Abstract
The x-ray crystal structure of dimeric (+)-bornyl diphosphate synthase, a metal-requiring monoterpene cyclase from Salvia officinalis, is reported at 2.0-Å resolution. Each monomer contains two α-helical domains: the C-terminal domain catalyzes the cyclization of geranyl diphosphate, orienting and stabilizing multiple reactive carbocation intermediates; the N-terminal domain has no clearly defined function, although its N terminus caps the active site in the C-terminal domain during catalysis. Structures of complexes with aza analogues of substrate and carbocation intermediates, as well as complexes with pyrophosphate and bornyl diphosphate, provide “snapshots” of the terpene cyclization cascade.
From antiquity, aromatic plants have been used for their pleasing fragrances, culinary flavorings, and medicinal applications. These useful properties derive largely from monoterpene natural products that plants generate for diverse molecular functions in regulation, communication, and defense (1, 2). Diversity in function requires diversity in structure, and myriad monoterpenes characterized to date reveal a broad array of variations in their monocyclic or bicyclic cyclohexanoid skeletons. However, despite the exquisite structural and stereochemical diversity of cyclic monoterpenes in biology, each derives from the same substrate, geranyl diphosphate (GPP). The cyclization of this C10-isoprenoid precursor through sequential carbocation intermediates is catalyzed by metal-dependent monoterpene synthases (cyclases) (1, 2). Thus, the manifold synthetic chemistry of terpene biosynthesis is a consequence of the evolution of diversity in terpene cyclases (3, 4), such that different active site templates evolved to chaperone reactive carbocation intermediates through different cyclization pathways (5, 6).
To date, no crystal structure of a monoterpene synthase has been available to understand the mechanistic details of GPP cyclization. However, the recently determined structures of four sesquiterpene synthases provide relevant mechanistic inferences regarding the cyclization of the larger C15-isoprenoid substrate, farnesyl diphosphate (FPP) (7–10). The sesquiterpene cyclases, along with farnesyl diphosphate synthase (11), share a common, α-helical “terpene synthase fold” (7), despite insignificant amino acid sequence relationships. The structure of trichodiene synthase complexed with co-product pyrophosphate (PPi) and three magnesium ions reveals a substantial conformational change that completely sequesters the hydrophobic active site cavity from solvent; this conformational change is likely triggered by the diphosphate group of the substrate as well (10). A less-extensive conformational change is reported for epi-aristolochene synthase on the binding of substrate analogues (8). These structures also reveal two metal binding motifs conserved among monoterpene, sesquiterpene, and diterpene cyclases: the so-called “aspartate-rich” motif DDXXD, and the motif (N,D)D(L,I,V)X(S,T)X3E, in which boldface residues coordinate to magnesium ions (8, 10, 12).
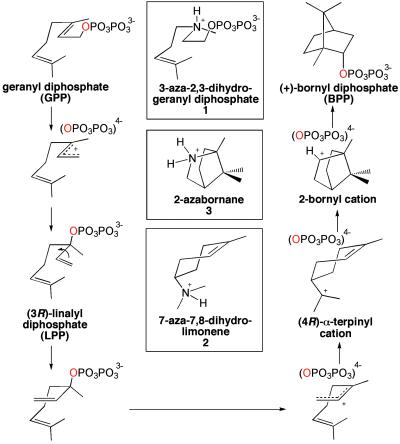
Here, we report an x-ray crystal structure of a monoterpene cyclase, recombinant (+)-bornyl diphosphate synthase (BPPS) from Salvia officinalis (culinary sage) (13). The 128-kDa BPPS homodimer catalyzes a particularly noteworthy cyclization cascade, in that the PPi leaving group of GPP is reincorporated into the final cyclization product, (+)-bornyl diphosphate (BPP), with retention of the original ester oxygen atom (Fig. 1). We also report the structures of several BPPS–ligand complexes that, taken together, map out the reaction coordinate of isoprenoid diphosphate cyclization.
Fig 1.
Cyclization of GPP as catalyzed by BPPS; note that the same diphosphate oxygen (red) is contained in the prenyl phosphoester linkages of GPP and BPP. Aza analogues of substrate and carbocation intermediates are shown in boxes.
Materials and Methods
Crystal Structure Determination.
The cDNA encoding BPPS (accession number AF051900), cloned as described (13), was subcloned into a pSBET expression vector (14). A pseudomature form of the enzyme truncated at E50 was expressed and purified (see Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org). Enzyme was crystallized in hanging drops by vapor diffusion at 21°C. Typically, a 2-μl drop of protein solution [8 mg/ml enzyme/1 mM DTT/20 mM bis-Tris (pH, 6.8)] was mixed with a 2-μl drop of precipitant solution [10% (wt/vol) polyethylene glycol 8000/15% (vol/vol) glycerol/2 mM DTT or 1 mM potassium mercury thiocyanate/220 mM MgCl2/0.1 M bis-Tris (pH = 6.6)] and equilibrated against 1 ml of precipitant solution in the well reservoir. Crystals appeared within 7–10 days and grew to maximal dimensions of 0.6 × 0.3 × 0.3 mm3. These crystals diffracted x-rays to 2.0-Å resolution at the Stanford Synchrotron Radiation Laboratory and belonged to space group P212121 with unit cell dimensions a = 101.4 Å, b = 116.9 Å, and c = 120.1 Å (one dimer in the asymmetric unit).
The structure of native BPPS was solved by multiwavelength anomalous dispersion (MAD) (15) using a single crystal of the selenomethionine-derivatized protein prepared by the methionine pathway inhibition method (ref. 16; see Supporting Methods). X-ray diffraction data were collected at the Cornell High Energy Synchrotron Source at wavelengths corresponding to the absorption peak and edge of selenium as well as a remote reference wavelength (Table 1). Selenium sites were located using software routines implemented in cns, and initial 2.65-Å phases were improved by solvent-flipping (17). Electron density maps were fit using O (18). The structure was refined against 2.0-Å resolution data (σ cutoff = 0.0) collected from the wild-type enzyme by using cns (ref. 17; Table 2). Strict noncrystallographic symmetry constraints were used initially and relaxed into appropriately weighted restraints as guided by Rfree. The noncrystallographic symmetry axis is the dyad axis of the dimer. The N termini (E50-A63) are disordered in both monomers, and loop segments D227–D234, T500–D509, and F578–S583 are disordered in both monomers. Partially occupied mercury ions associate with six of the eight cysteines in each monomer as a consequence of crystallization in the presence of potassium mercury thiocyanate. The final model has excellent geometry, with 93.0% and 6.6% of residues adopting most favored and additionally allowed conformations, respectively.
Table 1.
Data collection statistics
| Wild type | Remote | Edge | Peak | |
|---|---|---|---|---|
| Wavelength, Å | 1.080 | 0.9642 | 0.9794 | 0.9791 |
| Resolution, Å | 2.00 | 2.60 | 2.65 | 2.60 |
| Total reflections | 633,848 | 375,887 | 293,325 | 311,749 |
| Unique reflections | 97,291 | 89,037 | 80,805 | 85,168 |
| Completeness, % | 99.8 | 99.9 | 99.7 | 99.6 |
| Rmerge | 0.051 | 0.054 | 0.080 | 0.063 |
| Mean figure of merit for multiple anomalous dispersion phasing (2.65 Å) | 0.692 | |||
Friedel mates are treated as unique reflections for data reduction in multiple anomalous dispersion experiments.
Rmerge = ∑|I − 〈I〉 |/∑I, where I is the observed intensity and 〈I〉 is the average intensity calculated for replicate data.
Mean figure of merit = ∑cos〈Δαi〉/n, where Δαi is the error in the phase angle for reflection i, and n is the number of reflections.
Table 2.
Refinement statistics of BPPS structures
| Structure | BPPS | BPPS–PPi | BPPS–1 | BPPS–1 | BPPS–2 | BPPS–3 | BPPS–BPP |
|---|---|---|---|---|---|---|---|
| Limiting resolution, Å | 2.0 | 2.3 | 2.3 | 3.1 | 2.4 | 2.4 | 2.3 |
| No. of reflections, work/test | 89,853/3,756 | 61,228/3,269 | 57,096/2,410 | 24,228/2,072 | 53,718/2,862 | 53,674/2,864 | 58,692/2,465 |
| R/Rfree | 0.203/0.234 | 0.213/0.242 | 0.206/0.229 | 0.206/0.233 | 0.220/0.255 | 0.208/0.246 | 0.210/0.238 |
| Protein atoms | 8,549 | 8,699 | 8,717 | 4,405 | 8,719 | 8,742 | 8,751 |
| Water molecules | 588 | 290 | 269 | 26 | 226 | 222 | 348 |
| Metal ions/ligand atoms | 15/0 | 6/18 | 6/38 | 3/19 | 7/38 | 6/38 | 6/38 |
| rms deviations | |||||||
| Bond lengths, Å | 0.006 | 0.006 | 0.006 | 0.006 | 0.007 | 0.007 | 0.007 |
| Bond angles, ° | 1.1 | 1.1 | 1.1 | 1.2 | 1.1 | 1.1 | 1.2 |
| Proper dihedral angles, ° | 18.5 | 18.7 | 18.9 | 19.2 | 18.7 | 18.8 | 18.8 |
| Improper dihedral angles, ° | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.9 |
| PDB ID code |
Complexes: analogues 1–3 indicated in Fig. 1. All complexes are isomorphous with native BPPS in space group P212121.
The structure of the BPPS–1 complex was also solved in space group P6322.
R = ∑||Fo|−|Fc∥/∑|Fo|, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively. Rfree is calculated in the same manner for reflections in the test set excluded from refinement.
Crystals of enzyme–inhibitor complexes were prepared either by cocrystallization or by soaking crystals of the native enzyme in buffer solutions containing 2–10 mM concentrations of each inhibitor; equimolar concentrations of PPi were also included in crystal soaking experiments with 2 and 3; aza analogues 1-3 were prepared as described in Supporting Methods and are illustrated in Fig. 1. The structure of each complex was determined by the difference Fourier method, and refined to convergence by using cns (σ cutoff = 0.0; ref. 17; Table 2). Additionally, the structure of the complex with 1 was solved in hexagonal space group P6322 (unit cell dimensions a = b = 198.4 Å, c = 122.8 Å, one monomer in the asymmetric unit) by molecular replacement, and refined with CNS (17). Prepared by cocrystallization, the structure of the complex in the hexagonal crystal form is identical to that prepared by soaking the substrate analogue into orthorhombic crystals of the native enzyme. Atomic coordinates of BPPS and its inhibitor complexes have been deposited in the Protein Data Bank under the ID codes indicated in Table 2.
Modeling.
In constructing models of BPPS complexes with actual substrate and reaction intermediates, we assumed that the binding conformations of aza analogues approximate the binding conformations of the corresponding substrate or reaction intermediates. The linalyl diphosphate (LPP) intermediate was modeled in a conformation that was consistent with the required 3R stereochemistry, that prevented steric clashes with residues lining the active site, and that placed C1–C2 adjacent to the bound diphosphate moiety. Because the binding orientation of 2 appeared to be inverted relative to substrate analogue and product complexes, we manually modeled the (4R)-α-terpinyl cation intermediate in a conformation consistent with cyclization to the bornane skeleton as represented by the structures of the BPPS–3 and BPPS–BPP complexes. Following manual positioning of intermediates, the structures were subjected to 200 steps of positional refinement by using energy minimization routines implemented in cns (17). Protein atoms, magnesium ions, and water molecules were constrained to positions observed in the corresponding structures of enzyme–aza analogue complexes.
Results and Discussion
Structure of the Native Enzyme.
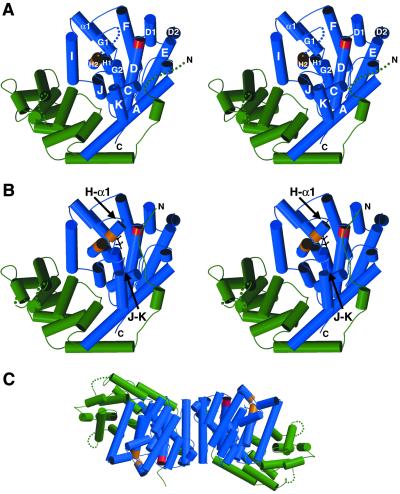
Each monomer is comprised of two α-helical domains (Fig. 2). The N-terminal domain forms an α-barrel similar to that of the sesquiterpene cyclase epi-aristolochene synthase (8), and partially similar to the (αα)6-barrel domains of protein farnesyltransferase (19) and the triterpene synthase squalene-hopene cyclase (5, 20). Although the N-terminal α-barrel domains of BPPS and epi-aristolochene synthase have no established catalytic function, the disordered N termini of these enzymes become ordered and cap their respective active sites in enzyme–ligand complexes (ref. 8; Fig. 2B). The N terminus of BPPS in ligand complexes is stabilized by multiple interdomain hydrogen bond interactions, including two with the aspartate-rich motif (R56–D355 carbonyl and Y60–D352). Truncation studies of limonene synthase show that tandem arginines in the N terminus may play a role in the mandatory isomerization step of the reaction cascade (21). If the same holds true for BPPS, then the R56–D355 carbonyl hydrogen bond, as well as other hydrogen bonds between the N terminus and the catalytic domain, may stabilize the diphosphate moiety during catalysis.
Fig 2.
(A) Stereoplot of the native BPPS monomer; the viewer is looking into the active site in the C-terminal domain (blue); helical segments are designated by the convention used for farnesyl diphosphate synthase (11). The aspartate-rich motif is red, and the second metal-binding motif starting at D496 is orange. The N-terminal domain is green. Disordered segments (E50–A63, E228–I233, T500–D509, and G579–S583) are indicated by dotted lines. (B) Stereoplot of the BPPS monomer complexed with PPi (black); orientation is the same as in A. The diphosphate group of GPP similarly triggers conformational changes that order three polypeptide segments to cap the active site: most of the N terminus (I54–A63), the C-terminal portion of helix H and the H-α1 loop (T500–D509), and a portion of the J–K loop (G579–S583). (C) Structure of the BPPS dimer; the left-hand monomer is oriented ≈90° down from the orientation shown in A. The dimer interface is formed by the C terminus of helix A, and helices D1, D2, and E (1,167 Å2 surface area per monomer is excluded from solvent).
The C-terminal domain of BPPS contains 12 α-helices, 6 of which (C, D, F, G, H, and J) largely define a hydrophobic active site cleft. Critical amino acid segments that bind catalytically required metal ions are conserved in this monoterpene cyclase. The aspartate-rich motif DDIYD starting at D351 is located at the C-terminal end of helix D (Fig. 2). In native BPPS, the carboxylate Oδ1 atoms of D351 and D355 coordinate to a single magnesium ion, with the carboxylate Oδ2 atoms hydrogen bonding to magnesium-coordinated water molecules; two additional water molecules complete a distorted octahedral metal coordination polyhedron (data not shown). This magnesium ion corresponds to Mg2+A observed in sesquiterpene cyclases (8, 10). The carboxylate of D352 is salt-linked with the side chain of R314. On the opposite wall of the active site cleft, at the C-terminal end of helix H, is the Mg2+B binding motif containing D496, T500, and E504. The T500–D509 segment is disordered in native BPPS, and Mg2+B is accordingly absent.
Structures of Enzyme–Ligand Complexes.
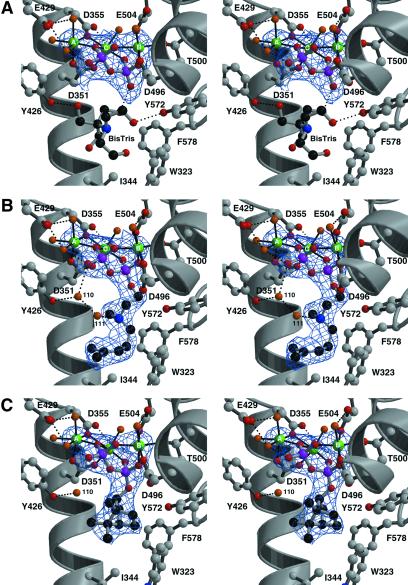
The electron density map of the BPPS–PPi complex (Fig. 3A) reveals that the binding of PPi–Mg2+3 is similar, but not identical, to that observed in sesquiterpene cyclases. The first and third aspartate residues in the aspartate-rich motif, D351 and D355, coordinate to Mg2+A. This is similar to the binding of Mg2+A by epi-aristolochene synthase (13); however, in trichodiene synthase, the third aspartate does not coordinate metal ions (15). As with the sesquiterpene cyclases (8, 10), Mg2+B is complexed by D496, T500, and E504, which stabilizes the previously disordered T500–D509 segment (Fig. 2B). Finally, Mg2+C is coordinated by D351 and D355. Octahedral coordination polyhedra are completed by water molecule(s) for all magnesium ions. The PPi anion accepts hydrogen bonds from protein residues R314 (A–C loop), R493 (helix H), and K512 (helix α1). Hydrogen bond interactions with PPi are not conserved among terpenoid cyclases.
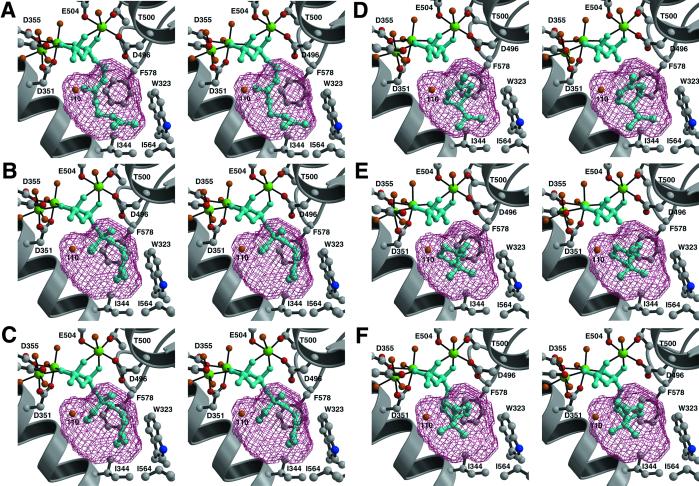
Fig 3.
Simulated annealing omit electron density maps of BPPS complexes contoured at 4σ; all inhibitor atoms and Mg2+ ions were omitted from structure factor calculations. Carbon, black (ligand) or gray (protein); oxygen, red; nitrogen, blue; phosphorus, magenta; Mg2+ ions, green; water molecules, orange. Metal coordination interactions are indicated by solid black lines and hydrogen bond interactions are indicated by dashed black lines. (A) Inorganic pyrophosphate (PPi). In addition to interactions described in the text, E429 from helix F accepts hydrogen bonds from magnesium-bound water molecules. An ordered buffer molecule (bis-Tris) occupies the active site. (B) 3-Aza-2,3-dihydrogeranyl diphosphate (1). Two water molecules, #110 and #111, bind in the active site cavity along with the inhibitor. (C) BPP. Note that water #110 remains trapped in the active site along with the cyclization product.
Complexation of PPi–Mg2+3 by BPPS triggers several conformational changes that cap and sequester the active site: disordered loop segments flanking the active site become ordered, and the N terminus becomes ordered and packs across the top of the active site cleft (Fig. 2B). The largest PPi-induced conformational changes occur in segments that cap the active site with Cα positions moving by as much as 5.3 Å; the rms deviation of 306 Cα atoms in the catalytic domain is 0.60 Å. These conformational changes sequester a 222-Å3 active site cavity from bulk solvent, in which a single buffer molecule (bis-Tris) is trapped. This cavity is smaller than the 324-Å3 cavity observed in the PPi–Mg2+3 complex with the sesquiterpene cyclase trichodiene synthase (10, 22). Several amino acid substitutions and slight structural differences in helix–helix packing account for the smaller active site volume of BPPS. Because the volume of the GPP hydrocarbon chain is 174 Å3, this implies that the packing density of the enzyme–substrate complex is 78%, i.e., comparable to that calculated for trichodiene synthase (10, 22) and comparable to the average packing density in the interiors of folded proteins (23). Thus, BPPS evolved to optimize the packing density of the substrate (and reaction intermediates) in the enzyme active site.
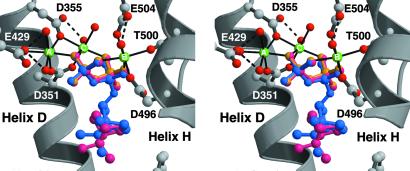
The structures of BPPS complexed with aza analogues of carbocationic intermediates (Fig. 1) provide key mechanistic inferences regarding the terpenoid cyclization cascade. In all complexes except the BPPS–BPP complex, the position, orientation, and intermolecular contacts of the diphosphate group, either as PPi or as the prenyl diphosphate ester, are identical to those observed in the BPPS–PPi complex. In the BPPS–BPP complex, the diphosphate group rotates slightly (17°) about the β-phosphate group such that the α-phosphate group moves 0.6 Å toward the active site cavity (Fig. 4). However, despite this slight movement, all intermolecular contacts of the diphosphate group are maintained. This observation suggests that the principal molecular recognition event between enzyme and substrate is mediated by the firmly anchored diphosphate group, established as the primary determinant of substrate binding (24, 25).
Fig 4.
BPPS–diphosphate recognition is mediated by metal coordination and hydrogen bond interactions. The binding positions and conformations of the diphosphate group, as PPi (yellow), the prenyl diphosphate ester of 1, or the PPi counterion to the carbocation intermediate analogues 2 and 3, are identical (for clarity, only the 3 PPi complex is indicated in magenta). The binding position of the product diphosphate group is slightly different (blue), but all intermolecular interactions of the diphosphate group are conserved.
The C4–N–C2–C1 dihedral angle of the substrate analogue 3-aza-2,3-dihydrogeranyl diphosphate 1 (26) adopts a conformation that is midway between the transoid and cisoid rotamers of the intermediate 3(R)-LPP, despite the fact that the diphosphate group is still attached to C1 (Fig. 3B). Presuming that the 3-aza nitrogen atom is protonated, 1 also partially mimics the allylic cation intermediates in catalysis (see Fig. 1). Of course, the substitution of pyramidal sp3-hybridized atoms for the planar C = C π linkage makes the analogue an imperfect mimic of GPP. However, it is interesting that the binding conformation of 1 partially mimics the conformational change in the C4–C3–C2–C1 dihedral angle of LPP that brings C1 and C6 closer together for cyclization (see Fig. 1).
The isoprenoid group of 1 makes van der Waals contacts with W323, I344, and F578. Surprisingly, two water molecules, #110 and #111, are trapped in the active site along with the substrate analogue (Fig. 3B). Water #111 appears to be hydrogen bonded to the 3-aza nitrogen atom, so its presence may be artifactual. However, water #110 donates hydrogen bonds to the diphosphate group of 1 and the backbone carbonyl of S451, and it hydrogen bonds with the side chain of Y426. Given the deduced orientations of hydrogen bonds with water #110, a lone electron pair is oriented directly toward 1. Because water #110 is also observed in the native enzyme and other BPPS complexes, including that with the actual cyclization product BPP, its presence in the analogue complexes is unlikely to be artifactual (Fig. 3C). Possibly, water #110 could serve as a diphosphate-assisted general base to account for the generation by this enzyme of cyclic olefin co-products (pinenes, camphene, and limonene) derived by direct deprotonation of carbocation intermediates (13, 27). Studies with noncyclizable analogues of GPP reveal significant formation of terpenol products, indicating that the substrate has greater access to water in the active site of BPPS than in most other monoterpene cyclases (28): water #110 may well be implicated by these studies.
The binding of 7-aza-7,8-dihydrolimonene 2 (29) in the presence of PPi, intended to mimic the (R)-α-terpinyl cation-PPi intermediate pair, appears to be backwards in comparison with the binding of the substrate and product (data not shown). We hypothesize that this peculiar binding mode results from overwhelmingly favorable electrostatic and hydrogen bond interactions between the positively charged 7-aza nitrogen atom of 2 and negatively charged PPi. Thus, we suggest that this binding conformation is nonproductive. This binding conformation nevertheless highlights the strength of electrostatic interactions between PPi and cationic species contained in the active site cavity.
The ternary complex of BPPS with (1R,4S)-2-azabornane 3 (30, 31) and PPi is highly informative. Relative to the binding of 1, the hydrocarbon moiety is closer to the PPi binding position. The O3 atom of the α-phosphate group is oriented toward the 2-aza nitrogen atom of 3 (O—N separation = 3.0 Å), such that if 3 were the actual bornyl cation shown in Fig. 1, the trajectory of the endo-oriented recapture of PPi that terminates the cyclization cascade would be clearly delineated. Importantly, the α-phosphate oxygen atom bound to C1 of substrate analogue 1 corresponds exactly to the PPi oxygen atom that is oriented toward the 2-aza nitrogen atom of 3. This orientation is consistent with the results of studies with 18O-labeled substrate, indicating a complete lack of positional isotope exchange, i.e., the same oxygen atom is contained in the prenyl diphosphate ester linkages of both GPP and BPP (32, 33). Water #110 also makes an exo-oriented hydrogen bond with the 2-aza nitrogen atom of 3; presuming that this water is also present in the complex with the bornyl cation, it is probably sterically hindered from making an exo-oriented approach to the bornyl cation by the syn-geminal methyl substituent of the one-carbon bridge. The secondary bornyl cation also must be sufficiently restrained by the active site template to prevent a classical Wagner–Meerwein rearrangement that would form the much more stable tertiary camphenyl carbocation (34).
The BPPS–BPP complex (Fig. 3C) reveals a slight movement of the diphosphate group relative to the structures of other complexes (Fig. 4), as previously noted. The presence of water #110 in the enzyme–product complex strongly suggests that this water is present during catalysis. Accordingly, water #110 comprises an integral part of the active site contour. This is rather surprising, given the risk that such a water molecule could prematurely quench a carbocationic intermediate. Because water #110 occupies the same location in BPPS complexes with 1, 3, and BPP, we conclude that this water molecule is restrained from aborting the cyclization cascade by three hydrogen bond interactions with protein and PPi.
Mechanistic Implications.
Structural comparison of all BPPS complexes suggests that the active site template chaperones the positions and conformations of the isoprenoid moiety, while the diphosphate position remains essentially anchored in a single location and conformation. These structures can be used to model the complete reaction coordinate of GPP cyclization, including key substrate-induced conformational changes in the active site that initiate the cyclization cascade (Fig. 5). The stereochemistry of this cyclization cascade is outlined in previous enzymological studies (32, 33, 35–38). On diphosphate recognition, diphosphate-triggered conformational changes trap GPP and water molecule #110 in the active site cavity. Active site residues W323, I344, V452, and F578, as well as water #110, enforce the binding of the left-handed helical conformation of GPP. We emphasize that water #110, present in the native enzyme and firmly anchored by hydrogen bonds, comprises an integral part of the active site contour required for template function. Following initial metal-activated diphosphate departure, isomerization to linalyl diphosphate (LPP) allows rotation about the newly formed C2–C3 bond to the cisoid conformer, after which reionization facilitates SN′cyclization by C1–C6 closure to form the (R)-α-terpinyl cation-PPi ion pair. Cation-π interactions with F578 and W323 likely stabilize the positive charge on C8. Further anti-Markovnikov cyclization yields the 2-bornyl cation, which is neutralized by stereo specific C–O bond formation with PPi on the endo face to yield BPP.
Fig 5.
GPP cyclization pathway in the active site of BPPS, represented by its solvent-accessible surface area. Note that water #110 comprises an integral part of the active site template. The modeling of GPP (A) is based on the binding conformation of inhibitor 1, which may not be a faithful representation of the native substrate. GPP can also bind in a more compact conformer that requires less rearrangement to form the (3R)-LPP (B and C).
As observed in the structures of sesquiterpene cyclases (7–10), aromatic residues in the BPPS active site may stabilize carbocation intermediates through cation-π interactions. However, evaluation of BPPS–inhibitor complexes suggests that the bound PPi moiety also plays an important role in stabilizing and orienting carbocations in the active site: if electrostatic interactions with PPi are sufficient to cause analogue 2 to bind backwards, then such interactions may also significantly stabilize transiently formed carbocations during catalysis. These interactions, as well as the rigid positioning of PPi adjacent to C2 of the (R)-α-terpinyl and bornyl cations, may help to favor the anti-Markovnikov cyclization and stereo-specific endo recapture of PPi and disfavor off-pathway rearrangements leading to the formation of more stable tertiary carbocation intermediates. That PPi is reincorporated only into the major product and none of the other co-products indicates that PPi is oriented exclusively for recapture by the endo face of the bornyl C2 cation.
In conclusion, the structural data acquired from BPPS and its ligand complexes allow for the first detailed comparison of structure–mechanism relationships between monoterpene and sesquiterpene cyclases. Comparison of BPPS, epi-aristolochene synthase (8), and trichodiene synthase (10) reveals that active site closure is triggered by a network of metal coordination and hydrogen bond interactions with the rigidly complexed substrate diphosphate group: metal coordination interactions are largely conserved, whereas hydrogen bond interactions are less conserved, among the three cyclases. Given the topological similarities among the cyclases, it is not surprising that a common secondary structural element participates in active site closure mechanisms (the J–K loop), but there are more differences than similarities. For example, the H–α1 loop participates in active site closure mechanisms in BPPS and trichodiene synthase, whereas it does not in epi-aristolochene synthase; the N termini of BPPS and epi-aristolochene synthase cap their respective active sites, whereas trichodiene synthase requires neither an N-terminal domain nor the N terminus for active site closure; trichodiene synthase contains a D101–R304 “switch” that triggers active site closure, but this switch is not conserved in the other cyclases. These features are consistent with the divergent evolution of each cyclase from a common primordial ancestor.
Following active site closure in each cyclase, the hydrophobic active site cavity serves as a template to chaperone the conformations of the flexible isoprenoid substrates GPP or farnesyl diphosphate (FPP). Surprisingly, in BPPS a firmly anchored water molecule comprises part of the template. In accord with this template function, the packing density of the enzyme–substrate complex is similarly efficient in monoterpene and sesquiterpene cyclases (75–78%), in that the enclosed active site cavities bind their respective substrates with little wasted space. Reactive carbocation intermediates in both GPP and FPP cyclization cascades are stabilized by the PPi counterion and cation-π interactions with (conserved) aromatic residues. Further studies of terpenoid cyclase structures, and the ultimate structure determination of a diterpene cyclase, will provide increasingly detailed comparisons among these enzymes catalyzing the most structurally and stereochemically complex reactions in biology.
Supplementary Material
Acknowledgments
We thank the Stanford Synchrotron Radiation Laboratory, the Cornell High Energy Synchrotron Source, and BioCARS Sector 14 of the Advanced Photon Source at Argonne National Laboratories for beamline access. We also thank R. K. Boeckman, Jr., University of Rochester, for helpful advice on the azabornane synthesis. This work was supported by National Institutes of Health Grants GM56838 (to D.W.C.), GM31354 (to R.B.C.), and GM13956 (to R.M.C.), and National Institutes of Health Postdoctoral Fellowship AI10648 (to D.A.W.).
Abbreviations
BPP, (+)-bornyl diphosphate
BPPS, BPP synthase
GPP, geranyl diphosphate
PPi, pyrophosphate
Data deposition: The atomic coordinates and structure factors for bornyl diphosphate synthase and its ligand complexes have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes , , and –).
References
- 1.McCaskill D. & Croteau, R. (1997) Adv. Biochem. Eng. Biotechnol. 55 107-146. [DOI] [PubMed] [Google Scholar]
- 2.Davis E. M. & Croteau, R. (2000) Top. Curr. Chem. 209 53-95. [Google Scholar]
- 3.Bohlmann J., Meyer-Gauen, G. & Croteau, R. (1998) Proc. Natl. Acad. Sci. USA 95 4126-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trapp S. C. & Croteau, R. B. (2001) Genetics 158 811-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wendt K. U. & Schulz, G. E. (1998) Structure (London) 6 127-133. [DOI] [PubMed] [Google Scholar]
- 6.Lesburg C. A., Caruthers, J. M., Paschall, C. M. & Christianson, D. W. (1998) Curr. Opin. Struct. Biol. 8 695-703. [DOI] [PubMed] [Google Scholar]
- 7.Lesburg C. A., Zhai, G., Cane, D. E. & Christianson, D. W. (1997) Science 277 1820-1824. [DOI] [PubMed] [Google Scholar]
- 8.Starks C. M., Back, K., Chappell, J. & Noel, J. P. (1997) Science 277 1815-1820. [DOI] [PubMed] [Google Scholar]
- 9.Caruthers J. M., Kang, I., Rynkiewicz, M. J., Cane, D. E. & Christianson, D. W. (2000) J. Biol. Chem. 275 25533-25539. [DOI] [PubMed] [Google Scholar]
- 10.Rynkiewicz M. J., Cane, D. E. & Christianson, D. W. (2001) Proc. Natl. Acad. Sci. USA 98 13543-13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarshis L. C., Yan, M., Poulter, C. D. & Sacchettini, J. C. (1994) Biochemistry 33 10871-10877. [DOI] [PubMed] [Google Scholar]
- 12.Cane D. E. & Kang, I. (2000) Arch. Biochem. Biophys. 376 354-364. [DOI] [PubMed] [Google Scholar]
- 13.Wise M. L., Savage, T. J., Katahira, E. & Croteau, R. (1998) J. Biol. Chem. 273 14891-14899. [DOI] [PubMed] [Google Scholar]
- 14.Schenk P. M., Baumann, S., Mattes, R. & Steinbis, H.-H. (1995) BioTechniques 19 196-200. [PubMed] [Google Scholar]
- 15.Hendrickson W. A. (1991) Science 254 51-58. [DOI] [PubMed] [Google Scholar]
- 16.Doublié S. (1997) Methods Enzymol. 276 523-530. [PubMed] [Google Scholar]
- 17.Brünger A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54 905-921. [DOI] [PubMed] [Google Scholar]
- 18.Jones T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47 110-119. [DOI] [PubMed] [Google Scholar]
- 19.Park H.-W., Boduluri, S. R., Moomaw, J. F., Casey, P. J. & Beese, L. S. (1997) Science 275 1800-1804. [DOI] [PubMed] [Google Scholar]
- 20.Wendt K. U., Poralla, K. & Schulz, G. E. (1997) Science 277 1811-1815. [DOI] [PubMed] [Google Scholar]
- 21.Williams D. C., McGarvey, D. J., Katahira, E. J. & Croteau, R. (1998) Biochemistry 37 12213-12220. [DOI] [PubMed] [Google Scholar]
- 22.Rynkiewicz M. J., Cane, D. E. & Christianson, D. W. (2002) Biochemistry 41 1732-1741. [DOI] [PubMed] [Google Scholar]
- 23.Creighton T. E., (1993) Proteins: Structure and Molecular Properties (Freeman, New York), pp. 229.
- 24.Wheeler C. J. & Croteau, R. (1987) J. Biol. Chem. 262 8213-8219. [PubMed] [Google Scholar]
- 25.Wheeler C. J. & Croteau, R. (1988) Arch. Biochem. Biophys. 260 250-256. [DOI] [PubMed] [Google Scholar]
- 26.Szabo C. M., Matsumura, Y., Fukura, S., Martin, M. B., Sanders, J. M., Sengupta, S., Cieslak, J. A., Loftus, T. C., Lea, C. R., Lee, H.-J., et al. (2002) J. Med. Chem. 45 2185-2196. [DOI] [PubMed] [Google Scholar]
- 27.Wise M. L. & Croteau, R. (1999) in Comprehensive Natural Products Chemistry: Isoprenoids Including Carotenoids and Steroids, ed. Cane, D. E. (Elsevier Science, Oxford), Vol. 2, pp. 97–153. [Google Scholar]
- 28.Schwab W., Williams, D. C., Davis, E. M. & Croteau, R. (2001) Arch. Biochem. Biophys. 392 123-136. [DOI] [PubMed] [Google Scholar]
- 29.McGeady P., Pyun, H.-J., Coates, R. M. & Croteau, R. (1992) Arch. Biochem. Biophys. 299 63-72. [DOI] [PubMed] [Google Scholar]
- 30.Gassman P. G. & Shudo, K. (1971) J. Am. Chem. Soc. 93 5899-5901. [DOI] [PubMed] [Google Scholar]
- 31.Buston, J. E. H., Coldham, I. & Mulholland, K. R. (1999) J. Chem. Soc. Perkin Trans. 1, 2327–2334.
- 32.Cane D. E., Saito, A., Croteau, R., Shaskus, J. & Felton, M. (1982) J. Am. Chem. Soc. 104 5831-5833. [Google Scholar]
- 33.Croteau R. B., Shaskus, J. J., Renstrøm, B., Felton, N. M., Cane, D. E., Saito, A. & Chang, C. (1985) Biochemistry 24 7077-7085. [DOI] [PubMed] [Google Scholar]
- 34.Berson J. A. (1963) in Molecular Rearrangements, ed. de Mayo, P. (Interscience, New York), Vol. 1, pp. 111–231. [Google Scholar]
- 35.Croteau R. & Felton, M. (1981) Arch. Biochem. Biophys. 207 460-464. [DOI] [PubMed] [Google Scholar]
- 36.Croteau R., Felton, N. M. & Wheeler, C. J. (1985) J. Biol. Chem. 260 5956-5962. [PubMed] [Google Scholar]
- 37.Croteau R., Satterwhite, D. M., Wheeler, C. J. & Felton, N. M. (1989) J. Biol. Chem. 264 2075-2080. [PubMed] [Google Scholar]
- 38.Wise M. L., Pyun, H.-J., Helms, G., Assink, B., Coates, R. M. & Croteau, R. B. (2001) Tetrahedron 57 5327-5334. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.