Abstract
The 20 aminoacyl-tRNA synthetases catalyze the first step of protein synthesis and establish the rules of the genetic code through aminoacylation reactions. Biological fragments of two human enzymes, tyrosyl-tRNA synthetase (TyrRS) and tryptophanyl-tRNA synthetase, connect protein synthesis to cell-signaling pathways including angiogenesis. Alternative splicing or proteolysis produces these fragments. The proangiogenic N-terminal fragment mini-TyrRS has IL-8-like cytokine activity that, like other CXC cytokines, depends on a Glu-Leu-Arg motif. Point mutations in this motif abolish cytokine activity. The full-length native TyrRS lacks cytokine activity. No structure has been available for any mammalian tRNA synthetase that, in turn, might give insight into why mini-TyrRS and not TyrRS has cytokine activities. Here, the structure of human mini-TyrRS, which contains both the catalytic and the anticodon recognition domain, is reported to a resolution of 1.18 Å. The critical Glu-Leu-Arg motif is located on an internal α-helix of the catalytic domain, where the guanidino side chain of R is part of a hydrogen-bonding network tethering the anticodon-recognition domain back to the catalytic site. Whereas the catalytic domains of the human and bacterial enzymes superimpose, the spatial disposition of the anticodon recognition domain relative to the catalytic domain is unique in mini-TyrRS relative to the bacterial orthologs. This unique orientation of the anticodon-recognition domain can explain why the fragment mini-TyrRS, and not full-length native TyrRS, is active in cytokine-signaling pathways.
Aminoacyl-tRNA synthetases are ancient proteins, presumably arising during the establishment of the genetic code in making the transition from the putative RNA world to the theater of proteins (1–3). The enzymes link amino acids to their cognate tRNAs in aminoacylation reactions that establish the connection between a specific amino acid and a nucleotide triplet anticodon imbedded in the tRNA. The 20 enzymes are divided into two classes of 10 enzymes each (4–7). This division is defined by the unique architectures associated with the catalytic domains specific to each class. During their long evolution, certain of the enzymes have acquired additional functions, including roles in RNA splicing (8, 9), RNA trafficking (10, 11), regulation of transcription and translation (12, 13), and cell signaling (14–21). These activities can result from the fusion or insertion of specialized domains. In addition, a number of synthetase-like proteins have been described and, in some instances, a biological role for these proteins has been demonstrated (22).
The class I close homologs human tyrosyl- and tryptophanyl-tRNA synthetases (TyrRS and TrpRS) are active in cytokine-signaling pathways (14–21). In the case of TyrRS, the human enzyme has a C-terminal domain not found in TyrRS orthologs of lower eukaryotes, archebacteria, or prokaryotes (Fig. 1). This domain is a homolog of endothelial monocyte-activating polypeptide II (EMAP II), a well characterized mammalian cell cytokine that also has RNA-binding properties (23). Although full-length, native TyrRS has no cell-signaling activity, the enzyme is secreted during apoptosis in cell culture and can be cleaved with an extracellular enzyme such as leukocyte elastase (14). The two released fragments, the N-terminal mini-TyrRS and the EMAP II-like C-terminal domain, are active cytokines. In the case of catalytically active mini-TyrRS, the fragment has IL-8-like activity, including stimulating migration of polymorphonuclear cells, binding to the IL-8 receptor CXCR1, and acting as a proangiogenic factor (as shown by its stimulation of chemotaxis of endothelial cells in culture and by stimulation of angiogenesis in two animal models; refs. 14, 16, and 17). This angiogenic activity can be blocked by antiangiogenic chemokines such as IP-10. Thus, fragmentation of human TyrRS links protein synthesis to cell-signaling pathways including angiogenesis.
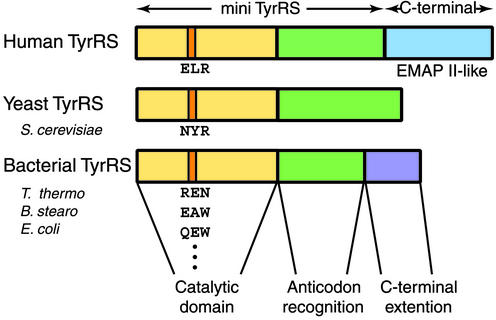
Fig 1.
Illustration of organization of domains of human, yeast, and bacterial TyrRS. All TyrRSs have a catalytic Rossmann fold domain (yellow) and an anticodon-recognition domain (green). In addition, human TyrRS has an EMAP II-like C-terminal domain (blue). In contrast, bacterial TyrRS has a smaller C-terminal extension (purple), which is distinct from the human TyrRS C domain. The critical ELR tripeptide for cytokine activity is highlighted in orange on human TyrRS. The corresponding tripeptides also are highlighted on yeast and bacterial TyrRSs. These tripeptides are not conserved among yeast and bacterial TyrRSs.
Proangiogenic CXC chemokines such as IL-8 have a conserved Glu-Leu-Arg (ELR) motif that is essential, for example, for polymorphonuclear leukocyte binding and activation (24, 25). Human TyrRS has an ELR motif that is conserved among organisms with a fully developed vasculature but is not found in orthologs of other eukaryotes or any prokaryotes (19). This motif is located within the catalytic domain (Fig. 1) whose architecture is the Rossmann nucleotide-binding fold that characterizes all class I tRNA synthetases (4–7). A single R→Q substitution in the ELR motif eliminates the ability of human mini-TyrRS to stimulate migration of human umbilical vein endothelial cells (16). Importantly, the Saccharomyces cerevisiae TyrRS has a NYR motif and is inactive as a cytokine (Fig. 1). Substitution of ELR for NYR in native S. cerevisiae TyrRS (which is an ortholog of mini-TyrRS) is sufficient to confer mammalian-like cytokine function on the yeast enzyme (20). Thus, in these respects, human mini-TyrRS is like other CXC cytokines, where ELR is correlated with neutrophil activation and stimulation of angiogenesis. Indeed, a DLQ substitution of ELR in IL-8 inactivates cytokine function (24).
The structural basis for activation of the cytokine activity of human TyrRS is not known. Although the structures of the catalytic and anticodon-recognition domains of Bacillus stearothermophilus and Staphylococcus aureus and of the native Thermus thermophilus TyrRS (including the C-terminal extension) have been determined (26–28), no structures are available for any mammalian aminoacyl-tRNA synthetase, including TyrRS. Mammalian enzymes such as TyrRS commonly have extra domains and fusions not found in bacterial orthologs. The joining of these mammalian-specific extra domains potentially can affect the conformations of the domains that are orthologous to the synthetases of lower organisms. With these considerations in mind, we set out to determine the crystal structure of human mini-TyrRS. Our main objectives were to understand better how removal of the EMAP II-like domain activated the cytokine function of the released N-terminal mini-TyrRS and to visualize the position and spatial organization of the critical ELR motif. A crystal diffracting to 1.18 Å was obtained and yielded a structure that had significantly distinct features relative to those found in the structures of the three aforementioned orthologs and relevant to the cytokine activities of the human protein. The results illustrate the subtle variations in synthetase structures that are correlated with functions other than aminoacylation.
Materials and Methods
Protein Expression, Purification, and Characterization.
The plasmid containing the gene for mini-TyrRS was the one described (14). The protein was expressed in Escherichia coli and purified to homogeneity (>95% pure by SDS/PAGE) by Ni-NTA, ion exchange (Mono-Q), and gel filtration (Superose 12) chromatography (Amersham Pharmacia). During the purification, all buffers contained 2 mM 2-mercaptoethanol. Selenium-methionine (Se-Met)-labeled protein was prepared as described (29). The replacement of methionine by Se-Met was checked by MS, using matrix-assisted laser desorption ionization time-of-flight analysis. About 95% of the seven methionines were replaced by Se-Met.
Crystallization by Using High-Throughput Technology.
Mini-TyrRS (22 mg/ml) was maintained in a stock solution of 10 mM Hepes (pH 7.5)/20 mM KCl/0.02% NaN3/2 mM 2-mercaptoethanol, with or without puromycin (1 mM). Initial crystallization trials of mini-TryRS were conducted by using the proprietary high-throughput protein crystallization platform developed at Syrrx (La Jolla, CA). The crystallization experiments were set up on the Agincourt crystallization robot by using patented submicroliter crystallization technology. For both protein samples, a sparse matrix approach was used with a suite of 480 unique crystallization conditions at 20 and 4°C. In addition, 480 systematic conditions, using different precipitants such as (NH4)2SO4 (0.5–2.8 M), polyethylene glycol 500 monomethyl ether (PEG MME 500; 10–40%), PEG MME 2000 (6–24%), PEG MME 5000 (5–20%), PEG 8000 (5–20%), and 2-methyl-2,4-pentanediol (6–40%) at ranges of pH 4–9, were set up at 20 and 4°C. Each crystallization plate contained 96 sitting-drop experiments on which 50 nl of protein sample was mixed with an equal volume of reservoir solution. The setups subsequently were incubated and automatically imaged at regular intervals. By using this technology, a total of 1,920 crystallization samples were set up over a 2-h period with a total of 200 μl of mini-TryRS protein sample.
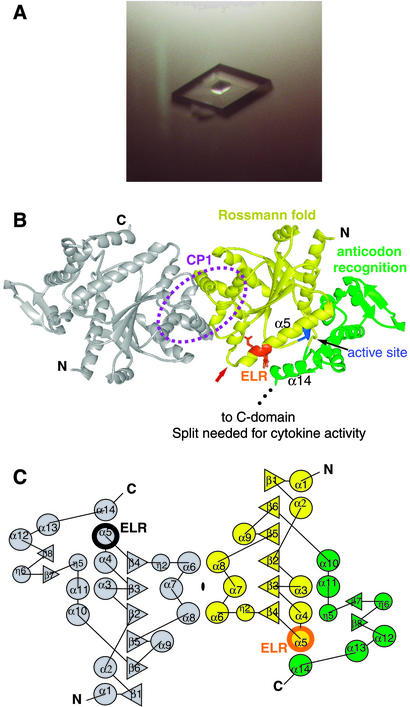
Productive crystallization conditions then were repeated by using 2 μl + 2 μl sitting drops. After further optimization, rhombic crystals of either mini-TyrRS or Se-Met mini-TyrRS grew in 3 days from 2.1 M (NH4)2SO4, 0.1 M NaH2PO4/K2HPO4 (pH 6.4), and 2% acetone at 4°C, with a maximal size of 0.4 mm × 0.3 mm × 0.3 mm in the presence of puromycin (Fig. 2A). In the absence of puromycin, crystals grew under the same solvent conditions at either pH 6.0 or pH 7.3.
Fig 2.
(A) A rhombic crystal of mini-TyrRS grown with puromycin. (B) Structure of mini-TyrRS dimer. Each monomer consists of a catalytic Rossmann fold domain (yellow), with the “HIGH” tetrapeptide in the active site highlighted in blue and an anticodon-recognition domain (green). The CP1 domain is circled. The critical ELR motif (orange) for cytokine activity is located on α5 of the catalytic domain and is close to α14 of the anticodon-recognition domain. The partially disordered loop before the ELR motif is indicated with a red arrow. Because the C-terminal end of the anticodon-recognition domain is close to ELR, adding the C-terminal EMAP II-like domain (as in native TyrRS) is likely to mask the ELR tripeptide. (C) Topology diagram of mini-TyrRS, with the secondary structure elements labeled sequentially, starting from the N terminus. The diagram contains two symmetry-related mini-TyrRS molecules. α-Helices (α, circles), 310 helices (η, circles), and β-strands (β, triangles) are shown.
Data Collection, Structure Determination, and Refinement.
Molecular replacement methods initially were attempted to solve the structure of mini-TyrRS, using B. stearothermophilus TyrRS structure as the search model. This attempt was unsuccessful. A de novo structure determination then was done with a three-wavelength Se-MAD (multiple-wavelength anomalous diffraction) data set that had a 1.9-Å resolution by using the Se-Met mini-TyrRS crystals. Data were collected with beamline X12B at the National Synchrotron Light Source at Brookhaven National Laboratory (Upton, NY). Data were integrated and scaled with HKL2000 (30). Five Se sites were identified by using SOLVE (31) and refined further by using SHARP (32), with two additional Se sites identified. After density modification in SOLOMON (33) by using solvent flattening with 47% solvent content, the overall figure of merit at 1.9 Å increased from 0.65 to 0.95, at which point the electron density map was well resolved. ARP/WARP (34) then successfully traced and built 280 of 364 residues. The remaining model with a total of 332 residues was built manually in o (35). The refinement then was performed against a 1.18-Å, high-resolution native data set collected with beamline 11-1 at the Stanford Synchrotron Radiation Laboratory. The refinement started in cns (36) and finished by using SHELXL (37), with final Rcryst = 17.37% and Rfree = 21.59% for Fo > 4 σ data (Rcryst = 17.97% and Rfree = 22.28% for all data). Data collection and refinement statistics of mini-TyrRS are summarized in Table 1.
Table 1.
Data collection and refinement statistics
| Native | Sepeak | Seinflection | Seremote | |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength, Å | 0.9800 | 0.9785 | 0.9793 | 0.9611 |
| Resolution, Å | 1.18 | 1.90 | 1.90 | 1.90 |
| Unique reflections | 134,129 | 35,113 | 35,080 | 35,263 |
| Completeness, % | 93 (59) | 100 (100) | 100 (100) | 100 (100) |
| Redundancy | 6.4 | 13.9 | 13.1 | 14.4 |
| Rmerge, % | 5.4 (69.9) | 8.2 (51.3) | 7.2 (49.9) | 8.7 (64.2) |
| 〈I/σ(I)〉 | 55.2 (1.0) | 41.2 (5.8) | 41.9 (5.9) | 38.1 (4.7) |
| Refinement statistics | ||||
| Resolution range, Å | 20–1.18 | |||
| Number of reflections, working/free | 118,016/6,310 | |||
| Number of atoms (protein/water) | 2,641/324 | |||
| Rwork/Rfree, % | 17.37/21.59 | |||
| rms deviation bond lengths, Å | 0.014 | |||
| rms deviation bond angle, ° | 2.5 | |||
| Ramachandran plot, % | ||||
| Favored | 92.8 | |||
| Allowed | 6.5 | |||
| Generously allowed | 0.3 | |||
| Disallowed | 0.3 | |||
| Average B factors for protein, Å2 | 29.4 | |||
| Average B factors for waters, Å2 | 40.4 | |||
Numbers in parentheses refer to the highest-resolution shell.
Rmerge = (ΣhΣi|Ii(h) − 〈I(h)〉|/ΣhΣiIi(h))× 100, where 〈I(h)〉 is the average intensity of i symmetry-related observations of reflections with Bragg index h.
Rwork = (Σhkl|Fo − Fc|/Σhkl|Fo) × 100, where Fo and Fc are the observed and the calculated structure factors, respectively, for 95% of the reflections used in the refinement. Rfree was calculated as for Rwork but on 5% of reflections excluded before refinement. Numbers refer to Fo > 4σ data.
Results
Structure Obtained from Crystal That Diffracted to 1.18 Å.
The high-resolution structure at 1.18 Å was obtained from crystals grown in the presence of puromycin (Fig. 2A). However, no puromycin density was identified in the structure. Because puromycin contains a tyrosyl moiety, we tested whether the drug would affect the aminoacylation or cell-migration activity of mini-TyrRS. Weak inhibition of both activities was observed at a puromycin concentration of 1 μM (J. Liu and P.S., unpublished data). Although no evidence could be obtained for a strong interaction between puromycin and mini-TyrRS, the data set obtained from crystals grown in the absence of puromycin had a lower resolution of 1.4–1.5 Å. The structure obtained from this data set was identical to the higher-resolution structure obtained from crystals grown in the presence of puromycin. Thus, puromycin appears to act as an additive (to the crystallization conditions) that is critical for obtaining the higher-resolution crystals.
General Description of Structure.
Mini-TyrRS was determined to have a homodimeric structure in solution (by gel filtration; X.-L.Y. and P.S., unpublished data), like all other TyrRS orthologs whose quaternary structures have been investigated so far (26–28). In the crystal structure, mini-TyrRS is a dimer with a 2-fold axis coincident with the crystallographic 2-fold axis of the P21212 unit cell. Therefore, only one monomeric subunit is in the asymmetric unit. The fragment is composed of 364 amino acids, of which 332 were resolved. The three N-terminal amino acids and 22 residues from Asp-343 to Ile-364 were disordered. In addition, the segment Lys-222 to Glu-228 encompassing the KMSSS loop, which is one of the signatures of class I enzymes (38), was not resolved, probably because no substrate was bound to the protein.
The structure of mini-TyrRS is organized along the lines similar to those seen in other class I tRNA synthetases, including B. stearothermophilus, S. aureus, and T. thermophilus TyrRS (26–28). There are two major domains: an N-terminal catalytic domain of ≈230 residues followed by an anticodon-recognition domain (Fig. 2B). The catalytic domain is a Rossmann nucleotide-binding fold of alternating β-strands and α-helices. The connectivity of these structural elements is displayed in Fig. 2C, where each α-helix and β-strand is numbered sequentially, starting at the N terminus. The anticodon-recognition domain is composed largely of α-helices, where its first α-helix (α10) is joined to the C-terminal β-strand (β6) of the catalytic domain via the KMSSS loop. The structure at the C-terminal end of mini-TyrRS is well resolved up to Pro-342 of α14, after which the last 22 residues of mini-TyrRS were disordered. This disordered segment is the linker that joins the 169-aa, EMAP II-like C domain of native TyrRS to the body of the enzyme (mini-TyrRS).
The catalytic domain is interrupted by an insertion (after the β-strand β4 and the 310 helix η2) known as connective polypeptide 1 (CP1; refs. 39 and 40). This insertion is found in all class I enzymes and, for some of them, contains the active site for editing of misactivated amino acids and the determinants for binding the acceptor helix of the tRNA structure (41–49). Tyrosyl-tRNA synthetases have no known editing activity, and it is for that reason that the CP1 insertion is small (54 residues) compared with those found in class I enzymes such as isoleucyl-, valyl-, and leucyl-tRNA synthetases that have robust editing activities. However, human TyrRS has determinants within CP1 for binding the acceptor helix and, in particular, the 39-aa peptide motif (Leu-125–Gly-163) within CP1 is essential for discrimination of the first base pair of that helix (45). In addition, the structure of human mini-TyrRS shows that, like the bacterial orthologs, the CP1 insertion of each monomer makes contacts at the dimer interface (Fig. 2B).
Structural Alignments of Mini-TyrRS with Bacterial TyrRSs.
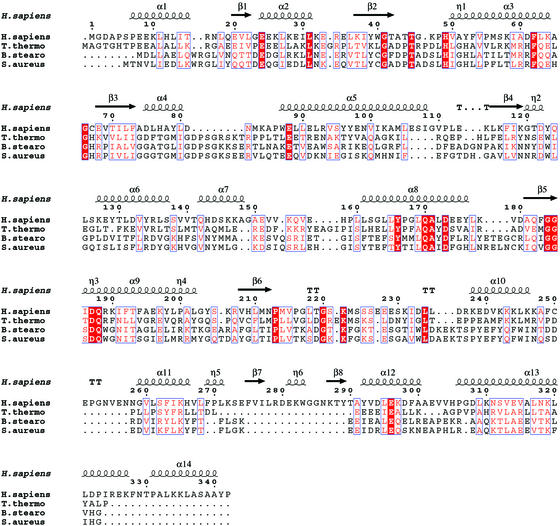
A structural alignment of mini-TyrRS with T. thermophilus, B. stearothermophilus, and S. aureus was made by using the program dali (ref. 50; Fig. 3). The labeling of the secondary structure for mini-TyrRS is adopted for the three bacterial TyrRSs to minimize confusion. This alignment extends from the N terminus to Ile-324 on helix α13, with the rest of the C-terminal sequence of mini-TyrRS unaligned with the bacterial orthologs. The Rossmann fold domain of mini-TyrRS is similar to that of the bacterial TyrRSs. The central β-sheets are virtually superimposable, whereas some of the α-helices that have surface locations adopt slightly different orientations. However, the structure of the anticodon-recognition domain of mini-TyrRS is more diverged from its bacterial counterpart (Fig. 4). Surprisingly, the anticodon-recognition domain of mini-TyrRS is more similar to that of bacterial TrpRS (52). With the program dali, the anticodon-recognition domain of mini-TyrRS superimposes on B. stearothermophilus TrpRS with a Z score of 6.2, whereas that of mini-TyrRS superimposes on B. stearothermophilus TyrRS with a score of only 3.7.
Fig 3.
Structural alignment of human mini-TyrRS with TyrRSs from three bacterial organisms that have known structures. The secondary structure of mini-TyrRS is superimposed on top. Human TyrRS can be aligned with the three bacterial TyrRSs from the N terminus to Ile-324. The alignment was generated by dali (50) and drawn with espript (51). Identical residues are highlighted in red and similarities are shown in pink.
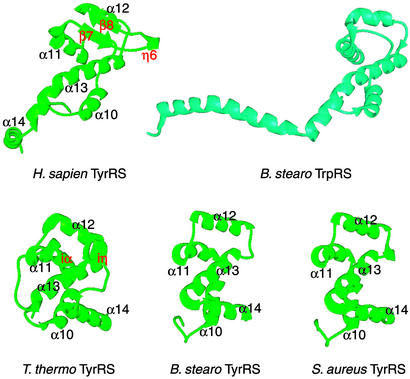
Fig 4.
Anticodon-recognition domains of human, T. thermophilus, B. stearothermophilus, and S. aureus TyrRS (green) and of B. stearothermophilus TrpRS (cyan). The structure of the anticodon-recognition domain of mini-TyrRS is more similar to that of B. stearothermophilus TrpRS than to that of the bacterial TyrRSs. The labeling of the secondary structure for mini-TyrRS is adopted for the three bacterial TyrRSs for comparison. A hairpin insertion in human mini-TyrRS between α11 and α12 is labeled in red. Similarly, an insertion (iα and iη) in T. thermophilus TyrRS is also labeled in red. Although located differently in their respective primary sequence, the insertions in human mini-TyrRS and in T. thermophilus TyrRS overlap in three-dimensional space.
An ≈20-residue “hairpin” structure is inserted between helices α11 and α12 of the anticodon-recognition domain (Fig. 4). This insertion is unique for the human enzyme and consists of two antiparallel β-strands (β7 and β8) and a short 310 helix η6. T. thermophilus TyrRS also has an insertion (between helices α13 and α14) that is made up of α-helix iα and 310 helix iη (Fig. 4). Interestingly, although located differently in their primary sequences, these two insertions overlap in three-dimensional space.
Orientation of the C-Terminal End of the Anticodon-Recognition Domain in Relation to Cytokine Activation.
When taking a closer look at the anticodon-recognition domains of human mini-TyrRS and the bacterial TyrRSs, it is obvious that the first three α-helices, α10, α11, and α12, are superimposable among the four proteins (Fig. 4). However, helix α13 of human mini-TyrRS starts from the same place as it does in the bacterial TyrRSs, but it is much longer in mini-TyrRS, with a small hinge in the middle. Most striking, helix α14 of mini-TyrRS has a completely different spatial position. In particular, it is close to the ELR motif that is essential for cytokine activity (Fig. 2 B and C). Thus, in native, full-length TyrRS, the end of α14 is positioned so that the fusion of the EMAP II-like domain would mask the ELR tripeptide. This spatial arrangement for α14 could explain why the mini-TyrRS, not full-length TyrRS, has cytokine activity.
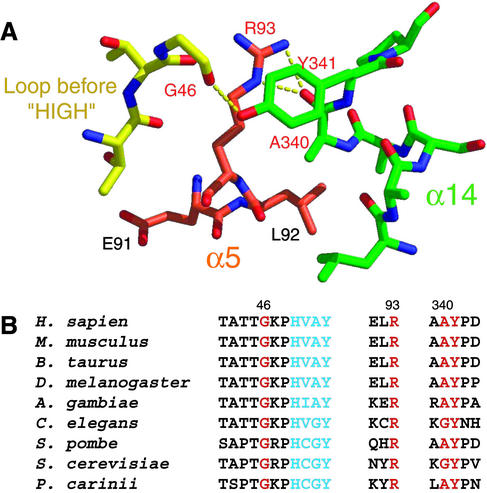
Moreover, this helix (α14) in mini-TyrRS is tethered to the ELR motif through a hydrogen-bonding network (Fig. 5A). The critical ELR (residues 91–93) motif for the cytokine activity is located on the α5 helix of the catalytic domain. As well as its proximity to helix α14, the ELR tripeptide is close to a loop containing G46 (see below) that is part of the canonical 11-aa signature sequence of class I enzymes that ends in the HIGH tetrapeptide (HAVY in human TyrRS; refs. 4 and 5). The R93 side chain makes two hydrogen bonds with the main chain carboxyl oxygen of A340 in helix α14. The aromatic side chain of Y341 of helix α14 stacks over the guanidino R93 side chain and makes a hydrogen bond with the main chain carboxyl oxygen of G46. This hydrogen-bonding network leaves R93 only partially exposed to solvent.
Fig 5.
(A) Close-up view of the ELR motif. R93 is in the middle of a hydrogen-bonding network involving G46, A340, and Y341. G46 is located on a loop that is only three residues before the HIGH tetrapeptide (HAVY in human TyrRS). A340 and Y341 are from α14 of the anticodon-recognition domain. (B) Partial sequence alignment of TyrRS from eukaryotes (except plants) indicating all four residues (red) involved in the above hydrogen-bonding network are conserved. HAVY is colored in blue.
Significantly, the four residues involved in this hydrogen-bonding network, G46, R93, A340, and Y341, are conserved among eukaryotic TyrRSs (A340 is replaced by another small amino acid G in Caenorhabditis elegans and S. cerevisiae), including mammals, insects, nematodes, and yeast (except for plants; Fig. 5B). Thus, the orientation of the anticodon-recognition domain with respect to the catalytic site probably is similar in these eukaryotic TyrRSs. In contrast, these four residues are not conserved in the bacterial TyrRSs, where the orientation of the C-terminal end of the anticodon-recognition domain is quite different. Significantly, an R93-to-Q mutation in human mini-TyrRS abolishes cytokine activity (14, 16). This mutation potentially could disrupt the hydrogen-bonding platform and untether helix α14 from the region of ELR. Although not considered previously, helix α14 potentially could be involved in receptor binding and signaling, because of the proximity (revealed in our structure) of this helix to the ELR tripeptide.
Discussion
Because residues needed for the hydrogen-bonding network described above are present in S. cerevisiae TyrRS, the position of the critical tripeptide with respect to the rest of the structure probably is closely similar to that of human mini-TyrRS. However, the ELR motif is replaced by NYR in the S. cerevisiae enzyme. Thus, the lack of cytokine activity of the yeast enzyme most likely is caused by the absence of the EL dipeptide and not to the spatial position of the tripeptide. With this insight, the acquisition of cytokine function by the yeast protein upon substitution of ELR for NYR is easier to understand (20). Moreover, this result, in light of the structure presented here, suggests that it is the E and the L that are strong candidates for a direct involvement in receptor signaling. This is especially the case for E91, which is more exposed and flexible, with an average B factor of 53 compared with an average B factor of 29 for all protein atoms in mini-TyrRS. The average B factor for R93 is 41 and for L92 is 43.
There are two partially disordered regions in the catalytic domain of mini-TyrRS. The first is a loop before helix α5 and extending into the first three residues of α5 (Lys-84—Leu-89; see red arrow in Fig. 2B). This segment is close to the ELR motif. We speculate that this loop region might become ordered in a receptor complex. The second partially disordered region is the loop (Val-153–His-158) between helix α7 and α8 that is part of the CP1 domain. This segment is distal to the ELR motif and, because it is part of a specificity element for recognition of the tRNATyr acceptor helix (45), probably becomes ordered on binding to tRNATyr.
Because mini-TyrRS has IL-8-like cytokine activity, we compared the structure of mini-TyrRS structure with that of IL-8 and related ELR-containing CXC cytokines such as Gro-α and Nap-2 (53–55). Overall, the three CXC cytokines have similar structures, which are completely different from that of mini-TyrRS. For example, IL-8 is a small protein of 77 amino acids that is constructed from a series of turns and loops in the N-terminal region followed by three β-strands and a C-terminal α-helix (53). The ELR motif that is critical for the IL-8 cytokine activity is located in a loop near the N terminus, where it is part of a flexible region that is exposed to solvent. Therefore, the ELR motifs of mini-TyrRS and the three aforementioned CXC cytokines are in different structural environments. This difference in environments raises the possibility that mini-TyrRS simultaneously binds to a coreceptor in addition to the CXCR1 receptor, perhaps as part of a larger signaling complex.
In conclusion, our structure provides more clarification of the role of the ELR motif in mini-TyrRS and suggests why mini-TyrRS, and not TyrRS, is active in cytokine signaling. The intricate interplay of structural elements described here, and their role in masking and unmasking cytokine activity, may be clarified further by additional mutational analysis.
Acknowledgments
We thank Dr. Howard Robinson at the National Synchrotron Light Source for collecting the Se-MAD data, Drs. Xiaoping Dai and Xueyong Zhu for help with collection of the high-resolution native data at the Stanford Synchrotron Radiation Laboratory, Dr. Marc Elsliger for assistance with computer software, Dr. Jianming Liu for doing the cytokine and charging activity assays with mini-TyrRS in the presence of puromycin, Dr. Manal Swairjo for helpful discussions throughout the work, and Prof. Alexander Rich and Drs. Karla Ewalt and Chris Myers for comments on the manuscript. This work was supported by National Institutes of Health Grant GM15539 and a fellowship from the National Foundation for Cancer Research.
Abbreviations
TyrRS, tyrosyl-tRNA synthetase
TrpRS, tryptophanyl-tRNA synthetases
EMAP II, endothelial monocyte-activating polypeptide II
Se-Met, selenium-methionine
PEG MME, polyethylene glycol monomethyl ether
CP1, connective polypeptide 1
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code ).
References
- 1.Giegé R., Puglisi, J. D. & Florentz, C. (1993) Prog. Nucleic Acid Res. Mol. Biol. 45 129-206. [DOI] [PubMed] [Google Scholar]
- 2.Carter C. W. (1993) Annu. Rev. Biochem. 62 715-748. [DOI] [PubMed] [Google Scholar]
- 3.Ibba M. & Söll, D. (2000) Annu. Rev. Biochem. 69 617-650. [DOI] [PubMed] [Google Scholar]
- 4.Webster T., Tsai, H., Kula, M., Mackie, G. A. & Schimmel, P. (1984) Science 226 1315-1317. [DOI] [PubMed] [Google Scholar]
- 5.Ludmerer S. W. & Schimmel, P. (1987) J. Biol. Chem. 262 10801-10806. [PubMed] [Google Scholar]
- 6.Eriani G., Delarue, M., Poch, O., Gangloff, J. & Moras, D. (1990) Nature 347 203-206. [DOI] [PubMed] [Google Scholar]
- 7.Cusack S., Berthet-Colominas, C., Hartlein, M., Nassar, N. & Leberman, R. (1990) Nature 347 249-255. [DOI] [PubMed] [Google Scholar]
- 8.Dujardin G. & Herbert, C. J. (1997) in Ribosomal RNA and Group I Introns, eds. Green, R. & Schroeder, R. (Lands Bioscience, Austin, TX), pp. 179–198.
- 9.Rho S. B. & Martinis, S. A. (2000) RNA 6 1882-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lund E. & Dahlberg, J. (1998) Science 282 2082-2085. [DOI] [PubMed] [Google Scholar]
- 11.Sarkar S. & Hopper, A. K. (1998) Mol. Biol. Cell 9 3041-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Putney S. D. & Schimmel, P. (1981) Nature 291 632-635. [DOI] [PubMed] [Google Scholar]
- 13.Putzer H., Grunbery-Manago, M. & Springer, M. (1995) in tRNA: Structure, Biosynthesis and Function, eds. Söll, D. & RajBhandary, U. (Am. Soc. Microbiol., Washington, DC), pp. 293–333.
- 14.Wakasugi K. & Schimmel, P. (1999) Science 284 147-151. [DOI] [PubMed] [Google Scholar]
- 15.Wakasugi K. & Schimmel, P. (1999) J. Biol. Chem. 274 23155-23159. [DOI] [PubMed] [Google Scholar]
- 16.Wakasugi K., Slike, B. M., Hood, J., Ewalt, K. L., Cheresh, D. A. & Schimmel, P. (2002) J. Biol. Chem. 277 20124-20126. [DOI] [PubMed] [Google Scholar]
- 17.Wakasugi K., Slike, B. M., Hood, J., Otani, A., Ewalt, K. L., Friedlander, M., Cheresh, D. A. & Schimmel, P. (2002) Proc. Natl. Acad. Sci. USA 99 173-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otani A., Slike, B. M., Dorrell, M. I., Hood, J., Kinder, K., Ewalt, K. L., Cheresh, D., Schimmel, P. & Friedlander, M. (2002) Proc. Natl. Acad. Sci. USA 99 178-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ewalt, K. L. & Schimmel, P. (2002) Biochemistry 41, in press. [DOI] [PubMed]
- 20.Liu, J., Yang, X.-L., Ewalt, K. L. & Schimmel, P. (2002) Biochemistry 41, in press. [DOI] [PubMed]
- 21.Otani A., Kinder, K., Ewalt, K., Otero, F. J., Schimmel, P. & Friedlander, M. (2002) Nat. Med. 8 1004-1010. [DOI] [PubMed] [Google Scholar]
- 22.Ribas de Pouplana L. & Schimmel, P. (1999) Proc. Natl. Acad. Sci. USA 96 327-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleeman T. A., Wei, D., Simpson, K. L. & First, E. A. (1997) J. Biol. Chem. 272 14420-14425. [DOI] [PubMed] [Google Scholar]
- 24.Clark-Lewis I., Dewald, B., Geiser, T., Moser, B. & Baggiolini, M. (1993) Proc. Natl. Acad. Sci. USA 90 3574-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daly T. J., LaRosa, G. J., Dolich, S., Maione, T. E., Cooper, S. & Broxmeyer, H. E. (1995) J. Biol. Chem. 270 23282-23292. [DOI] [PubMed] [Google Scholar]
- 26.Brick P., Bhat, T. N. & Blow, D. M. (1988) J. Mol. Biol. 208 83-98. [DOI] [PubMed] [Google Scholar]
- 27.Qiu X., Janson, C. A., Smith, W. W., Green, S. M., McDevitt, P., Johanson, K., Carter, P., Hibbs, M., Lewis, C., Chalker, A., et al. (2001) Protein Sci. 10 2008-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yaremchuk A., Kriklivyi, I., Tukalo, M. & Cusack, S. (2002) EMBO J. 21 3829-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendrickson W. A., Horton, J. R. & LeMaster, D. M. (1990) EMBO J. 9 1665-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otwinowski Z. & Minor, W. (1997) Methods Enzymol. 276 307-326. [DOI] [PubMed] [Google Scholar]
- 31.Terwilliger T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55 849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de La Fortelle E. & Bricogne, G. (1997) Methods Enzymol. 276 472-494. [DOI] [PubMed] [Google Scholar]
- 33.Abrahams J. P. & Leslie, A. G. W. (1996) Acta Crystallogr. D 52 30-42. [DOI] [PubMed] [Google Scholar]
- 34.Perrakis A., Morris, R. & Lamzin, V. S. (1999) Nat. Struct. Biol. 6 458-463. [DOI] [PubMed] [Google Scholar]
- 35.Jones T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47 110-119. [DOI] [PubMed] [Google Scholar]
- 36.Adams P. D., Rice, L. M. & Brunger, A. T. (1998) Curr. Opin. Struct. Biol. 8 606-611. [DOI] [PubMed] [Google Scholar]
- 37.Sheldrick G. M. & Schneider, T. R. (1997) Methods Enzymol. 277 319-343. [PubMed] [Google Scholar]
- 38.Hountondji C., Dessen, P. & Blanquet, S. (1986) Biochimie 68 1071-1078. [DOI] [PubMed] [Google Scholar]
- 39.Starzyk R. M., Webster, T. A. & Schimmel, P. (1987) Science 237 1614-1618. [DOI] [PubMed] [Google Scholar]
- 40.Hou Y. M., Shiba, K., Mottes, C. & Schimmel, P. (1991) Proc. Natl. Acad. Sci. USA 88 976-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rould M. A., Perona, J. J., Söll, D. & Steitz, T. A. (1989) Science 246 1135-1142. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt E. & Schimmel, P. (1994) Science 264 265-267. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt E. & Schimmel, P. (1995) Biochemistry 34 11204-11210. [DOI] [PubMed] [Google Scholar]
- 44.Lin L., Hale, S. P. & Schimmel, P. (1996) Nature 384 33-34. [DOI] [PubMed] [Google Scholar]
- 45.Wakasugi K., Quinn, C. L., Tao, N. & Schimmel, P. (1998) EMBO J. 17 297-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nureki O., Vassylyev, D. G., Tateno, M., Shimada, A., Nakama, T., Fukai, S., Konno, M., Hendrickson, T. L., Schimmel, P. & Yokoyama, S. (1998) Science 280 578-582. [DOI] [PubMed] [Google Scholar]
- 47.Silvian L. F., Wang, J. & Steitz, T. A. (1999) Science 285 1074-1077. [PubMed] [Google Scholar]
- 48.Fukai S., Nureki, O., Sekine, S., Shimada, A., Tao, J., Vassylyev, D. G. & Yokoyama, S. (2000) Cell 103 793-803. [DOI] [PubMed] [Google Scholar]
- 49.Larkin D. C., Williams, A. M., Martinis, S. A. & Fox, G. E. (2002) Nucleic Acids Res. 30 2103-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holm L. & Sander, C. (1993) J. Mol. Biol. 233 123-138. [DOI] [PubMed] [Google Scholar]
- 51.Gouet P., Courcelle, E., Stuart, D. I. & Metoz, F. (1999) Bioinformatics 15 305-308. [DOI] [PubMed] [Google Scholar]
- 52.Ilyin V. A., Temple, B., Hu, M., Li, G., Yin, Y., Vachette, P. & Carter, C. W., Jr. (2000) Protein Sci. 9 218-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clore G. M., Appella, E., Yamada, M., Matsushima, K. & Gronenborn, A. M. (1990) Biochemistry 29 1689-1696. [DOI] [PubMed] [Google Scholar]
- 54.Qian Y. Q., Johanson, K. O. & McDevitt, P. (1999) J. Mol. Biol. 294 1065-1072. [DOI] [PubMed] [Google Scholar]
- 55.Malkowski M. G., Wu, J. Y., Lazar, J. B., Johnson, P. H. & Edwards, B. F. (1995) J. Biol. Chem. 270 7077-7087. [DOI] [PubMed] [Google Scholar]