Abstract
We here describe the cloning and characterization of the functionally active Drosophila melanogaster (Drm) FMRFamide receptor, which we designated as DrmFMRFa-R. The full-length ORF of a D. melanogaster orphan receptor, CG 2114 (Berkeley Drosophila Genome Project), was cloned from genomic DNA. This receptor is distantly related to mammalian thyroid-stimulating hormone-releasing hormone receptors and to a set of Caenorhabditis elegans orphan receptors. An extract of 5,000 central nervous systems from the related but bigger flesh fly, Neobellieria bullata (Neb), was used to screen cells expressing the orphan receptor. Successive purification steps, followed by MS, revealed the sequence of two previously uncharacterized endogenous peptides, APPQPSDNFIRFamide (Neb-FIRFamide) and pQPSQDFMRFamide (Neb-FMRFamide). These are reminiscent of other insect FMRFamide peptides, having neurohormonal as well as neurotransmitter functions. Nanomolar concentrations of the Drm FMRFamides (DPKQDFMRFamide, TPAEDFMRFamide, SDNFMRFamide, SPKQDFMRFamide, and PDNFMRFamide) activated the cognate receptor in a dose-dependent manner. To our knowledge, the cloned DrmFMRFa-R is the first functionally active FMRFamide G protein-coupled receptor described in invertebrates to date.
The publication of the complete euchromatic portion of the Drosophila melanogaster (Drm) genome by Adams et al. (1) enabled the analysis of the full repertoire of Drm G protein-coupled receptors (GPCRs) for the first time. The Drm genome encodes about 160 GPCR genes, including at least 21 GPCRs for classic neurotransmitters and neuromodulators and between 39 and 45 peptide receptors (2, 3). To date, only five Drm peptide GPCRs have been fully characterized functionally (4–8). The others are orphan receptors, meaning that neither their function nor their ligand is known.
Based on sequence similarities revealed by blast analysis and phylogenetic tree construction, the orphan receptor CG2114 was classified within the thyroid-stimulating hormone (TSH)-releasing hormone (TRH) receptors (3). This similarity is of particular interest, because the presence of TRH or thyroid hormones has as yet not been demonstrated in insects or invertebrates in general. Vertebrate thyroid hormones T3 and T4 could play a role in the reproduction of arthropods, because they have effects similar to juvenile hormone on the follicle cells during vitellogenesis (9).
Because we found that this orphan receptor was not efficiently activated by bovine TRH, the natural ligand in Drm was searched for by means of a “reverse physiology” approach (10). The functionally expressed orphan Drm receptor was screened against peptide fractions purified from a CNS tissue extract from the related but much bigger flesh fly, Neobellieria bullata (Neb). In the present paper, we show that FMRFamides are the cognate ligands for this orphan receptor, which we annotated as the Drm FMRFamide receptor or DrmFMRFa-R.
Materials and Methods
Cloning of the DrmFMRFa-R.
The ORF of the orphan GPCR was amplified by PCR performed on the genomic Drm bacterial artificial chromosome clone, RPCI98-21A2 (GenBank accession no. AC010561), which contains the entire uninterrupted coding sequence of the CG2114 gene (1). Oligonucleotide PCR primers were designed to encompass the ORF. The forward and reverse primers had the following sequences: forward primer, 5′-GGAATTCGCCACCATGAGTGGTACAGCGGTTGCG-3′ and reverse primer, 5′-GCTCTAGAGCCCGGACACAATCTCAGAATC-3′. The forward primer also incorporates the Kozak sequence (GCCACC) to optimize the translation initiation (11), as well as an EcoRI restriction site. The reverse primer contains an XbaI restriction site to allow for directional cloning. The Advantage 2 PCR kit (CLONTECH) was used to amplify the receptor-coding sequence under the following PCR conditions: 95°C, 3 min; 94°C, 60 s; 55°C, 30 s; 68°C, 3 min; 30 cycles. After TOPO TA cloning (TOPO TA cloning kit, Invitrogen) of the obtained PCR product, the integrity of the cloned insert was verified using automated DNA sequencing. Clones containing the correct fragment were used to subclone the PCR product into the pcDNA3 mammalian expression vector (Invitrogen) to yield the CG2114/pcDNA3 plasmid, and the DNA sequence was reanalyzed. Plasmid DNA for transfection was prepared by using Qiagen (Valencia, CA) ion exchange columns.
Cell Culture and Creation of a Stable Cell Line Expressing DrmFMRFa-R.
To find a cognate ligand for the orphan receptor, we stably transfected the receptor in Chinese hamster ovary (CHO) cells [a kind gift of M. Detheux (Euroscreen, Brussels)] that also stably expressed the human Gα16 protein (G16), as well as mitochondrially targeted aequorin (mtAEQ). The CHO/mtAEQ/G16 cells were cultured in 75-cm2 flasks at 37°C in a humidified atmosphere of 5% CO2 in Ham's F12 medium supplemented with 500 mg of l-glutamine (BioWhittaker)/10% FBS (Invitrogen)/250 μg/ml Zeocin (Invitrogen). The CG2114/pcDNA3 construct was stably transfected into the CHO/mtAEQ/G16 cells by using the FuGENE 6 reagent (Roche Diagnostics) according to the manufacturer's instructions to yield CHO/mtAEQ/G16/CG2114. Two days after transfection, the cells were selected for 2.5 weeks with 400 μg/ml G418 (Invitrogen), and clones were selected for further experiments. Subsequently, the expression of CG2114 was determined by RT-PCR as described below. The clone that showed the highest level of CG2114 expression was used for screening.
Expression Analysis by RT-PCR.
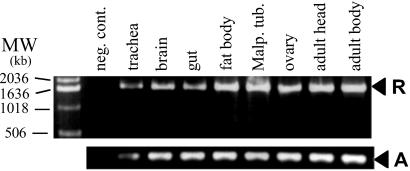
To evaluate tissue expression, tissue-specific (polyA)+mRNA was extracted from the CNS, fat body, intestine, Malpighian tubules, and trachea of Drm wandering larvae and from ovaries and from both heads and headless bodies of adult flies by using the QuickPrep Micro mRNA Purification kit (Roche Diagnostics) (see Fig. 2). First-strand cDNA was synthesized from 0.5 μg of each mRNA sample by using the RevertAid M-MuLV Reverse Transcriptase kit (Fermentas, St. Leon-Rot, Germany) with an oligo(dT)18 primer, according to the manufacturer's protocol. Receptor expression in transfected cells was demonstrated in the same manner. Negative control reactions, in which reverse transcriptase was omitted, were tested in parallel. As a positive control, primers were designed to amplify a 340-bp fragment of Drm actin (forward primer, 5′-GGGCATGTGCAAAGCCG-3′; reverse primer, 5′-GAAGGTCTCGAACATGATCTGGG-3′). Expression patterns were determined by analyzing the resulting RT-PCR product on an agarose gel.
Fig 2.
DrmFMRFa-R expression in different Drm larval organs (trachea, brain, gut, fat body, and Malpighian tubules) and adult samples (ovaries, heads, and headless bodies). Primers were chosen to amplify the full coding region of the receptor (R) or a 340-bp fragment of Drm actin (A).
Measurement of Intracellular Ca2+ Mobilization.
An aequorin bioluminescence assay was used to measure the intracellular Ca2+ concentration. After trypsinization with PBS-EDTA (5 mM EDTA), cells were incubated with coelenterazine h (Molecular Probes) in BSA medium (DMEM/Ham's F12 medium supplemented with 15 mM of Hepes and 1% BSA, without phenol red) at a final concentration of 5 μM for 4 h to reconstitute the holoenzyme apoaequorin. Thirty minutes before screening, the cells were diluted 10-fold. Test fractions (HPLC fractions and synthetic peptides) were dried, reconstituted in 50 μl of sterile BSA medium, and pipetted into the wells of a 96-microtiter plate. For each measurement, 50 μl of cell suspension (final volume, 100 μl) was added to the test samples by injection, by using the Microlumat plus microplate luminometer (EG&G Berthold; Perkin–Elmer; Life Sciences, St. Petersburg, FL). Triton X-100 (0.1%) in BSA medium was used as a positive control, BSA medium only as a negative control, and 1 μM of ATP was used to test the functionality of the assay. The emitted light was recorded for 30 seconds, immediately after injection. Data were analyzed by using excel (Microsoft) and sigmaplot (SPSS, Chicago).
Tissue Extraction and HPLC Purification.
A total of 5,000 CNSs were dissected from the gray flesh fly and extracted in methanol/water/acetic acid (90:9:1; vol/vol/vol). After centrifugation, the supernatant was delipidated with ethyl acetate and subsequently with n-hexane, by adding equal volumes of solvent to the supernatant. The aqueous residue was used for C18 solid phase extraction on MegabondElute C18 cartridges (Varian). The material eluted by 0–60% acetonitrile in aqueous 0.1% trifluoroacetic acid was further fractionated by reversed-phase HPLC (Table 1). Columns 1 and 2 were purchased from Waters; column 3 was purchased from Vydac (Hesperia, CA).
Table 1.
HPLC purification of the Neb ligands for the DrmFMRFa-R
| Column | Gradient of acetonitrile in 0.1% trifluoroacetic acid | Retention time of active fractions, min |
|---|---|---|
| 1. Xterra C8 (7.8 × 300 mm; 7 μm; 125 Å) | A. 0–80% (120 min; 3 ml/min) | 40 |
| B. 0–80% (120 min; 3 ml/min) | 43 | |
| 2. Symmetry C18 (4.6 × 250; 5 μm; 100 Å) | A. 0–10% (10 min; 1 ml/min), 10–35% (60 min; 1 ml/min) | 47 |
| B. 0–20% (10 min; 1 ml/min), 20–35% (60 min; 1 ml/min) | 31 | |
| 3. Diphenyl (2.1 × 250; 5 μm; 300 Å) | A. 0–5% (5 min; 0.2 ml/min), 5–30% (60 min; 0.2 ml/min) | 58 |
| B. 0–17.5% (10 min; 0.2 ml/min), 17.5–35% (50 min; 0.2 ml/min) | 48 |
Conditions for the purification of Neb-FIRFamide are indicated after A; purification conditions for Neb-FMRFamide are presented after B.
Ligand Identification.
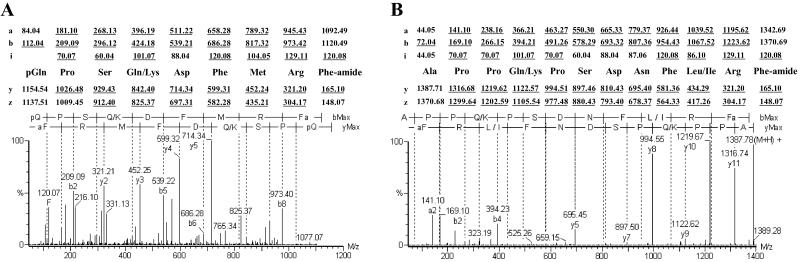
The positive ion electrospray ionization (ESI) mass spectra of purified fractions were recorded on a Q-TOF mass spectrometer (Micromass, Manchester, U.K.), equipped with a nano-ESI ion source (Fig. 4). The sequence was determined by MS/MS, or tandem MS. Fragment ions were generated from a selected precursor ion by collision-induced dissociation. When amino acids could not be unequivocally identified by MS/MS, sequencing was also performed on an automated Procise protein sequencer (Applied Biosystems).
Fig 4.
(A) Collision-induced dissociation (CID) spectrum of the peptide at m/z 569.28 yields pQPSQ/KDFMRFamide as sequence. (B) CID spectrum of the peptide at m/z 694.39 yields APPQ/KPSDNFL/IRFamide as sequence, a-, b-, y-, or z-type, and immonium (i) fragment ions are indicated. The theoretical fragment–ion masses found in the spectrum are underlined. pQ, pyroglutamic acid. In B, automated Edman amino acid sequencing revealed a Gln at position 4 and an Ile residue at position 10. In A, the Gln-4 was established by MS/MS of tryptic digests of both possible sequences.
Pharmacological Characterization.
To verify that the purified Neb peptides are indeed ligands for the receptor, synthetic analogues were analyzed for activity. Orthologous FMRFamide peptides in Drm were also synthesized and tested for activity. The pharmacological specificity of the FMRFamide receptor was evaluated by exposure of receptor-expressing cells to various other insect and crustacean peptides as well as truncated FMRFamide analogues (Table 2). Peptides were either custom-synthesized (Invitrogen) or made in house by using conventional fluorenylmethoxycarbonyl (Fmoc) chemistry.
Table 2.
Pharmacological profiling of DrmFMRFa-R
| Peptide | Sequence | EC50, nM |
|---|---|---|
| Drm-FMRFamide-1 | DPKQDFMRF-NH2 | 2.0 |
| Drm-FMRFamide-2 | TPAEDFMRF-NH2 | 2.8 |
| Drm-FMRFamide-3 | SDNFMRF-NH2 | 1.9 |
| Drm-FMRFamide-4 | SPKQDFMRF-NH2 | 2.5 |
| Drm-FMRFamide-5 | PDNFMRF-NH2 | 1.8 |
| [Ala4] Drm-FMRFa3 | SDNAMRF-NH2 | 102 |
| [Ala4] Drm-FMRFa5 | PDNAMRF-NH2 | 64 |
| Neb-FIRFamide | APPQPSDNFIRF-NH2 | 3.5 |
| Neb-FMRFamide | pQPSQDFMRF-NH2 | 2.0 |
| Drm-sNPF-1 | AQRSPSLRLRF-NH2 | 270 |
| Drm-SK-1 | FDDYGHMRF-NH2 | 38 |
| Drm-SK-2 | GGDDQFDDYGHMRF-NH2 | 105 |
| Drm-MS | TDVDHVFLRF-NH2 | 91 |
| Scg-FLRFamide | PDVDHVFLRF-NH2 | 25 |
| Pev-SK | AGGSGGVGGEYDDYGHLRF-NH2 | 85 |
| FMRFamide | FMRF-NH2 | 28 |
| AMRFamide | AMRF-NH2 | 3,217 |
| YMRFamide | YMRF-NH2 | 31 |
| MRFamide | MRF-NH2 | 6,416 |
| RFamide | RF-NH2 | n.a. |
| Lom-MIP | AWQDLNAGW-NH2 | n.a. |
| Pev-PK 2 | ADFAFNPRL-NH2 | n.a. |
| Corazonin | pQTFQWSHGWTN-NH2 | n.a. |
n.a., not active up to 10 μM. The Y residues of the sulfakinins indicated in bold are sulfated.
Results
Cloning of the Full-Length cDNA of DrmFMRFa-R.
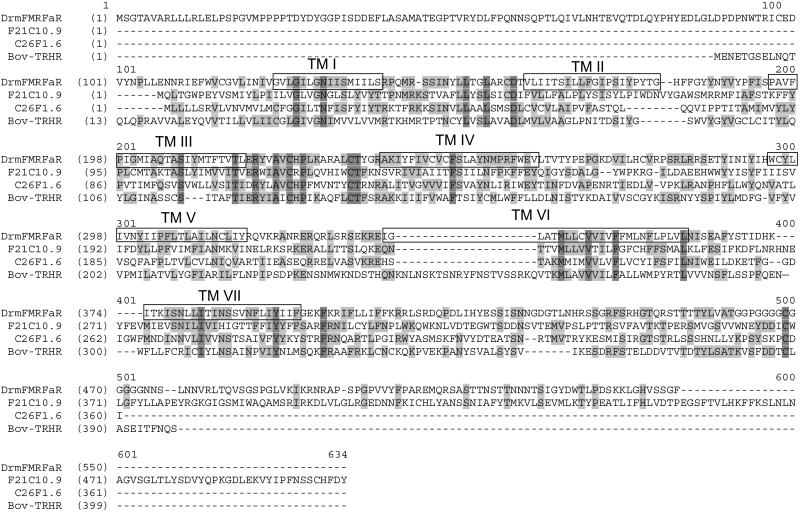
Alignment of the predicted mRNA sequence of the CG2114 receptor with the genomic DNA of Drm revealed that the corresponding gene was intronless. Therefore, the ORF of the gene could be cloned from genomic Drm DNA. The DNA sequence was found in a Drm bacterial artificial chromosome (BAC) clone (RPCI98-21A2) with GenBank accession no. AC010561. Amplification with sequence-specific primers confirmed that the BAC clone encoded the full-length ORF of the orphan receptor. Prediction of the presence of seven-membrane-spanning domains of the corresponding amino acid sequence was confirmed before amplification, by using the tmhmm prediction program (www.cbs.dtu.dk/services/TMHMM-2.0/). The Drm receptor displays 16.7–20.8% overall amino acid identity with some C. elegans receptors and 16% sequence identity with the bovine TRH receptor (Fig. 1). All alignments were performed by using the alignx program (Informax, Oxford).
Fig 1.
Alignment of the DrmFMRFa-R with the two most closely related C. elegans orphan receptors (F21C10.9 and C26F1.6) and with the bovine TRH receptor. Identical amino acids are highlighted in dark gray, conservative amino acids are in light gray, and the seven-membrane-spanning domains of DrmFMRFa-R are numbered I–VII. Dashed lines are spaces to optimize alignment.
Distribution of DrmFMRFa-R.
The receptor is present in all analyzed Drm larval organs, as well as in ovaries, heads, and bodies of adult fruit flies (Fig. 2). Tracheae also express the receptor. Therefore, expression in all tested organs may be attributed (at least partially) to the presence of internal tracheoles, which could not be removed during dissection. All samples in which reverse transcriptase was omitted were negative.
Identification of a Neuropeptide Ligand.
Cells expressing the Drm orphan receptor were challenged with fractions of the flesh fly CNS extract. Flesh fly, rather than fruit fly, extracts were used because of the starting material required: 4⋅105 Drm whole bodies (8), in contrast to 5,000 CNSs from Neb larvae, which are relatively easy to dissect and hence require fewer purification steps.
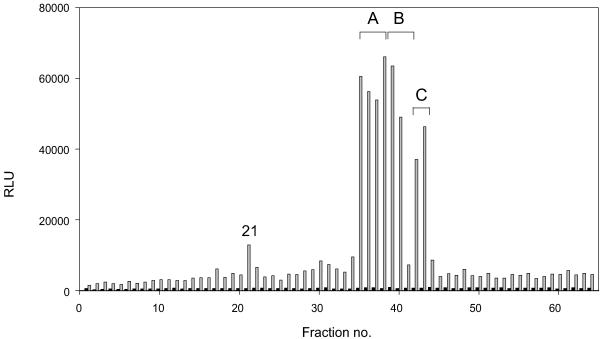
The closest related receptor for which a cognate ligand had been identified was the bovine TRH receptor (only ≈16% sequence homology). Thyroid hormones (T3 and T4) have not yet been described in insects, and the receptor-expressing cells did not respond to bovine TRH in concentrations up to 10 μM (data not shown). We used CNS extracts because we expected the ligand to be related to TRH, and TRH is predominantly present in the hypothalamus. After assessing activity in the 0–60% acetonitrile fraction, we fractionated the peptide extract on an Xterra C8 column and tested the obtained 70 fractions for their ability to elicit a bioluminescent calcium response in CG2114-expressing CHO cells. Three areas of activity were found in eight fractions, suggesting the presence of more than one active ligand (Fig. 3). This response was not seen in CHO cells that were transfected with the empty pcDNA3 vector. Bioactive fractions were subjected to two further HPLC fractionations (Table 1) and testing until a single active peak was obtained.
Fig 3.
Bioluminescence response in relative light units (RLU) of the DrmFMRFa-R-expressing CHO cells (gray bars) and of CHO/G16 cells that were transfected with the empty pcDNA3 vector (black bars) after addition of 0.3% of first column (C8) HPLC fractions (≈16 Neb CNS equivalents). Three areas of activity can be distinguished (fractions 35–37, 38–40, and 42–43), and these fractions were mutually pooled for further purification. The weak activity in fraction 21 was lost after further purification.
Two fractions were obtained from which the two most prominent peaks at m/z 569.28 and m/z 694.39 were selected for fragmentation. The amino acid sequence of the peptides was determined to be pQPSQ/KDFMRFamide and APPQ/KPSDNFI/LRFamide (Fig. 4). Because MS/MS sequencing cannot distinguish between Leu and Ile (identical masses) or between Lys and Gln (mass difference of 0.04 Da), the second peptide was also subjected to automated Edman-based sequencing, which yielded a Gln at position 4 and an Ile at position 10. Because the first mentioned peptide is blocked by a pyroGlu at the N terminus, it cannot be sequenced directly by using Edman chemistry. An overnight tryptic digest experiment, followed by MS/MS, revealed that the synthetic Lys-4 isoform was cleaved into pQPSK (m/z 442.20) and DFMRFamide (m/z 374.65), whereas the Gln-4 isoform and the natural peptide remained intact. The primary structure of the native peptide is hence pQPSQDFMRFamide.
Both identified peptides have an FXRFamide C terminus (X being M or I) and were therefore assigned as Neb-FMRFamide and Neb-FIRFamide, respectively. Identical retention times of the native and synthetic peptides further confirm the proposed sequences.
Pharmacological Characterization of the DrmFMRFa-R (Table 2).
blast analysis, followed by testing of the synthetic peptides, revealed that the Drm FMRFamides are the native ligands in Drm. All five different Drm-FMRFamides derived from the dFMRFa gene as well as Neb-FMRFamide and Neb-FIRFamide (Table 2) tested positive on the transformed CHO cells in a dose-dependent manner (Fig. 5). Cells did not respond to the addition of BSA medium alone. The positive controls, 1 μM of ATP and 0.1% Triton X-100 in BSA medium, also yielded a clear calcium response. The EC50 values of all tested Drm-FMRFamides varied around 2 nM.
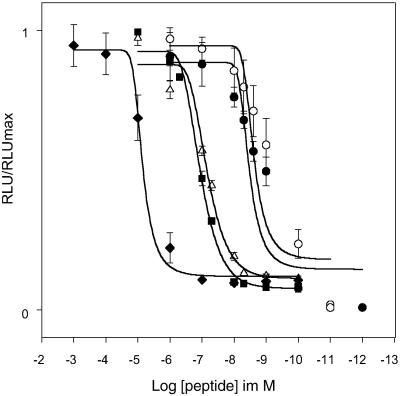
Fig 5.
Dose–response curves of the effects of synthetic Drm-FMRFamide-2 (•), Drm-FMRFamide-3 (○), Drm-myosuppressin (▪), PDNAMRFamide (Δ), and MRFamide (♦) on the DrmFMRFa-R-expressing CHO cells. Bioluminescence is expressed as relative light units (RLU/RLUmax), and error bars are indicated. All EC50 values were determined after nonlinear regression analysis with SIGMAPLOT.
Drosophila peptides ending in −RF-NH2: Drm-myosuppressin (EC50: 9.1⋅10−8 M), Drm-sulfakinin-1 (Drm-SK-1) (3.8⋅10−8 M), Drm-SK-2 (1.1⋅10−7 M), and Drm-short neuropeptide F (2.7⋅10−8 M) were found to be less potent than the Drm FMRFamides. The importance of the Phe residue at position 4 from the C terminus was assessed by testing mutant peptides with Ala substitutions such as AMRFamide, PDNAMRFamide, and SDNAMRFamide. All these displayed lower potency (Table 2). Substitution of Phe by Tyr did not result in an appreciable loss of activity, suggesting that an aromatic residue at that position is essential for full activity. Furthermore, it was found that all active insect FMRFamides have a D residue at position 5 or 6 from the C terminus. The truncated MRFa was the shortest form that was still active, with an EC50 value of 6.4⋅10−6 M.
FMRF-related peptides from other insects, i.e., Schistocerca gregaria-FLRFamide (desert locust) (12), and from crustaceans, i.e., Penaeus vannamei (Pev)-sulfakinin (white shrimp) (13), were also able to induce a bioluminescence response, again with lesser potency. Neuropeptides that are not related to FMRFamide, such as Pev-pyrokinins (14), corazonin (15), and Locusta-myoinhibiting peptide (16), were inactive.
Discussion
The above data clearly demonstrate that we have cloned and functionally characterized the first, to our knowledge, G protein-coupled FMRFamide receptor in invertebrates. Two previously undescribed peptides, Neb-FMRFamide and Neb-FIRFamide, from the gray flesh fly were identified as agonists of the FMRFamide receptor. Neb-FIRFamide is identical to CalliFIRFamide from the blowfly Calliphora vomitoria (17). As revealed by blast, Neb-FIRFamide is most similar to Drm-FMRFamide 3, whereas Neb-FMRFamide is most similar to Drosophila virilis FMRFamides (18). The Drm FMRFamide gene (dFMRFa) was one of the first insect neuropeptide genes to be cloned (19, 20). It encodes multiple putative peptide sequences, four of which (Drm-FMRFamide-1, -2, -3, and -5) have been confirmed either by traditional purification (20, 21) or by peptidomics (22). The different Drm-FMRFamides (Drm-FMRFamides 1–5, Table 2), all yield a clear calcium response in the receptor-expressing cells. The responses are dose-dependent, with EC50 values of ≈2 nM, similar to the responses obtained with the purified Neb-FMRFamide and Neb-FIRFamide (EC50 of 2 and 3.5 nM, respectively).
The first FMRF-amide was isolated in 1977 as a cardioexcitatory molecule from the clam Macrocallista nimbosa (23). Subsequent to this discovery, FMRFamide-related peptides have been found throughout the animal kingdom (24, 25), where they have many physiological functions, including muscular control (17, 25, 26), cardioregulation (23, 27), pain modulation (28), and learning (29).
In Drm, expression of dFMRFa is found in a variety of cell types, including interneurones and neurosecretory cells of the CNS and midgut (30, 31). The presence of FMRFamide-like immunoreactivity in insect neurohemal organs and in the hemolymph suggests a hormonal role (32). In addition, FMRFamides act as neurotransmitters/neuromodulators within the larval and adult CNS (33, 34), as well as at selected peripheral targets. The latter include, for example, tissues associated with feeding (gut, salivary glands), reproduction (accessory glands, spermatheca, and oviducts), movement (skeletal muscle), circulation (aorta), and ecdysis (coordinated modulation of visceral and skeletal muscles) (33–35) (for a recent review, see ref. 32). In vivo experiments have shown the inhibitory effect of Drm-FMRFamide-3 on heart rate (35). Hewes et al. (36) show that Drm FMRFamides strongly enhance twitch tension of larval body-wall muscle, in a very similar and functionally redundant way. The EC50 values reported in that study (≈40 nM) are 20-fold higher than the values we found using the aequorin assay (≈2 nM) and are probably due to the higher sensitivity of the aequorin assay. This result also provides further evidence that the presently identified orphan receptor is the authentic FMRFamide receptor.
It has been reported that FMRFamides exert their effect by directly activating FMRFamide-gated sodium channels without involvement of a G protein (37, 38). However, earlier electrophysiological studies in molluscs suggested that FMRFamide could also activate a GPCR (39). In 1998, a GPCR from Lymnaea heart cDNA was identified and found to be activated by TPHWRPQGRFamide (Lymnaea cardioexcitatory peptide or LyCEP) (40). Whereas the LyCEP receptor shows homology to the Drm PR4 or neuropeptide Y receptor (41), our Drm FMRFamide receptor does not display substantial sequence similarities to the LyCEP receptor. Pharmacological profiling indicated that the LyCEP receptor did not respond to FMRFamide, SDPFLRFamide, or GDPFLRFamide. The presently cloned Drm receptor is preferentially activated by peptides ending either in −FMRFamide or −FI(L)RFamide. Therefore, this is the first report, to our knowledge, of invertebrates demonstrating that FMRFamides, in addition to their interaction via Na+ channels, are also able to activate a specific GPCR (at least in vitro), thereby causing changes in the cell of longer duration.
This work provides tools to study the secondary messenger pathway that is turned on upon activation of DrmFMRFa-R in Drm tissues. Our results also provide important leads to identify additional FMRFamide receptors in other insect species, i.e. Anopheles gambiae, as well as in other invertebrates. Especially interesting is the similarity of the DrmFMRFa-R with a set of orphan C. elegans receptors that show between 22% and 33% mutual overall amino acid sequence similarity and of which homology within the 7TM regions varies between 32% and 53% (Fig. 1). It is possible that these C. elegans orphan receptors are also FMRFamide receptors. The possibility of C. elegans having more than one such receptor is not very surprising, given the number of FMRFamide-type peptides (at least 56) present in this nematode (42). Interesting to note in this respect is that in the nematode C. elegans, the numerous FMRFamides do not seem to be functionally redundant, because deletion of one of the 20 FMRFamide-like peptide genes (flp) resulted in numerous behavioral deficits (43).
Alignment of the Drm FMRFamide receptor with the closest related Drm receptor genes (CG5911, CG13803, CG8985, and CG5936) yielded only 11–14% sequence identity (data not shown), suggesting that only one FMRFamide receptor is encoded by the Drm genome.
Taken together, the identification of the insect FMRFamide receptor opens new opportunities for researchers to evaluate the physiological role of these peptides. Throughout the Metazoa, FMRFamides play a vital role in neuroendocrinological processes. In addition, the FMRFamide receptor may be a pharmacologically interesting target for the selection and design of insect control agents.
Acknowledgments
We thank Dr. M. Detheux (Euroscreen, Gosselies, Belgium) for supplying cell line CHO/mtAEQ/G16 and for help with the aequorin assay, and L. Vanden Bosch and S. Van Soest for excellent technical assistance. The Flemish Science Foundation (G.0356.98, G.0187.00, and G.0175.02), the Interuniversity Poles of Attraction Programme P5/30, and the National Institutes of Health (MH62177) sponsored this project.
Abbreviations
CHO, Chinese hamster ovary
Drm, Drosophila melanogaster
DrmFMRFa-R, Drm FMRFamide receptor
G16, G protein-16
GPCR, G protein-coupled receptor
Neb, Neobellieria bullata
TRH, thyroid-stimulating hormone-releasing hormone
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The amino acid sequences for both Neb-FIRFamide and Neb-FMRFamide have been deposited in the SWISS-PROT protein sequence database (accession nos. P83349 and P83350, respectively). The complete nucleotide sequence of DrmFMRFa-R has been deposited in the GenBank database (accession no. BK000442).
References
- 1.Adams M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., et al. (2000) Science 287 2185-2195. [DOI] [PubMed] [Google Scholar]
- 2.Brody T. & Cravchik, A. (2000) J. Cell Biol. 150 F83-F88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hewes R. S. & Taghert, P. H. (2001) Genome Res. 11 1126-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenz C., Williamson, M., Hansen, G. N. & Grimmelikhuijzen, C. J. (2001) Biochem. Biophys. Res. Commun. 286 1117-1122. [DOI] [PubMed] [Google Scholar]
- 5.Auerswald L., Birgul, N., Gade, G., Kreienkamp, H. J. & Richter, D. (2001) Biochem. Biophys. Res. Commun. 282 904-909. [DOI] [PubMed] [Google Scholar]
- 6.Larsen M. J., Burton, K. J., Zantello, M. R., Smith, V. G., Lowery, D. L. & Kubiak, T. M. (2001) Biochem. Biophys. Res. Commun. 286 895-901. [DOI] [PubMed] [Google Scholar]
- 7.Kubiak T. M., Larsen, M. J., Burton, K. J., Bannow, C. A., Martin, R. A., Zantello, M. R. & Lowery, D. E. (2002) Biochem. Biophys. Res. Commun. 291 313-320. [DOI] [PubMed] [Google Scholar]
- 8.Staubli F., Jorgensen, T. J., Cazzamali, G., Williamson, M., Lenz, C., Sondergaard, L., Roepstorff, P. & Grimmelikhuijzen, C. J. (2002) Proc. Natl. Acad. Sci. USA 99 3446-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davey K. G. & Gordon, D. R. (1996) Arch. Insect Biochem. Physiol. 32 613-622. [DOI] [PubMed] [Google Scholar]
- 10.Reinscheid R. K., Nothacker, H. P., Bourson, A., Ardati, A., Henningsen, R. A., Bunzow, J. R., Grandy, D. K., Langen, H., Monsma, F. J., Jr. & Civelli, O. (1995) Science 270 792-794. [DOI] [PubMed] [Google Scholar]
- 11.Kozak M. (1989) J. Cell Biol. 108 229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoofs L., Holman, G. M., Paemen, L., Veelaert, D., Amelinckx, M. & De Loof, A. (1993) Peptides (Tarrytown, NY) 14 409-421. [DOI] [PubMed] [Google Scholar]
- 13.Schoofs L., Nieto, J., Cerstiaens, A., Derua, R., Waelkens, E., Calderon, J. & De Loof, A. (2002) in Crustaceans and the Biodiversity Crisis: Proceedings of the Fourth International Crustacean Congress, eds. Schram, F. R. & von Vaupel Klein, J. C. (Brill, Leiden, The Netherlands), pp. 951–960.
- 14.Torfs P., Nieto, J., Cerstiaens, A., Boon, D., Baggerman, G., Poulos, C., Waelkens, E., Derua, R., Calderon, J., De Loof, A., et al. (2001) Eur. J. Biochem. 268 149-154. [DOI] [PubMed] [Google Scholar]
- 15.Veenstra J. A. (1991) Peptides (Tarrytown, NY) 12 1285-1289. [DOI] [PubMed] [Google Scholar]
- 16.Schoofs L., Holman, G. M., Hayes, T. K., Nachman, R. J. & De Loof, A. (1991) Regul. Pept. 36 111-119. [DOI] [PubMed] [Google Scholar]
- 17.Duve H., Johnsen, A. H., Sewell, J. C., Scott, A. G., Orchard, I., Rehfeld, J. F. & Thorpe, A. (1992) Proc. Natl. Acad. Sci. USA 89 2326-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taghert P. H. & Schneider, L. E. (1990) J. Neurosci. 10 1929-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider L. E. & Taghert, P. H. (1988) Proc. Natl. Acad. Sci. USA 85 1993-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nambu J. R., Murphy-Erdosh, C., Andrews, P. C., Feistner, G. J. & Scheller, R. H. (1988) Neuron 1 55-61. [DOI] [PubMed] [Google Scholar]
- 21.Nichols R. (1992) J. Mol. Neurosci. 3 213-218. [DOI] [PubMed] [Google Scholar]
- 22.Baggerman G., Cerstiaens, A., De Loof, A. & Schoofs, L. (2002) J. Biol. Chem. 277 40368-40374. [DOI] [PubMed] [Google Scholar]
- 23.Price D. A. & Greenberg, M. J. (1977) Science 197 670-671. [DOI] [PubMed] [Google Scholar]
- 24.Greenberg M. J. & Price, D. A. (1992) Prog. Brain Res. 92 25-37. [DOI] [PubMed] [Google Scholar]
- 25.Raffa R. B. (1988) Peptides (Tarrytown, NY) 9 915-922. [DOI] [PubMed] [Google Scholar]
- 26.Maule A. G., Geary, T. G., Marks, N. J., Bowman, J. W., Friedman, A. R. & Thompson, D. P. (1996) Int. J. Parasitol. 26 927-936. [DOI] [PubMed] [Google Scholar]
- 27.Kuhlman J. R., Li, C. & Calabrese, R. L. (1985) J. Neurosci. 5 2310-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang J., Yang, H. Y. & Costa, E. (1984) Proc. Natl. Acad. Sci. USA 81 5002-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackey S. L., Glanzman, D. L., Small, S. A., Dyke, A. M., Kandel, E. R. & Hawkins, R. D. (1987) Proc. Natl. Acad. Sci. USA 84 8730-8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider L. E., Roberts, M. S. & Taghert, P. H. (1993) Neuron 10 279-291. [DOI] [PubMed] [Google Scholar]
- 31.Nichols R., McCormick, J. & Lim, I. (1999) J. Neurobiol. 39 347-358. [DOI] [PubMed] [Google Scholar]
- 32.Orchard I., Lange, A. B. & Bendena, W. G. (2001) in Advances in Insect Physiology, ed. Evans, P. (Academic, London), pp. 268–329.
- 33.Nichols R., McCormick, J. B. & Lim, I. A. (1999) Ann. N.Y. Acad. Sci. 897 264-272. [DOI] [PubMed] [Google Scholar]
- 34.Taghert P. H. (1999) Microsc. Res. Tech. 45 80-95. [DOI] [PubMed] [Google Scholar]
- 35.Nichols R., McCormick, J., Cohen, M., Howe, E., Jean, C., Paisley, K. & Rosario, C. (1999) J. Neurogenet. 13 89-104. [DOI] [PubMed] [Google Scholar]
- 36.Hewes R. S., Snowdeal, E. C., III, Saitoe, M. & Taghert, P. H. (1998) J. Neurosci. 18 7138-7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeziorski M. C., Green, K. A., Sommerville, J. & Cottrell, G. A. (2000) J. Physiol. (London) 526 13-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lingueglia E., Champigny, G., Lazdunski, M. & Barbry, P. (1995) Nature 378 730-733. [DOI] [PubMed] [Google Scholar]
- 39.Green K. A., Falconer, S. W. & Cottrell, G. A. (1994) Pflügers Arch. 428 232-240. [DOI] [PubMed] [Google Scholar]
- 40.Tensen C. P., Cox, K. J., Smit, A. B., van der Schors, R. C., Meyerhof, W., Richter, D., Planta, R. J., Hermann, P. M., van Minnen, J., Geraerts, W. P., et al. (1998) J. Neurosci. 18 9812-9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X. J., Wu, Y. N., North, R. A. & Forte, M. (1992) J. Biol. Chem. 267 9-12. [PubMed] [Google Scholar]
- 42.Li C., Kim, K. & Nelson, L. S. (1999) Brain Res. 848 26-34. [DOI] [PubMed] [Google Scholar]
- 43.Nelson L. S., Rosoff, M. L. & Li, C. (1998) Science 281 1686-1690. [DOI] [PubMed] [Google Scholar]