting amount of a template and its TC during real-time PCR. Given known starting amounts of the target nucleic acid, a standard curve can be constructed by plotting the log of starting amounts versus the corresponding TCs. This standard curve can then be used to determine the starting amount for each unknown template based on its TC and the efficiency of reaction.
For the purposes of our experiments we used a standard DNA stock solution extracted from one of the transgenic tomato lines. The DNA concentration of this solution was approximately 300 ng/μl (estimated by UV-spectrophotometry). However, since this measurement is not precise per se, and, in the case of relative quantitation, this data is not relevant, we preferred to use arbitrary units (a.u.) in this work. From the standard DNA stock solution accurate three-fold serial dilutions were prepared and utilized to obtain the standard curves necessary for relative quantification of an endogenous gene and two transgenes.
For quantitation normalized to an endogenous control, standard curves are prepared for both the transgene and the endogenous gene. For each tomato line to be tested (experimental sample), the amount of transgene and endogenous gene is determined from the appropriate standard curve. Then the amount of transgene is divided by the amount of endogenous gene and a normalized transgene value is obtained (rline).
Primers and probes
Three systems were developed, the first for the apx tomato endogenous gene to quantitate tomato DNA; the others for the transgenes TSWV-N and nptII. Primers and TaqMan probes were designed on the basis of sequences present in the GenBank database. The sequences and sizes of amplicons are detailed in Table 5. The 5' and 3' ends of the probes were labelled with fluorescent dyes FAM (6-carboxyfluorescein, excitation wavelength = 494 nm, emission wavelength = 521 nm) and TAMRA (6-carboxy-tetramethyl-rhodamine), respectively. All primers and probes were synthesized by Eurogentec (Belgium).
Table 5.
Primers and probes used for quantitative real-time PCR assays. Primers and TaqMan probes were designed on the GenBank sequences indicated. Probes were labelled with FAM and TAMRA at the 5' and 3' ends, respectively.
| apx gene, tomato endogenous (93 bp amplicon) | (from GenBank acc. no. Y16773) |
| Q-TOMAPX-2818(+) | 5'-TTTTACTTGTATATTCGAAGTGTGCCA-3' |
| Q-TOMAPX-2910(-) | 5'-ACAACTGCAAAATTAGAATCTAGTTGGTA-3' |
| Q-TOMATO probe | 5'-TTTCACCTATCTGGACCAAGTTTTGGCAAG-3' |
| TSWV-N transgene (79 bp amplicon) | (from GenBank acc. no. Z36882) |
| Q-TSWVN-492(+) | 5'-TGTCTTGGCTATATATCAGGATGCA-3' |
| Q-TSWVN-570(-) | 5'-TAAGGCTTCCCTGGTGTCATACTT-3' |
| Q-TSWVN probe | 5'-ACAAGGACCTTGGGATCGACCCAAA-3' |
| nptII transgene (60 bp amplicon) | (from GenBank acc. no. U09365) |
| Q-NPT-250(+) | 5'-AGGAAGCGGTCAGCCCAT-3' |
| Q-NPT-309(-) | 5'-GCGTTGGCTACCCGTGATAT-3' |
| Q-NPT probe | 5'-CGCCGCCAAGCTCTTCAGCAA-3' |
Real-time PCR reactions
The real-time PCR reactions were performed in the iCycler iQ Real-Time PCR Detection System (Bio-Rad Laboratories, USA) and were carried out in 96-well reaction plates. PCR reactions consisted of 1 × Platinum Quantitative PCR SuperMix UDG (Life Technologies-Invitrogen) which contains dUTP and the enzyme uracyl-N-glycosilase (UNG) to prevent contamination deriving from previous PCR reactions, 2 μM of specific TaqMan probe, specific primers at optimal concentration, DNA sample and water to a 25 μl final volume. For each tomato line, a single DNA extract was used in all experiments, to optimize the reproducibility between the two experiments conducted on each gene, and also to allow a correct relative quantification.
The cycling parameters used were as follows: one cycle at 50°C for 3 min for activation of UNG, one cycle a 95°C for 5 min for DNA polymerase activation, and 45 cycles of 95°C for 15 sec (denaturation) and 60°C for 1 min (annealing and extension). All reactions were run in triplicate.
Optimization of primer concentrations
For each primer pair, the primer concentrations were optimized in preliminary experiments, to account for unpredictable differences in annealing efficiency. PCR reactions were run with different combinations of primer concentrations. All nine combinations of 75, 150 and 300 nM (final concentrations) were tested for the TSWV-N and the nptII systems. Similarly, all combinations of 150, 300 and 600 nM were tested for the apx system.
For each system, the lowest concentration of forward and reverse primers giving a high endpoint fluorescence and low TC value, was chosen as the optimal primer concentrations: 300 nM of Q-TSWVN-492(+) and 150 nM of Q-TSWVN-570(-) for N-TSWV system; 150 nM each primer for nptII system; 600 nM each primer for the apx system.
Calculation of copy number and statistical analysis
For each line and each gene we had 6 evaluations of the starting quantity, except for the TSWV-N of line 97–1 for which we had only 5 evaluations, since one experimental point was obviously flawed. The uncertainties on the starting quantities, corresponding to 95% confidence interval, were evaluated by using the t-distribution with 5 (or 4) degrees of freedom. From the starting quantities we constructed, for each line and for the two transgenes, the ratio:
rline = SQtrans/SQend
where SQtrans and SQend are the starting quantities of the transgene and the endogenous gene, respectively. The uncertainty (δrline) was propagated from the uncertainties on the starting quantities with the standard formula:
δrline = rline [(δ SQtrans/SQtrans)2 + (δ SQend/SQend)2]1/2
Such ratios are proportional to the copy number of the transgene, since the endogenous gene is present in one copy.
To determine the copy number for each line, a possibility would be to choose a transgenic line whose copy number is known to be one as the calibrator; the rline ratio for the calibrator line (rcal) would then be associated with one copy of transgene, therefore the copy numbers for the other lines would be determined as rline/rcal. However this procedure is likely to produce biased results, since any fluctuation in the determination of the starting quantities for the calibrator line would affect the copy number for all the other lines. Therefore we chose a different strategy, based on the idea of constructing a "virtual calibrator" which takes into account all the available lines. Our aim is to determine a value r1, corresponding to copy number 1, in such a way that the copy numbers determined for all the lines are as close to integers as possible: this value will be used instead of the calibrator value rcal in determining the copy numbers of the various lines. For each possible value of r1 we define the quantity
F(r1) = ∑lines [rline/r1 -N(rline/r1)]2 / (δrline)2
where N(rline/r1) is the nearest integer to rline/r1.
F(r1) gives a measure of how distant the determined copy numbers are from integer numbers, if r1 is chosen as representing copy number 1. The denominator ensures that the lines that were measured with highest accuracy weigh more in the construction of the virtual calibrator. The best value of r1 is the one for which the quantity F(r1) reaches a minimum, meaning that the determined copy numbers are as close to integers as possible. In practice one should start with a value of r1 higher than all the measured values of rline and gradually decrease r1 until the first local minimum is found (if one were to explore still lower values of r1, more local minima would be found, which however should be discarded as they correspond to fractional copy numbers).
The values of r1 were 0.31 for nptII and 0.21 for TSWV-N transgenes, respectively. Once r1 has been determined, the copy number for each line is determined as rline/r1. In this way the copy number is determined as a real number: as an estimate of the actual, integer copy number we quote the range of integers that are included in the 95% confidence interval around the ratio rline/r1. For two lines in the case of the nptII transgene there is actually no integer included in such interval, so we quote as our estimate of the copy number the nearest integer to rline/r1.
Authors' contributions
GM carried out all real-time PCR experiments and edited the results. PP designed and performed the statistical analysis of the data. AMV carried out molecular analysis. GPA was involved in data analysis and was responsible for the coordination of the study. All authors participated in the design of the experiments, read and approved the final manuscript.
Acknowledgments
Acknowledgements
We are deeply grateful to E. Mozzon, T. Mancuso and L. Ruocco for help in designing primers and probes.
Contributor Information
Giovanna Mason, Email: g.mason@ifa.to.cnr.it.
Paolo Provero, Email: provero@to.infn.it.
Anna Maria Vaira, Email: a.vaira@ifa.to.cnr.it.
Gian Paolo Accotto, Email: g.accotto@ifa.to.cnr.it.
References
- Khachatourians GG, McHughen A, Scorza R. Transgenic plants and crops. New York (USA), Marcel Dekker. 2002.
- Flavell RB. Inactivation of gene expression in plants as a consequence of specific sequence duplication. Proc Natl Acad Sci USA. 1994;91:3490–3496. doi: 10.1073/pnas.91.9.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Mourrain P, Palauqui JC, Vernhetters S. Transgene-induced gene silencing in plants. Plant J. 1998;16:651–659. doi: 10.1046/j.1365-313x.1998.00337.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Flood SJ, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–62. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5'→3' exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–80. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–93. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Song P, Cai CQ, Skokut M, Kosegi BD, Petolino JF. Quantitative real-time PCR as a screening tool for estimating transgene copy number in WHISKERS™-derived transgenic maize. Plant Cell Rep. 2002;20:948–954. doi: 10.1007/s00299-001-0432-x. [DOI] [Google Scholar]
- Applied Biosystems Relative Quantitation of Gene Expression. User Bulletin #2 ABI PRISM 7700 Sequence Detection System. 2001.
- Ingham DJ, Beer S, Money S, Hansen G. Quantitative real-time PCR assay for determining transgene copy number in transformed plants. Biotechniques. 2001;31:132–140. doi: 10.2144/01311rr04. [DOI] [PubMed] [Google Scholar]
- Cirillo C, Accotto GP, Vaira AM, Vecchiati M, Acciarri N, Nervo G. Selection for resistance to Tomato Spotted Wilt Virus (TSWV) in Tomato. Petria. 1999;9:344–345. [Google Scholar]
- Vaira AM, Semeria L, Crespi S, Lisa V, Allavena A, Accotto GP. Resistance to tospoviruses in Nicotiana benthamiana transformed with the N gene of tomato spotted wilt virus: correlation between transgene expression and protection in primary transformants. Mol Plant Microbe Interact. 1995;8:66–73. doi: 10.1094/mpmi-8-0066. [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. Extraction of total cellular DNA from plants, algae and fungi. In: Gelvin SB, Schilperoot RA, editor. Plant molecular biology manual. D1. Dordrecht, Kluwer Academic Publ; 1994. pp. 1–8. [Google Scholar]