Abstract
Precise regulation of the evolutionarily conserved hypoxia-inducible transcription factor (HIF) ensures proper adaptation to variations in oxygen availability throughout development and into adulthood. Oxygen-dependent regulation of HIF stability and activity are mediated by hydroxylation of conserved proline and asparagine residues, respectively. Because the relevant prolyl and asparginyl hydroxylases use O2 to effect these posttranslational modifications, these enzymes are implicated as direct oxygen sensors in the mammalian hypoxic response pathway. Here we present the structure of factor-inhibiting HIF-1 (FIH-1), the pertinent asparaginyl hydroxylase involved in hypoxic signaling. Hydroxylation of the C-terminal transactivation domain (CTAD) of HIF by FIH-1 prevents CTAD association with transcriptional coactivators under normoxic conditions. Consistent with other structurally known hydroxylases, FIH-1 is comprised of a β-strand jellyroll core with both Fe(II) and the cosubstrate 2-oxoglutarate bound in the active site. Details of the molecular contacts at the active site of FIH-1 have been elucidated and provide a platform for future drug design. Furthermore, the structure reveals the presence of a FIH-1 homodimer that forms in solution and is essential for FIH activity.
Mammalian cells employ an evolutionarily conserved hypoxic response pathway to detect and adapt to changes in oxygen availability. Hypoxia-inducible factor (HIF) is a transcription factor that lies at the heart of this pathway. HIF is a heterodimer composed of oxygen-sensitive α (HIF-1α or HIF-2α) and constitutive β subunits (HIF-1β also known as the arylhydrocarbon receptor nuclear translocator, ARNT) (1). Multiple layers of regulation ensure tight control of HIF activity, as disregulation of the pathway can have profound effects on both development and the progression of cancer and ischemia (2). The HIF-α subunit contains two regions that modulate its activity as a function of O2 availability. Under normoxic conditions, conserved proline residues within the oxygen-dependent degradation domain (ODD) of the α-subunit are hydroxylated by a family of prolyl hydroxylases (3, 4). This posttranslational modification serves as a recognition element for the product of the von Hippel Lindau tumor suppressor gene (pVHL), a component of the protein–ubiquitin ligase complex that tags the α-subunit for rapid degradation (5–7). Under hypoxic conditions, hydroxylation is diminished, leading to α-subunit accumulation and subsequent promotion of target gene transcription. Full HIF induction requires the interaction of the C-terminal transactivation domain (CTAD) with coactivators such as CBP/p300 to recruit the transcriptional machinery (8–11). Under normoxic conditions, this interaction is inhibited by hydroxylation of an asparagine residue within the CTAD by factor-inhibiting HIF-1 (FIH-1), an asparaginyl hydroxylase enzyme (12–15). Again, this hydroxylation event is selectively decreased under hypoxic conditions (12, 16).
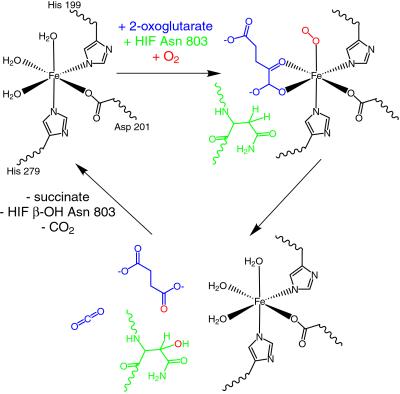
Hydroxylation catalyzed by FIH-1 requires Fe(II) and consumes molecular oxygen and 2-oxoglutarate (2-OG) (14). In the active sites of known 2-OG-dependent oxygenases, Fe(II) is coordinated by two histidine residues and the carboxylate of either an aspartate or a glutamate residue. The remaining three coordination sites on the metal initially are occupied by water molecules (Fig. 1) (17). Two of the water molecules are displaced by 2-OG, followed by binding of the HIF substrate and O2 (Fig. 1). Molecular oxygen must bind after the protein substrate to prevent a futile reaction cycle that is capable of generating harmful hydroxyl radicals in the cell (18). Multiple steps at the reactive metal center lead to the generation of a highly reactive ferryl intermediate that abstracts a hydrogen from the β-carbon of the target asparagines (Fig. 1) (19, 20). A radical rebound mechanism likely ensues with the final product, β-hydroxy-asparagine, as the end result (Fig. 1) (21, 22). 2-OG accepts the other oxygen atom from molecular oxygen and subsequently undergoes decarboxylation to produce succinate (23). Because of its utilization of O2 as a substrate, FIH-1 is viewed as a prime candidate for the cellular oxygen sensor.
Fig 1.
FIH-1 hydroxylates HIF-1α at Asn-803. The FIH-1 active site contains Fe(II) coordinated by three protein side chains: His-199, Asp-201, and His-279. The enzyme binds 2-OG, HIF peptide substrate, and molecular oxygen to facilitate hydroxylation of the β-carbon on Asn-803 of HIF-1α. In the course of the reaction, molecular oxygen is consumed and 2-OG is converted into succinate and carbon dioxide.
Given the crucial role of asparaginyl hydroxylases in the regulation of the hypoxic response pathway, the X-ray crystal structure of human asparaginyl hydroxylase FIH-1 was determined in the presence and absence of 2-OG and Fe(II). Based on sequence analysis, FIH-1 was predicted to contain an eight β-strand jellyroll core that surrounds an Fe(II)-binding site (14), consistent with known hydroxylase enzymes (15). Here we present these FIH-1 structural models and describe the molecular details of the active site architecture mediating Fe(II) and 2-OG binding. In addition, the FIH-1 crystal structure suggests and biochemical data confirm that FIH-1 forms a homodimer via the catalytically required C terminus.
Materials and Methods
Protein Expression and Purification.
Variants of the human FIH-1 coding region were generated by PCR and subcloned into the pHis-parallel-1 and pMBP-parallel-1 vector for structural and biochemical studies, respectively (24). To obtain milligram quantities of full-length human FIH-1 protein, N-terminally His-tagged FIH-1 (His-FIH-1) was expressed using the Escherichia coli rare codon strain Rosetta(DE3)pLysS (Invitrogen). Cells expressing His-FIH-1 were induced with 0.2 mM isopropyl β-d-thiogalactoside (IPTG) for 18 h at 20°C, lysed in buffer containing 20 mM Tris⋅HCl (pH 8.0), 300 mM NaCl, 1 mg/ml lysozyme, 5 μg/ml DNase I, and 5 mM 2-mercaptoethanol, and initially purified by using Ni-nitrilotriacetic acid-immobilized metal affinity chromatography techniques (25). Subsequent FIH-1 protein preparation included ion exchange chromatography using Mono Q resin (Amersham Pharmacia), treatment with tobacco etch virus (TEV) protease (26) to remove the hexahistidine tag, and gel filtration using a Superdex 75 16/60 column (Amersham Pharmacia). For the biochemical pull-down experiments, E. coli cells expressing maltose-binding protein (MBP)-FIH-1 fusions were induced with 0.2 mM IPTG for 15 h at room temperature. Cell pellets were lysed in buffer containing 20 mM Tris⋅HCl (pH 7.5), 200 mM NaCl, 10 mM 2-mercaptoethanol, 1 mg/ml lysozyme, and 1:200 dilution Protease Inhibitor Mixture (Sigma). Soluble protein was affinity purified over an amylose agarose column (New England Biolabs), and the MBP-FIH-1 protein was eluted with buffer containing 10 mM maltose.
FIH-1 Crystallization.
FIH-1 protein was concentrated to 10 mg/ml in 5 mM Tris⋅HCl pH 8.0, 50 mM NaCl, and 5 mM 2-mercaptoethanol. The protein was crystallized by hanging drop vapor diffusion at 21°C against 50 mM CAPSO pH 9.6, 0.9–1.2 M K tartrate, and 30% glycerol. Crystals containing 2-OG were obtained by adding 5 mM FeSO4 and 20 mM 2-OG to the crystallization solution. Selenomethionine (SeMet) FIH-1 crystallized under similar conditions at a protein concentration of 15 mg/ml. All crystals obtained for FIH-1 had P41212 space group symmetry with a one molecule in the asymmetric unit (see Table 1).
Table 1.
Summary of FIH-1 crystallographic data, refinement, and model statistics
| FIH-1/Fe | FIH-1/Fe/2-OG | SeMet FIH-1 | |
|---|---|---|---|
| Crystal data | |||
| Space group | P41212 | P41212 | P41212 |
| Unit-cell dimensions, Å | |||
| a | 86.62 | 86.46 | 86.57 |
| c | 147.76 | 147.81 | 147.72 |
| Wavelength, Å | 0.97916 | 1.0781 | 0.97916 |
| Resolution range, Å | 50–2.2 (2.28–2.20) | 50–2.4 (2.49–2.40) | 50–2.8 (2.90–2.80) |
| No. of observations | 322,469 | 172,309 | 80,741 |
| No. of unique reflections | 29,190 | 21,793 | 14,364 |
| Completeness | 99.9 (100.0) | 98.2 (99.9) | 95.8 (96.1) |
| Rsym, % | 6.2 (48.4) | 7.6 (44.3) | 5.5 (31.0) |
| 〈I/σ(I)〉 | 35.7 (4.9) | 21.8 (5.1) | 17.5 (3.2) |
| Phasing statistics | |||
| No. of sites | 7 | ||
| Phasing power | 2.2 | ||
| Figure of merit | NA/0.82 | 0.35/0.88 | |
| Refinement statistics | |||
| Resolution, Å | 30–2.2 | 30–2.4 | 30–2.8 |
| Rwork, % | 21.6 | 23.2 | |
| Rfree, % | 23.7 | 24.4 | |
| rms deviations | |||
| Bond lengths, Å | 0.010 | 0.006 | |
| Bond angles, ° | 1.45 | 1.28 | |
| B factors, Å2 | |||
| Main chain, bonded | 1.44 | 1.48 | |
| Side chain, bonded | 2.36 | 2.19 | |
| Main chain, angles | 2.31 | 2.51 | |
| Side chain, angles | 3.59 | 3.36 | |
| σA-coordinate error, Å | 0.24 | 0.29 | |
| Non-H atoms in model | |||
| Protein | 2,739 | 2,700 | |
| Fe | 1 | 1 | |
| Hetero-atoms | 10 (tartrate) | 10 (2-OG) | |
| Water molecules | 106 | 50 | |
| Disordered residues | 1–12; 304–305 | 1–12; 138–139 | |
| Percentage of residues in Ramachandran plot regions | |||
| Most favored | 91.5 | 89.4 | |
| Additional allowed | 8.5 | 10.6 | |
Values in parentheses are for the highest resolution shell. NA, not applicable.
Rsym = 100 × Σh|Il(h) − 〈I(h)〉|/ΣhΣlIl(h), where Il is the lth measurement of reflection h and 〈I(h)〉 is a weighted mean of all measurements of h.
Figure of merit values are indicated for phases generated before and after solvent flattening.
Rwork includes all reflections used during refinement, whereas Rfree is calculated from 5% of reflections not included in refinement.
Percentages are based on the procheck program (40).
Structure Determination.
To obtain phases for structure determination, selenomethionyl FIH-1 was expressed using standard techniques (27). Data from FIH-1 and SeMet FIH-1 crystals were collected at Argonne National Laboratory Structural Biology Center beamline 19ID, and diffraction data for determination of the complex between FIH-1 and 2-OG were collected at the Advanced Light Source beamline 8.2.2. All data were reduced using the HKL2000 software package (28). sharp (29) was used to locate and refine selenium sites before conducting SAD phasing (30). Initial phase information from SeMet FIH-1 data were combined with native FIH-1 amplitudes before solvent flipping with solomom (31), and the arp/warp program package (26, 32) was used to automatically build approximately two-thirds of the residues in the model. The remainder of the model was manually built with the program o (33) and refined with cns (34). The initial model of FIH-1 with Fe(II) and 2-OG was generated using rigid body refinement of the refined FIH-1 model. Geometry and B-factor minimization with cns completed the refinement of the FIH-1 Fe(II) 2-OG model. Final analysis of all models was performed by using composite simulated annealing omit maps.
GST Pull-Down Assays.
Thirty microliters of 35S-labeled Trx6H mouse HIF-2α 774–874 protein (14) generated using the TNT Coupled Reticulocyte Lysate System (Promega) was treated for 1 h at 30°C with 1.5 μg MBP-FIH-1 protein in reaction buffer containing 20 mM Tris⋅HCl (pH 7.5), 5 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 2 mM ascorbic acid, 2 mM 2-OG, and 250 μM FeSO4. Approximately 1 μg of immobilized GST-p300 CH1 domain (14) was added to each reaction in 500 μl of binding buffer (20 mM Tris⋅HCl, pH 8/150 mM NaCl/20 μM ZnCl2/0.5 mM DTT) and incubated for 1 h at 4°C. Protein-bound resin was washed four times with 1 ml of binding buffer containing 0.1% Nonidet P-40. Protein was eluted by boiling in SDS sample buffer, and samples were analyzed by SDS/PAGE
MBP Pull-Down Assays.
Twenty-five microliters of 35S-labeled Trx6H mouse HIF-2α 774–874 protein was incubated with MBP-FIH-1 protein immobilized on amylose resin in 200 μl of PBS containing 0.1% Tween-20 for 1 h at 4°C. Protein-bound resin was washed three times with 1 ml buffer and bound protein was eluted by boiling in SDS sample buffer and analyzed by SDS/PAGE.
Equilibrium Sedimentation.
Purified FIH-1 was subjected to equilibrium sedimentation at 4°C in a Beckman Model XL-I analytical ultracentrifuge at both 14,000 and 20,000 rpm in an An60Ti rotor. A final centrifugation at 42,000 rpm was executed to determine baseline quality for data analysis. Three FIH-1 concentrations, 12.4, 8.3, and 4.1 μM, were tested, and data were analyzed at both 280 and 290 nm. Data were fitted to both a monomer-dimer equilibrium and a dimer alone model using the Optima XLA/XLI program from Beckman Instruments.
Results
FIH-1 Overall Structure.
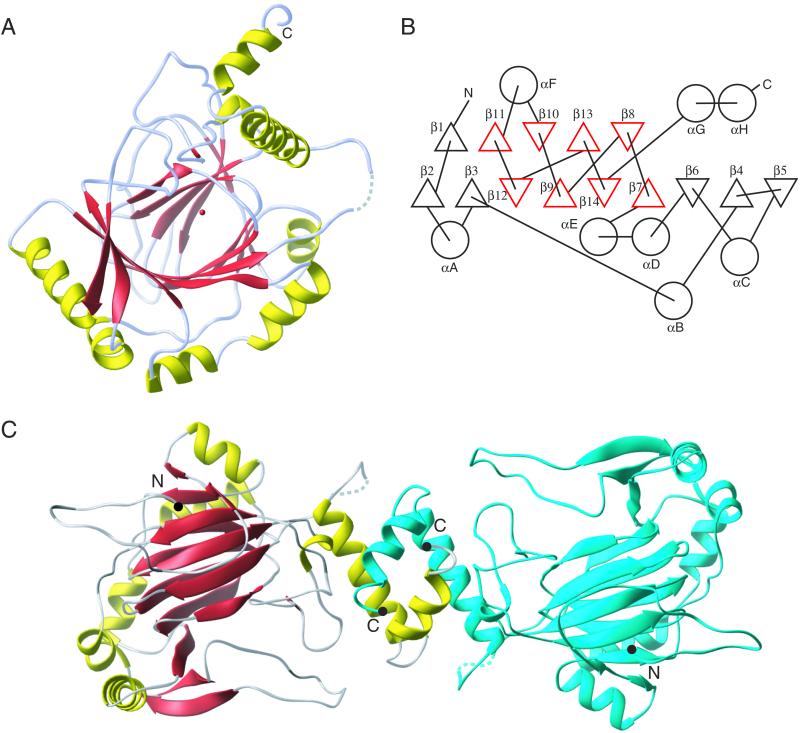
FIH-1 is a mixed α/β structure composed of 14 β-strands arranged in two sheets with nine strands in the first sheet and five strands in the second sheet (Figs. 2 and 3). The central core of FIH-1 is a C-terminal eight β-strand jellyroll (β7–β14) that is homologous to other 2-OG-dependent oxygenases and the cupin protein family (17). The N-terminal region of the protein provides six additional strands that expand the β-sheets away from the central jellyroll core. The eight helical regions are dispersed throughout the structure with the only significant helix–helix interactions occurring at an apparent FIH-1 dimer interface formed by the equivalent C-terminal helices of each monomer (αG and αH, Fig. 3C). Given the position of the active site metal and the solvent accessibility of the active site, a proposed surface for HIF substrate binding can be predicted and is shown in Fig. 3A.
Fig 2.
The primary structure of FIH-1 is labeled with secondary structure elements taken from the x-ray crystallographic model. β-strands and helices are depicted as red arrows and yellow boxes, respectively. Residues responsible for Fe(II) binding are highlighted in red, whereas residues in close contact with 2-OG are highlighted in green.
Fig 3.
FIH-1 structure contains a β-jellyroll core marked by an extension of one of the β-sheets away from the core and helices dotting the periphery. (A) A ribbon model of the FIH-1 monomer is positioned looking between the β-sheets comprising the jellyroll and into the active site cavity. The active site metal is shown as a red sphere. Structural elements are colored as in Fig. 2. (B) A secondary structure topology diagram shows the arrangement of the 14 β-strands (triangles) and 8 helices (circles) in FIH-1. The core jellyroll motif, structurally homologous to the cupin protein family, is colored in red. (C) FIH-1 exists as a functionally relevant dimer in the crystal. The first monomer of the dimer is colored as in A, whereas the second monomer is blue. N and C termini are marked as black circles. The figure was generated by using RIBBONS (41).
FIH-1 Active Site Contacts.
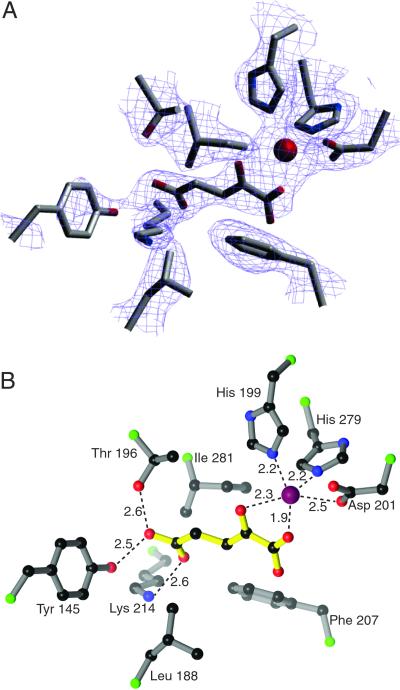
In the absence of exogenous iron and 2-OG, FIH-1 crystallizes with a metal properly coordinated by His-199, Asp-201, and His-279. A single tartrate molecule from the crystallization solution is partially ordered at the active site and occupies two of the metal coordination sites via its carboxylate. The potential sixth ligand site, trans to His-279, remains unresolved at the resolution of the current structure, but is likely to be occupied by a water molecule. When cocrystallized in the presence of both Fe and 2-OG, the 2-OG molecule displaces the tartrate molecule as judged by clear electron density (Fig. 4A). Other than the presence of 2-OG at the active site, the two structures vary only slightly, with a rms deviation of 0.30 Å with 333 of 337 Cα positions aligned.
Fig 4.
Multiple contacts are present in the FIH-1 active site. (A) A 2Fo − Fc simulated annealing omit map contoured at 1.1 σ reveals the presence of 2-OG in the active site. (B) 2-OG makes several hydrophilic contacts with residues of FIH-1, most notably hydrogen bonds to Lys-214 and Thr-196. Additional contacts include coordination to the Fe (red sphere) and hydrophobic interactions with Leu-188, Phe-207, and Ile-281. Ball-and-stick model in B is colored with 2-OG in yellow, Cα positions in green, carbon in black, nitrogen in blue, and oxygen in red. The figure was generated by using SETOR (42) and RIBBONS (41) modeling programs.
To accommodate 2-OG in the active site, the 2-OG-dependent oxygenases must stabilize the charge of the carboxylate at the C5 position. A subset of this class of enzymes have a conserved R-X-S motif from which hydrogen bonds can be made to both oxygens on the C5 carbon of 2-OG (35, 36). Sequence comparisons of FIH-1 with other oxygenases did not reveal the identity of orthologous residues, as FIH-1 uses residues from distant parts of its primary structure for this purpose. Lys-214 and Thr-196 in FIH-1 take on the role of the R-X-S motif by forming hydrogen bonds to the C5 carboxylate of 2-OG. In addition to Lys-214 and Thr-196, other residues including Tyr-145 and Asn-294 may form hydrogen bonds to the 2-OG, though the electron densities of the side chains for these residues are not well defined. Hydrophobic interactions may also occur between 2-OG and Leu-188, Phe-207, and Ile-281.
Functional Relevance of the FIH-1 Dimer.
The two C-terminal helices (αG and αH) of each monomer form tight interactions with a symmetry related molecule, thereby producing an antiparallel helix bundle that buries ≈1,600 Å2 of surface area per monomer. To determine whether the dimer formed in the crystal was also present in solution, purified FIH-1 was subjected to equilibrium sedimentation ultracentrifugation. The data collected from these experiments indicated that FIH-1 is a dimer in solution with a molecular mass of ≈80,300 Da, which agrees well with the molecular mass, 80,817 Da, calculated from the primary structure. Attempts to fit ultracentriguation data to a monomer–dimer equilibrium resulted in unrealistically high values for the monomer–monomer association constant, suggesting that no monomer was present at the concentrations tested.
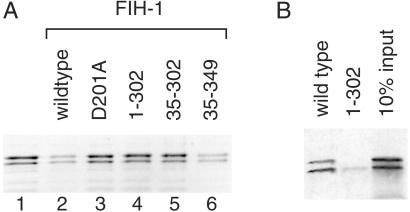
To test whether homodimer formation was required for FIH-1 activity, MBP fusion proteins lacking 34 aa from the N terminus or 47 aa from the C terminus of FIH-1 were expressed (FIH-1 constructs 35–349 and 1–302, respectively). Recombinant FIH-1 proteins were incubated with 35S-labeled CTAD from HIF-2α in the presence of Fe(II), 2-OG and O2. Active wild-type FIH-1 hydroxylated the HIF-2α CTAD as measured by an ability to inhibit CTAD association with the coactivator p300 in a pulldown assay (Fig. 5A, lane 2). A point mutant (D201A) predicted to disrupt the Fe(II) binding site did not interefere with the CTAD/p300 association as predicted for an inactive FIH-1 variant (Fig. 5A, lane 3). Likewise, deletion of the C-terminal 47 FIH-1 residues prevented the truncated protein's ability to hydroxylate the HIF CTAD (Fig. 5A, lanes 4 and 5). Deletion of the N-terminal 34 aa of FIH-1 had no effect on FIH-1 activity (Fig. 5A, lane 6).
Fig 5.
The C terminus of FIH-1 is required for activity. (A) In vitro hydroxylation of HIF-2α 774–874 by wild-type FIH-1 inhibits interaction with p300. 35S-labeled HIF-2α 774–874 was incubated in the absence (lane 1) or presence of various recombinant MBP-FIH-1 enzymes (lanes 2–6) followed by incubation with immobilized GST-p300 CH1. 35S-labeled HIF-2α 774–874 bound to the GST-p300 CH1 domain was visualized by phosphorimaging after SDS/PAGE. Mutations that interfere with Fe(II) binding (D201A) or delete residues 303–349 compromise FIH-1 ability to block p300 association with the HIF-2α CTAD. (B) Deletion of FIH-1 residues 303–349 prevents interaction with the CTAD. 35S-labeled HIF-2α 774–874 bound to immobilized wild-type or truncated (1–302) MBP-FIH-1 was visualized after SDS/PAGE. The right lane indicates 10% of the input 35S-labeled protein in the pull-down experiments.
Because deletion of residues 303–349 would not be expected to interfere with the catalytic core of the FIH-1 enzyme, we examined the effect of this deletion on the ability of FIH-1 to recognize the CTAD substrate. As shown in Fig. 5B, wild-type MBP-FIH-1 is able to interact with 35S-labeled HIF-2α CTAD; however, deletion of the FIH-1 C-terminal 47 aa prevents the FIH-1/CTAD interaction. These data indicate that the FIH-1 homodimer observed in the crystal structure is functionally relevant in solution and is required for FIH-1 substrate recognition.
Discussion
Regulation of the mammalian hypoxic response pathway is mediated by oxygen-dependent hydroxylases. To date, there are few examples of protein regulation dependent upon hydroxylation as a posttranslational modification (reviewed in ref. 37). Although the best-characterized hydroxylase enzymes modify collagen to promote proper folding (23), the number of known functional 2-OG oxygenases involved in other biological pathways is likely to grow (38). FIH-1 is the first asparaginyl hydroxylase to be examined in atomic detail and provides important structural clues regarding both HIF regulation and the differences between this enzyme and the more distantly related prolyl and lysyl hydroxylase enzymes. Although FIH-1 contains a β-sheet core similar to other hydroxylase enzymes, the identities of key residues involved in substrate binding are different.
The structure of FIH-1 revealed the formation of an FIH-1 homodimer that is crucial for its activity in solution. Homodimer formation is required for substrate recognition (Fig. 5), though further investigation is needed to examine possible effects of dimerization on the activity of the FIH-1 catalytic core. The elucidation of the FIH-1 structure also provides a framework to examine proposed interactions between FIH-1 and other components of the hypoxic response pathway. In vitro studies reveal interactions between FIH-1 and pVHL, which play a central role in the regulation of HIF stability, and between FIH-1 and transcriptional corepressors such as histone deacetylases (13). Although the physiological relevance of these interactions remains to be determined, the FIH-1 structure, in combination with that of the pVHL/elongin B/elongin C complex (39), provides a starting point to identify residues that mediate these interactions and to examine their relative contribution to coordinated HIF regulation.
Many inhibitors of Fe(II)-dependent oxygenases operate through direct competition with 2-OG (reviewed in ref. 23). The active site architecture of FIH-1 is similar to other 2-OG dependent oxygenases with respect to Fe(II) binding but differs in overall 2-OG binding, particularly in the residues responsible for binding to the carboxylate of 2-OG distal to the active site metal. Thus the possibility exists for identification of inhibitors that selectively target individual hydroxylases to specifically enhance HIF activity in the context of hypoxic diseases such as ischemia.
Acknowledgments
We thank Murray Whitelaw for providing the HIF-2α CTAD expression construct, the beamline staffs at Advanced Photon Source Structural Biology Center 19ID and Advanced Light Source 8.2.2 for support with data collection, and Derk Binns for assistance with the ultracentrifugation experiments. Use of the Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Biological and Environmental Research, under Contract No. W-31-109-ENG-38. J.D. is an Investigator of the Howard Hughes Medical Institute, and the research of R.K.B. is supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund.
Abbreviations
HIF, hypoxia-inducible factor
FIH-1, factor-inhibiting HIF-1
OG, oxoglutarate
pVHL, product of the von Hippel Lindau tumor suppressor gene
CTAD, C-terminal transactivation domain
MBP, maltose-binding protein
SeMet, selenomethionine
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org [PDB ID codes (FIH-1) and (FIH-1 Fe(II) 2-OG complex)].
References
- 1.Wang G. L., Jiang, B. H., Rue, E. A. & Semenza, G. L. (1995) Proc. Natl. Acad. Sci. USA 92 5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Semenza G. L. (2000) Genes Dev. 14 1983-1991. [PubMed] [Google Scholar]
- 3.Epstein A. C., Gleadle, J. M., McNeill, L. A., Hewitson, K. S., O'Rourke, J., Mole, D. R., Mukherji, M., Metzen, E., Wilson, M. I., Dhanda, A., et al. (2001) Cell 107 43-54. [DOI] [PubMed] [Google Scholar]
- 4.Bruick R. K. & McKnight, S. L. (2001) Science 294 1337-1340. [DOI] [PubMed] [Google Scholar]
- 5.Jaakkola P., Mole, D. R., Tian, Y.-M., Wilson, M. I., Gielbert, J., Gaskell, S. J., von Kriegsheim, A., Hebestreit, H. F., Mukherji, M., Schofield, C. J., et al. (2001) Science 292 468-472. [DOI] [PubMed] [Google Scholar]
- 6.Ivan M., Kondo, K., Yang, H., Kim, W., Valiando, J., Ohh, M., Salic, A., Asara, J. M., Lane, W. S. & Kaelin, W. G. (2001) Science 292 464-468. [DOI] [PubMed] [Google Scholar]
- 7.Yu F., White, S. B. & Lee, F. S. (2001) Proc. Natl. Acad. Sci. USA 98 9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ema M., Hirota, K., Mimura, J., Abe, H., Yodoi, J., Sogawa, K., Poellinger, L. & Fujii-Kuriyama, Y. (1999) EMBO J. 18 1905-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrero P., Okamato, K., Coumailleau, P., O'Brien, S., Tanaka, H. & Poellinger, L. (2000) Mol. Cell. Biol. 20 402-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kung A. L., Wang, S., Klco, J. M., Kaelin, W. G. & Livingston, D. M. (2000) Nat. Med. 6 1335-1340. [DOI] [PubMed] [Google Scholar]
- 11.Gu J., Milligan, J. & Huang, L. E. (2001) J. Biol. Chem. 276 3550-3554. [DOI] [PubMed] [Google Scholar]
- 12.Lando D., Peet, D. J., Whelan, D. A., Gorman, J. J. & Whitelaw, M. L. (2002) Science 295 858-861. [DOI] [PubMed] [Google Scholar]
- 13.Mahon P. C., Hirota, K. & Semenza, G. L. (2001) Genes Dev. 15 2675-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lando D., Peet, D. J., Gorman, J. J., Whelan, D. A., Whitelaw, M. L. & Bruick, R. K. (2002) Genes Dev. 16 1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hewitson K. S., McNeill, L. A., Riordan, M. V., Tian, Y. M., Bullock, A. N., Welford, R. W., Elkins, J. M., Oldham, N. J., Bhattacharya, S., Gleadle, J. M., et al. (2002) J. Biol. Chem. 277 26351-26355. [DOI] [PubMed] [Google Scholar]
- 16.Sang N., Fang, J., Srinivas, V., Leshchinsky, I. & Caro, J. (2002) Mol. Cell. Biol. 22 2984-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clifton I. J., Hsueh, L. C., Baldwin, J. E., Harlos, K. & Schofield, C. J. (2001) Eur. J. Biochem. 268 6625-6636. [DOI] [PubMed] [Google Scholar]
- 18.Zhou J., Gunsior, M., Bachman, B. O., Townsend, C. A. & Solomon, E. I. (1998) J. Am. Chem. Soc. 120 13539-13540. [DOI] [PubMed] [Google Scholar]
- 19.Hanauske-Abel H. M. & Gunzler, V. (1982) J. Theor. Biol. 94 421-455. [DOI] [PubMed] [Google Scholar]
- 20.McNeill, L. A., Hewitson, K. S., Claridge, T. D., Seibel, J. F., Horsfall, L. E. & Schofield, C. J. (2002) Biochem. J., 10.1042/BJ20021162. [DOI] [PMC free article] [PubMed]
- 21.Wu M., Moon, H. S., Begley, T. P., Myllyharju, J. & Kivirikko, K. I. (1999) J. Am. Chem. Soc. 121 587-588. [Google Scholar]
- 22.Baldwin, J. E., Adlington, R. M., Crouch, N. P., Keeping, J. W., Leppard, S. W., Pitlik, J., Schofield, C. J., Sobey, W. J. & Wood, M. (1991) J. Chem. Soc. Chem. Commun., 768–770.
- 23.Kivirikko K. I. & Pihlajaniemi, T. (1998) Adv. Enzymol. Relat. Areas Mol. Biol. 72 325-398. [DOI] [PubMed] [Google Scholar]
- 24.Sheffield P., Garrard, S. & Derewenda, Z. (1999) Protein Expression Purif. 15 34-39. [DOI] [PubMed] [Google Scholar]
- 25.Porath J., Carlsson, J., Olsson, I. & Belfrage, G. (1975) Nature 258 598-599. [DOI] [PubMed] [Google Scholar]
- 26.Carrington J. C. & Dougherty, W. G. (1988) Proc. Natl. Acad. Sci. USA 85 3391-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doublie S. (1997) Methods Enzymol. 276 523-530. [PubMed] [Google Scholar]
- 28.Otwinowski Z. & Minor, W. (1997) Methods Enzymol. 276 307-326. [DOI] [PubMed] [Google Scholar]
- 29.de La Fortelle E. & Bricogne, G. (1997) Methods Enzymol. 276 472-494. [DOI] [PubMed] [Google Scholar]
- 30.Hendrickson W. A. (1991) Science 254 51-58. [DOI] [PubMed] [Google Scholar]
- 31.Abrahams J. P. & Leslie, A. G. W. (1996) Acta Crystallogr. D 52 30-42. [DOI] [PubMed] [Google Scholar]
- 32.Lamzin V. S. & Wilson, K. S. (1993) Acta Crystallogr. D 49 129-149. [DOI] [PubMed] [Google Scholar]
- 33.Jones T., Zou, J.-Y., Cowan, S. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47 110-119. [DOI] [PubMed] [Google Scholar]
- 34.Brunger A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54 905-921. [DOI] [PubMed] [Google Scholar]
- 35.Valegard K., van Scheltinga, A. C., Lloyd, M. D., Hara, T., Ramaswamy, S., Perrakis, A., Thompson, A., Lee, H. J., Baldwin, J. E., Schofield, C. J., Hajdu, J. & Andersson, I. (1998) Nature 394 805-809. [DOI] [PubMed] [Google Scholar]
- 36.Roach P. L., Clifton, I. J., Hensgens, C. M., Shibata, N., Schofield, C. J., Hajdu, J. & Baldwin, J. E. (1997) Nature 387 827-830. [DOI] [PubMed] [Google Scholar]
- 37.Ryle M. J. & Hausinger, R. P. (2002) Curr. Opin. Chem. Biol. 6 193-201. [DOI] [PubMed] [Google Scholar]
- 38.Aravind L. & Koonin, E. V. (2001) Genome Biol. 2 1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stebbins C. E., Kaelin, W. G. & Pavletich, N. P. (1999) Science 284 455-461. [DOI] [PubMed] [Google Scholar]
- 40.Laskowski R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283-291. [Google Scholar]
- 41.Carson M. (1991) J. Appl. Crystallogr. 24 961-985. [Google Scholar]
- 42.Evans S. V. (1993) J. Mol. Graphics 11 134-8., 127–8. [DOI] [PubMed] [Google Scholar]