Abstract
We report studies of hydration dynamics at the surface of the enzyme protein bovine pancreatic α-chymotrypsin. The probe is the well known 1-anilinonaphthalene-8-sulfonate, which binds selectively in the native state of the protein, not the molten globule, as shown by x-ray crystallography. With femtosecond time resolution, we examined the hydration dynamics at two pHs, when the protein is physiologically in the inactive state (pH 3.6) or the active state (pH 6.7); the global structure and the binding site remain the same. The hydration correlation function, C(t), whose decay is governed by the rotational and translational motions of water molecules at the site, shows the behavior observed in this laboratory for other proteins, Subtilisin Carlsberg and Monellin, using the intrinsic amino acid tryptophan as a probe for surface hydration. However, the time scales and amplitudes vary drastically at the two pHs. For the inactive protein state, C(t) decays with an ultrafast component, close to bulk-type behavior, but 50% of the C(t) decays at a much slower rate, τ = 43 ps. In contrast, for the active state, the ultrafast component becomes dominant (90%) and the slow component changes to a faster decay, τ = 28 ps. These results indicate that in the active state water molecules in the hydration layer around the site have a high degree of mobility, whereas in the inactive state the water is more rigidly structured. For the substrate–enzyme complex, the function and dynamics at the probe site are correlated, and the relevance to the enzymatic action is clear.
Almost all biological macromolecules (proteins, enzymes, and DNA) are inactive in the absence of water. The hydration shell formed by water molecules in close vicinity of a protein/enzyme is particularly important for the stability of the structure and the function or recognition at a specific site. This role of hydration in enzyme catalysis is well known and has recently been reviewed in a number of publications (see, e.g., refs. 1–3). In one of these studies it was shown that the dehydration of a protein, which makes it more rigid and increases its denaturation temperature, is correlated with the loss of its physiological function (1). An understanding of the dynamics of water molecules at the surface of the protein, with spatial molecular and temporal femtosecond resolution of a single site, is important for elucidating a molecular picture and was the goal of a series of publications from this laboratory (4–6).
Using the intrinsic single-tryptophan amino acid as a probe (4, 5), we found that the hydration dynamics at the surface of two different proteins, Subtilisin Carlsberg (SC) (4) and Monellin (5), occur on two well separated time scales: 800 fs and 38 ps for SC and 1.3 ps and 16 ps for Monellin. The slow and fast components are of similar magnitude. From these results, we discussed two types of water hydration trajectories: one fast, bulk type and another much slower, surface-bound type. When these experiments were made on a probe away from the surface by ≈1 nm, bulk-type hydration was recovered. The observed hydration times were correlated with residence times (4), and from a theoretical point of view these times are governed by the exchange of hydrogen-bonded (to the surface) water with the bulk by translational and rotational diffusion (see the reviews in refs. 6 and 7 and references therein).
Surface probing is evident from x-ray and spectroscopic studies of the amino acid probe, as discussed in Results and Discussion. Studies of solvation using chromophores inside proteins or in their clefts show much longer time scales (8, 9). Inside the protein (8, 10) ultrafast decays are attributed to polar amino acid side chains and included water molecules. Molecular dynamics simulations have elucidated the degree of hydration, the involvement in recognition, and the various energetics of binding that give rise to different time scales (11–14).
Here, we report our study of protein hydration as a probe for the expression of functionality. Specifically, we examine the local hydration dynamics at the surface of the protein bovine pancreatic α-chymotrypsin (CHT). The fluorescence of the probe ANS (1-anilinonaphthalene-8-sulfonate), commonly used to monitor structural changes of proteins and membranes, was studied with femtosecond resolution in two states of the protein, the physiologically active and inactive states. The protein, isolated from bovine pancreas, is in a class of digestive enzymes and has the biological function of hydrolyzing polypeptide chains. However, physiological activity is determined by the pH.
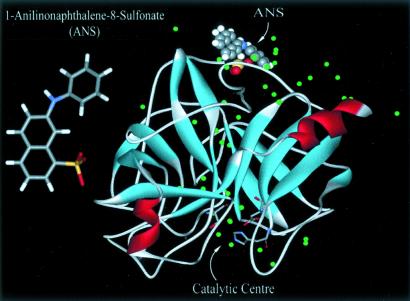
The enzyme is not active at lower pH, but the activity increases nonmonotonically with pH (15); in the duodenum (low pH), it is inactive, whereas in the lower small intestinal track (high pH), it becomes active (15, 16). The inactivity of the enzyme at lower pH is known to be related to the protonation of the residues of the catalytic triad (15). The crystal structure (Fig. 1), as discussed below, gives the active site for the enzyme and the site for recognition of ANS, which is unchanged at both pH ranges (17, 18). From spectroscopic studies, it was suggested that the degree of mobility of water molecules, not hydrophobicity, may be the important factor for the change with pH.
Fig 1.
The structure of the ANS is shown on the left. The x-ray structure of CHT is shown on the right. The catalytic center in the hydrophobic pocket is indicated in the lower part of the structure. In the upper part, we show the probe ANS at its known binding site (18). The structure was downloaded from the Protein Data Bank (ID code ) and handled with the program weblab viewerlite (http://molsim.vei.co.uk/weblab/).
Such change in activity of the protein with pH offers a unique opportunity for correlating the function with the dynamics, because the global structure and binding site for recognition are unchanged at either pH range. By careful study of hydration of the protein surface at different pHs, and ANS in bulk solvents (water, methanol, ethanol and n-hexane), we are able to relate such expression in functionality to the order and rigidity of the hydration shell and determine the rate at which hydration occurs, from the femtosecond to the picosecond scale.
Experimental Procedures
Time-Resolved Studies.
All of the transients were obtained by using the femtosecond-resolved fluorescence up-conversion technique. The detailed experimental setup has been described (5). Here, a femtosecond excitation pulse (200 nJ) was used at 320 nm. The probe pulse was set at 800 nm. The up-converted signal in the deep UV region (280–350 nm) was detected by a photomultiplier after dispersion through a double grating monochromator. The transients were taken at the magic angle (54.7°) of the pump polarization (320 nm) relative to that of the probe (800 nm) and the up-conversion crystal's acceptance axis. For anisotropy measurements at 510 nm the pump polarization was adjusted to be parallel or perpendicular to that of the probe, defining r(t) = (Ipara − Iperp)/(Ipara + 2Iperp). We measured fluorescence lifetimes by the time-correlated single-photon counting method using a Picoquant (Berlin) commercial setup with excitation wavelength of 400 nm.
To construct time-resolved emission spectra after the excitation pulse, we adopted the method of ref. 19. For every sample solution, the fluorescence transients were measured as a function of detection wavelength in the range of 430–610 nm in intervals of ≈20 nm. The observed fluorescence transients were fit to a function, which is the convolution of the instrument response function with a sum of exponentials, by using a nonlinear least-squares fitting procedure (software scientist). The purpose of this fitting is to obtain the decays in an analytic form suitable for further data analysis. For each detection wavelength, the transient was normalized by using the steady-state spectrum. The resulting time-resolved spectra were fitted with a Gaussian shape function to estimate the spectrum maximum ν(t). The temporal Stokes shift can be represented by the time dependence of the fit. By following the time-resolved emission, we constructed the hydration correlation function, C(t) = [ν(t) − ν(∞)]/[(ν(0) − ν(∞)].
Sample Preparation.
CHT (3× crystallized, dialyzed, and salt-free lyophilized powder) and ANS (ammonium salt, 95% pure) were purchased from Sigma. The solvents methanol and 100% ethanol were from Omnisolv (Gibbstown, NJ) and AAPER (Shelbyville, KY), respectively. Chemicals and the CHT were used as received. Aqueous protein solutions were prepared in an acetate buffer (0.1 M) at pH 3.6 and 6.7 in water from a Nanopure (Dubuque, IA) purification system.
The CHT–ANS complexes at different pH were prepared by mixing ANS (0.5 mM) with CHT (1 mM) in the corresponding buffer solutions with continuous stirring for 1 h. The stoichiometry of the protein/probe was maintained as described in ref. 17. The yield of 85% and 82% ANS–CHT complexes at our ANS concentration was calculated from knowledge of the binding constants, which were determined (1:1 complex) to be 1.00 × 104 M−1 and 0.71 × 104 M−1 at pH 6.7 and 3.6, respectively (17).
Results and Discussion
ANS in Bulk Solvents.
Steady-state studies.
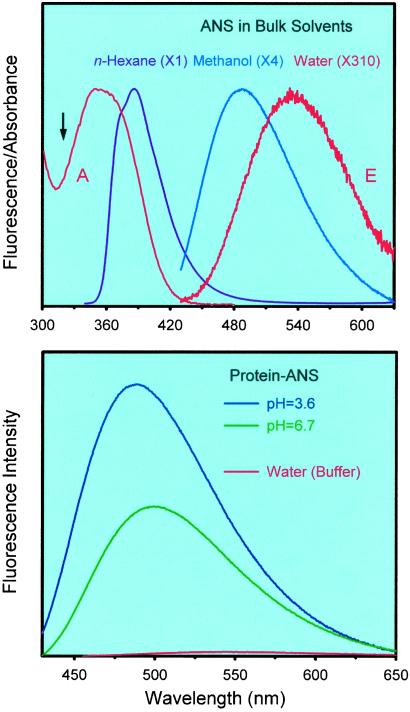
ANS is a well known solvation probe (20, 21). The steady-state fluorescence spectra of the ANS dye in three neat solvents are presented in Fig. 2 Upper. The spectra show a large red solvatochromic effect. The fluorescence maximum changes from 390 nm in n-hexane, to 490 nm in methanol, and to 540 nm in the aqueous solution (buffer, pH 3.6 and 6.7). The steady-state emission is quenched dramatically in polar solvents. Because of its bichromophoric structure, ANS is known (22) to undergo charge transfer (CT) from one aromatic moiety to the other ring and solvation. In steady state, in nonpolar solvents, the emission is strong and is mostly from the locally excited state, i.e., before charge separation. In polar solvents, the fluorescence decreases and is dominated by emission from the CT state. The solvent polarity and rigidity determine the wavelength and yield of emission, and that is why ANS is a useful biological probe.
Fig 2.
(Upper) Normalized steady-state fluorescence spectra of ANS in three solvents: n-hexane, methanol, and water. The decrease in fluorescence is indicated by the numbers in parentheses. For water, both absorption (A) and emission (E) are shown, and the arrow marks the excitation wavelength (320 nm). Note that the excitation in water is near the n-hexane and the solvation Stokes shift is relatively large; see text. (Lower) Fluorescence spectra of CHT–ANS in buffer solutions at pH 3.6 and 6.7. For comparison, we also include the fluorescence spectrum of ANS in water solution. The three spectra correspond to solutions with the same total concentration (optical density) of ANS.
A molecular picture of solvation for this molecule, and its sibling 2-(p-toluidino)naphthalene-6-sulfonate (TNS), has been given (23). Both the solvent coordinate and the intramolecular charge separation must be considered to construct the free energy landscape. Trajectories begin from the locally excited state, and solvent motion determines the dynamics toward the final CT state. This picture is entirely consistent with the theoretical calculation advanced in refs. 24 and 25 and the experimental correlation between excited-state lifetime and solvent relaxation time made in ref. 26. More on the molecular picture can be found in ref. 23.
Time-resolved measurements.
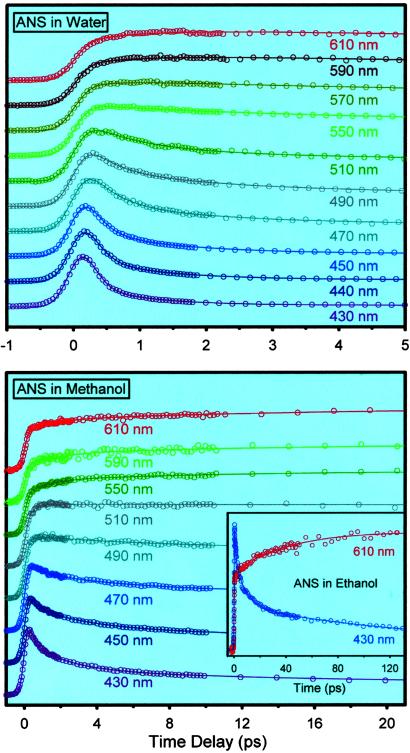
Time-resolved emission separates the contribution of ultrafast solvation from longer time, nonradiative processes. As shown in Fig. 3, the femtosecond transients are typical of those observed for other chromophores in water. On the blue edge of the spectrum, the signal is seen to decay on the time scale up to ≈1.5 ps, whereas on the red edge it rises on a similar time scale. From this family of transients we construct the C(t) in Fig. 4: ν(t = ∞) at 5 ps; C(t) shows two exponential decays of 185 fs (22%) and 1.2 ps (78%), indicating that hydration is complete on this time scale, as is the case with other solutes including coumarin dyes (27, 28) and tryptophan (4).
Fig 3.
Femtosecond-resolved fluorescence at a series of wavelengths for ANS in water (Upper), methanol (Lower), and ethanol (Lower, Inset). The excitation wavelength was 320 nm.
Fig 4.
Hydration correlation function for ANS in water (Upper) and methanol (Lower); note the difference in time scales. (Insets) The corresponding normalized time-resolved spectra. In water, the shift from the initial t = 0 spectrum to 5-ps spectrum completes the process of solvation; the spectrum at equilibrium (steady state) requires further time to reach, consistent with the longer time scale for posthydration-activated CT in ANS (see text).
To complete the picture regarding bulk dynamics of ANS in water and less polar solvents (for comparison with protein environment), we made similar studies in methanol and ethanol (see Fig. 3). In this case, the decays are slower than those in water and consistently show the time scale of solvation for the two solvents (Fig. 4). C(t) decays with 260 fs (58%) and 4.8 ps (42%) time constants, and our value of 4.8 ps is close to the 5-ps average time (using coumarin as a probe) reported in ref. 29. To further elucidate the nature of the spectral shift we also measured the time-resolved anisotropy, r(t). At t = 0, r(t) = 0.32 and decays to the base line with τ = 58 ps, the rotational relaxation time of ANS in methanol; in water τ was found to be 70 ps (30). Most importantly, the r(t) decay in methanol is on a much different time scale than that of solvation; conformational changes (which should alter the anisotropy) to yield the CT state are separable in their time scale from that of solvation (23). The results support our picture that the spectral shift we observe in the first 2 ps in water is caused by hydration dynamics.
Following this hydration time scale, all transients at different wavelengths have a contribution (up to ≈10%) that decays with a time constant of 5–10 ps, depending on wavelength, and is then followed by a decay to the baseline with a time constant of 150–300 ps, depending on wavelength. This subnanosecond decay is the lifetime of ANS in water; we confirmed this by measuring the lifetime in water and methanol by using single-photon counting techniques and obtained values of 0.25 and 5 ns, respectively. These decays describe the nonradiative pathways from the locally excited to the CT state and the total lifetime of the latter in water, as detailed in ref. 23.
Consistent with previous work (29) we found a relatively insignificant role for vibrational relaxation/redistribution, which occurs in these large molecules on the subpicosecond time scale (31). As shown in Fig. 2 Upper, the absorption spectrum of ANS in water cuts the emission spectrum in n-hexane (gas-phase type emission) at 373 nm, indicating a putative 0–0 transition close to the excitation wavelength at 320 nm. Thus, solvation redshift (≈7,000 cm−1) is dominant over the intramolecular Stokes shift. This dominance of solvation was confirmed by studies of two probes (tryptophan and 2-aminopurine) at low and high vibrational energy (4, 32, 33). These observations are further supported by the fact that the shapes of time-resolved emission spectra at early times are not significantly different from those obtained at later times and that the ANS transients in different solvents (water, methanol, and ethanol) give the characteristic solvation times obtained with other probes. It should be noted that in the study of the protein, the effect of the pH was observed under identical conditions of excitation and relaxation.
ANS Bound to the Protein CHT.
X-ray structures and equilibrium studies.
An extensive fluorescence study (17) followed by an x-ray study (18) of the CHT–ANS complex indicated that ANS binds rigidly (binding constant of the order of 104 M−1; see above for quantification) at a single site on the surface of the protein near the Cys-1–122 disulfide bond. This ANS binding site is almost opposite in position to the enzymatic center as shown in Fig. 1. From those studies, it was concluded that the local environment around ANS in the complex in solution and in the crystalline phase are identical. Also, the increase in fluorescence yield at low pH was associated with the reduced mobility of water molecules around the probe. The global structure and binding site to the protein CHT nearly do not change at the different pHs studied; at lower pH there are some local group protonations, or what was called different ionization states of the protein, which give rise to some local (group) conformational changes (17, 18).
In Fig. 2 Lower, we present the steady-state fluorescence spectra of CHT–ANS solutions at pH 3.6 and 6.7. For comparison, we also include a fluorescence spectrum of ANS in water. The total concentration of the ANS dye in the three solutions was kept the same (0.5 mM). In the protein solutions, the steady-state fluorescence intensity is much larger than in the water solution. At pH 3.6 and 6.7, the spectra of the CHT-ANS system is 100 times and 50 times, respectively, more intense than that of ANS in water. As mentioned earlier, in the protein solutions, approximately the same fraction of the dye is in the free state (18% at pH 3.6 and 15% at pH 6.7); therefore, the relative contribution from free ANS is not responsible for the changes in the fluorescence intensity at the two different pHs.
The fluorescence maximum wavelength in the protein also depends on the pH: It shifts from 485 nm at pH 3.6 to 500 nm at pH 6.7, whereas for the weak fluorescence in the buffer, the maximum is at 540 nm for both pHs. Both the spectral shift and the fluorescence intensity trends we observe are in agreement with the previous study of the CHT–ANS complex (17). We measured the fluorescence lifetime of the CHT–ANS solutions by using the single-photon counting method. The nanosecond fluorescence decays were fitted to multiexponential functions to reflect the decay of the complex in equilibrium with its components. At pH 3.6, we obtained the time constants (ns) 0.45 (28%), 3.5 (34%), and 11.0 (38%), and for pH 6.7, these constants were 0.27 (40%), 2.4 (30%), and 11.5 (30%). We note that the lifetime of free ANS in water is ≈0.25 ns, as discussed above, and in less polar ethanol it is 11.1 ns (30). In ref. 17 the authors reported a 12-ns component for CHT–ANS solutions at the two pHs, and our longtime component is in agreement with these results.
Dynamics and function.
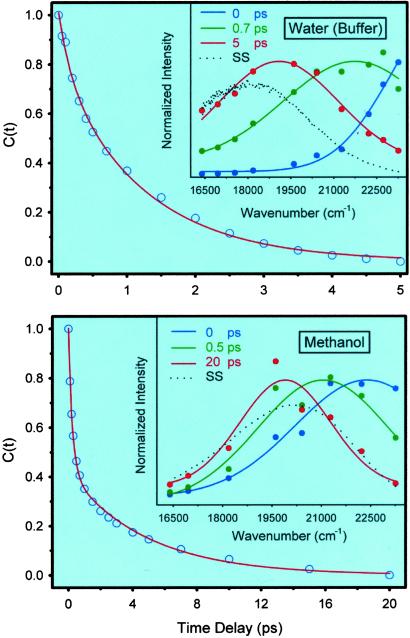
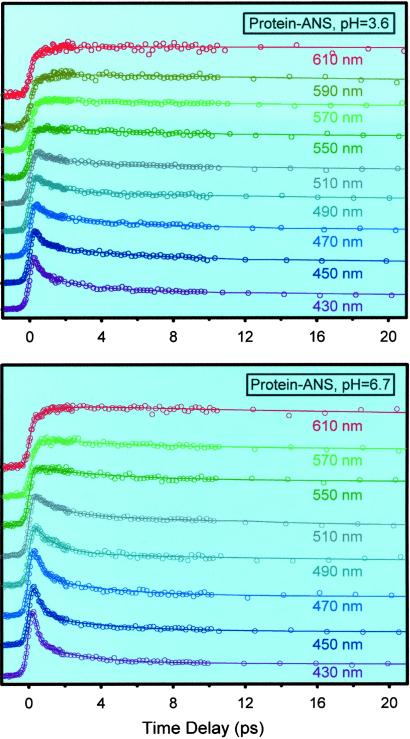
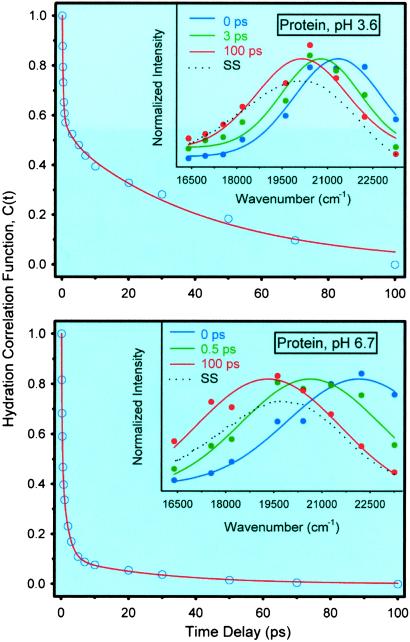
Our results from femtosecond up-conversion experiments on the CHT–ANS complexes are shown in Fig. 5. In both pHs, the signals have features typical of ultrafast solvation: subpicosecond decays on the blue edge of the spectra and corresponding rises in the red edge part of the spectra. The scans also contain decays in the few to tens of picoseconds with different amplitudes and time constants across the fluorescence spectra, as discussed before. The C(t) functions constructed from these data, together with normalized time-resolved spectra, are shown in Fig. 6. The functional decays give the following results for pH 3.6: 290 fs (40%), 2.9 ps (10%), and 43 ps (50%). For pH 6.7, we obtained 260 fs (51%), 1.8 ps (39%), and 28 ps (10%).
Fig 5.
Femtosecond-resolved fluorescence at a series of wavelengths for CHT–ANS in buffer solution at pH 3.6 (Upper) and pH 6.7 (Lower). The excitation wavelength was 320 nm. Note the decay in the blue side and the rise in the red side of the spectra.
Fig 6.
Hydration correlation function for CHT–ANS complexes in water at pH 3.6 (Upper) and pH 6.7 (Lower). (Insets) The corresponding normalized time-resolved spectra.
Clearly for the ANS–protein complexes we observe the slow component characteristic of hydration with a significant contribution, and the time scale is two orders of magnitude different from bulk-type solvation. Moreover, the ultrafast decay is dominant in the high pH case. From the equilibrium constant we know that ≈15% of ANS are free in the buffer and will contribute only to the ultrafast part of the decay. However, the trend observed for the increase in the amplitude of the ultrafast component with pH cannot be from the free ANS molecules because Keq is nearly the same in the two pHs. In fact, it is somewhat larger in higher pH, and if anything, the contribution of free ANS to this component is smaller.
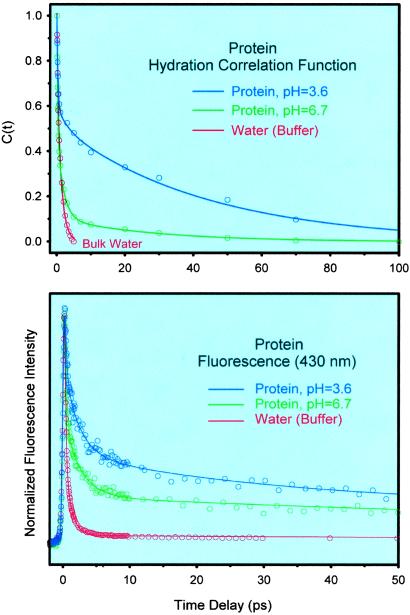
Given that the x-ray crystal structure is globally the same at different pHs, our observation that hydration dynamics of ANS bound to CHT at pH 6.7 has a much larger contribution from the ultrafast, bulk-type hydration (90% of the spectral shift vs. 50% at pH 3.6) indicates that at this pH the protein at the ANS-binding site exists in a state of hydration that is significantly different from that at lower pH; the site experiences much more of a bulk-type water at higher pH (Fig. 7). Also, at the higher pH, the time constant of the slow component (28 ps) is shorter than at pH 3.6 (43 ps), which again reflects a hydration from less rigid water layer than at lower pH. The ratio of amplitudes, bulk type to rigid layer behavior, has changed at least by an order of magnitude.
Fig 7.
(Upper) Comparison of the hydration correlation function for the CHT–ANS complexes at pH 3.6 and 6.7. We also include the result from ANS in water for comparison with bulk behavior. (Lower) Normalized femtosecond-resolved fluorescence for the three systems at the fluorescence wavelength of 430 nm. Note the similarity in behavior.
These studies of femtosecond dynamics of the ligand probe ANS with the protein CHT thus elucidate the nature of surface recognition process and the key time scales involved in the hydration layer. Surface hydration in this protein, and two others reported before (4, 5), is evident because of the following. First, from x-ray studies the probe is at the surface, and spectroscopic studies support the exposure to water; for example, in Monellin and SC, the emission peaks are at 342 and 351 nm (4, 5), whereas inside a protein the peak is at 308 nm (34).
Second, the time-dependent anisotropy decays on a much different time scale from that of the hydration correlation function in Monellin; e.g., the time constant for the former is 32 ps, with a longer time persistency, and the longer time constant for the latter is 16 ps [with C(t) decaying to its equilibrium value]. This finding suggests that tumbling and hydration of amino acid probe (Trp) are on a much different time scale. Even if there is local restricted fast motions of side chains, their collective solvation will require longer time scales. Third, the fact that the C(t) decay pattern and its two time scales are robust in three different proteins suggests the generality of their hydration picture, because the local side chain structure in all three proteins is different and we expect a very different time scale for solvation. The change is evident in bulk solvation. We compared the results in different solvents (water, methanol, and ethanol) and found, for Trp, that its solvation time changes by an order of magnitude. Finally, vibrational relaxation, which we addressed in ref. 5 and above, is relatively insignificant.
Conclusion
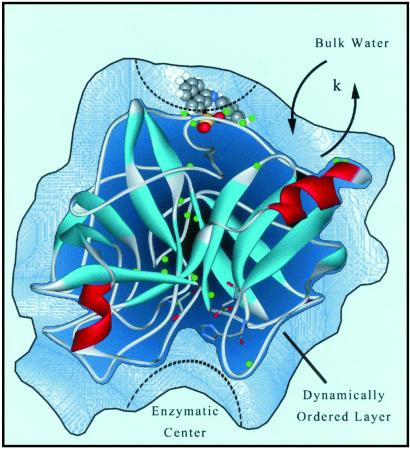
Three significant points can be made. First, the longer hydration times at pH 3.6 and 6.7 (43 ps and 28 ps) are an order of magnitude slower than that of bulk water (≈1 ps), consistent with the general trend we observed for two other proteins, SC and Monellin (4, 5). Second, at pH 3.6, the contribution of the slower component in the hydration correlation function (43 ps) is much larger than that found for pH 6.7 (10%, 28 ps), reflecting a more structured water on the surface at low pH when the enzyme is inactive. Third, the drastic increase in the mobility of water, from the hydration correlation function, in the structured water layer is correlated with the function of the protein at high pH. Fig. 8 presents a simplified picture of the role of hydration.
Fig 8.
A simplified picture of the model emerging from the studies reported here. The dynamically ordered water is a general feature of the three proteins studied (see text). The function of the protein at two pHs is illustrated by the degree of surface hydration and rigidity of water layer (see text).
At low pH, the protonation of amino groups at the site studied presumably enhances positive charge interaction with the water layer (17). Although the site of the protein we probed is not the reactive site, which is hydrophobic in nature (14) and near the enzymatic center, our findings may suggest a similar mechanism for the substrate–enzyme activity. The rigid water molecules at low pH (3.6) hinder the expression of protein functionality, whereas a mobile, less rigid water structure at high pH (6.7) makes the active recognition dynamically favorable. Recognition and dehydration requires rapid movement of water molecules. Currently, we are examining the site involving Ser-195, His-57, and Asp-102 at the catalytic center (35), using a molecular probe to study hydration as we did in an earlier series of articles (4–6). Theoretical studies aided by molecular dynamics simulations (14) will be part of our effort.
Acknowledgments
This work was supported by the National Science Foundation.
Abbreviations
SC, Subtilisin Carlsberg
CHT, α-chymotrypsin
ANS, 1-anilinonaphthalene-8-sulfonate
CT, charge transfer
References
- 1.Rupley J. A. & Careri, G. (1991) Adv. Protein Chem. 41 37-172. [DOI] [PubMed] [Google Scholar]
- 2.Kornblatt J. A. & Kornblatt, M. J. (2002) Int. Rev. Cytol. 215 49-73. [DOI] [PubMed] [Google Scholar]
- 3.Pocker Y. (2000) Cell. Mol. Life Sci. 57 1008-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pal S. K., Peon, J. & Zewail, A. H. (2002) Proc. Natl. Acad. Sci. USA 99 1763-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peon J., Pal, S. K. & Zewail, A. H. (2002) Proc. Natl. Acad. Sci. USA 99 10964-10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pal, S. K., Peon, J., Bagchi, B. & Zewail, A. H. (2002) J. Phys. Chem. B, in press.
- 7.Bizzarri A. R. & Cannistraro, S. (2002) J. Phys. Chem. B 106 6617-6633. [Google Scholar]
- 8.Barret P. C., Choma, C. T., Gooding, E. F., DeGrado, W. F. & Hochstasser, R. M. (2000) J. Phys. Chem. B 104 9322-9329. [Google Scholar]
- 9.Jordanides X., Lang, M., Song, X. & Fleming, G. R. (1999) J. Phys. Chem. B 103 7995-8005. [Google Scholar]
- 10.Homoelle B. J., Edington, M. D., Diffey, W. M. & Beck, W. F. (1998) J. Phys. Chem. B 102 3044-3052. [Google Scholar]
- 11. (2002) Acc. Chem. Res. 35 321-489. [DOI] [PubMed] [Google Scholar]
- 12.Henchman R. H. & McCammon, J. A. (2002) Protein Sci. 11 2080-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camacho C. J., Weng, Z., Vajda, S. & DeLisi, C. (1999) Biophys. J. 76 1166-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carey C., Cheng, Y.-K. & Rossky, P. J. (2000) Chem. Phys. 258 415-425. [Google Scholar]
- 15.Zubay G., (1983) Biochemistry (Addison–Wesley, Reading, MA), pp. 130–175.
- 16.Charman W. N., Porter, C. J. H., Mithani, S. & Dressman, J. B. (1997) J. Pharm. Sci. 86 269-282. [DOI] [PubMed] [Google Scholar]
- 17.Johnson J. D., El-Bayoumi, M. A., Weber, L. D. & Tulinsky, A. (1979) Biochemistry 18 1292-1296. [DOI] [PubMed] [Google Scholar]
- 18.Weber L. D., Tulinsky, A., Johnson, J. D. & El-Bayoumi, M. A. (1979) Biochemistry 18 1297-1303. [DOI] [PubMed] [Google Scholar]
- 19.Maroncelli M. & Fleming, G. R. (1987) J. Chem. Phys. 86 6221-6239. [Google Scholar]
- 20.DeToma R. P., Easter, J. H. & Brand, L. (1976) J. Am. Chem. Soc. 98 5001-5007. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J. & Bright, F. V. (1991) J. Phys. Chem. 95 7900-7907. [Google Scholar]
- 22.Kosower E. M. (1982) Acc. Chem. Res. 15 259-266. [Google Scholar]
- 23.Zhong D., Pal, S. K. & Zewail, A. H. (2001) Chem. Phys. Chem. 2 219-227. [DOI] [PubMed] [Google Scholar]
- 24.Kim J. H. & Hynes, J. T. (1997) J. Photochem. Photobiol. A 105 227-343. [Google Scholar]
- 25.Kim J. H. & Hynes, J. T. (1996) in Femtochemistry and Femtobiology, Nobel Symposium 101, ed. Sundstrom, V. (Imperial College Press, London), pp. 501.
- 26.Kosower E. M. & Huppert, D. (1983) Chem. Phys. Lett. 96 433-435. [Google Scholar]
- 27.Jarzeba W., Walker, G. C., Johnson, A. E., Kahlow, M. A. & Barbara, P. F. (1988) J. Phys. Chem. 92 7039-7041. [Google Scholar]
- 28.Jimenez R., Fleming, G. R., Kumar, P. V. & Maroncelli, M. (1994) Nature 369 471-473. [Google Scholar]
- 29.Horng M. L., Gardecki, J. A., Papazyan, A. & Maroncelli, M. (1995) J. Phys. Chem. 99 17311-17337. [Google Scholar]
- 30.Robinson G. W., Robbins, R. J., Fleming, G. R., Morris, J. M., Knight, A. E. W. & Morrison, R. J. S. (1978) J. Am. Chem. Soc. 100 7145-7150. [Google Scholar]
- 31.Elsaesser T. & Kaiser, W. (1991) Annu. Rev. Phys. Chem. 42 83-107. [Google Scholar]
- 32.Zhong D., Pal, S. K., Zhang, D., Chan, S. I. & Zewail, A. H. (2002) Proc. Natl. Acad. Sci. USA. 99 13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pal S. K., Peon, J. & Zewail, A. H. (2002) Chem. Phys. Lett. 363 57-63. [Google Scholar]
- 34.Lakowicz J. R., (1999) Principles of Fluorescence Spectroscopy (Kluwer, New York), pp. 445–486.
- 35.Haugland R. P. & Stryer, L. (1967) in Conformation of Biopolymers, ed. Ramachandran, G. M. (Academic, New York), Vol. 1, pp. 321–333. [Google Scholar]