Abstract
We report the x-ray crystal structure of human topoisomerase I covalently joined to double-stranded DNA and bound to the clinically approved anticancer agent Topotecan. Topotecan mimics a DNA base pair and binds at the site of DNA cleavage by intercalating between the upstream (−1) and downstream (+1) base pairs. Intercalation displaces the downstream DNA, thus preventing religation of the cleaved strand. By specifically binding to the enzyme–substrate complex, Topotecan acts as an uncompetitive inhibitor. The structure can explain several of the known structure–activity relationships of the camptothecin family of anticancer drugs and suggests that there are at least two classes of mutations that can produce a drug-resistant enzyme. The first class includes changes to residues that contribute to direct interactions with the drug, whereas a second class would alter interactions with the DNA and thereby destabilize the drug-binding site.
Eukaryotic DNA topoisomerase I (topo I) is an enzyme that acts to relax supercoils generated during transcription and DNA replication (1). Because of the size of the eukaryotic chromosome, removal of these supercoils can only be accomplished locally by introducing breaks into the DNA helix. Topo I mediates DNA relaxation by creating a transient single-strand break in the DNA duplex. This transient nick allows the broken strand to rotate around its intact complement, effectively removing local supercoils. Strand nicking results from the transesterification of an active-site tyrosine (Tyr-723) at a DNA phosphodiester bond forming a 3′-phosphotyrosine covalent enzyme–DNA complex. After DNA relaxation, the covalent intermediate is reversed when the released 5′-OH of the broken strand reattacks the phosphotyrosine intermediate in a second transesterification reaction. The rate of religation is normally much faster than the rate of cleavage, and this ensures that the steady-state concentration of the covalent 3′-phosphotyrosyl topo I–DNA complex remains low (2).
However, a variety of DNA lesions and drugs have been shown to stabilize the covalent 3′-phosphotyrosyl intermediate (3). For example, camptothecin (CPT) is a natural product that was originally discovered because of its antitumor activity (4) and was later demonstrated to cause the accumulation of topo I–DNA adducts in vitro and in vivo (5, 6). CPTs bind the covalent 3′-phosphotyrosyl intermediate and specifically block DNA religation (7), thus converting topo I into a DNA-damaging agent (8). Topo I is the sole intramolecular target of CPT, and the cytotoxic effects of CPT poisoning are S-phase specific (9). During DNA replication, the replication fork is thought to collide with the “trapped” topo I–DNA complexes, resulting in double-strand breaks and ultimately cell death (10).
It has been difficult to study the mechanism of CPT activity because the drug acts as an uncompetitive inhibitor and binds only the transient covalent enzyme–substrate complex (7, 11). To isolate the covalent topo I–DNA complex, we have used suicide DNA substrates containing a 5′-bridging phosphorothiolate (12). Topo I-mediated cleavage of these substrates generates a 5′-sulfhydryl, instead of a 5′-hydroxyl, which is inert in subsequent ligation reactions. These substrates have previously allowed the crystallization of a reconstituted version of human topo I covalently attached to DNA (13), which displays altered sensitivity to CPTs. Here, we report the structure of a fully active, CPT-sensitive form of human topo I covalently joined to duplex DNA in the absence (3.2 Å) and presence (2.1 Å) of Topotecan, a clinically approved CPT analog (trademark name Hycamtin).
Materials and Methods
The human topoisomerase I construct of residues Lys-175 to Phe-765 (topo70) was purified from a baculovirus–insect cell (SF9) expression system as described (14). Topo70 was concentrated to 4 mg/ml in 10 mM Tris⋅HCl, pH 7.5/1 mM EDTA/1 mM DTT. Blunt-ended duplex oligonucleotides were prepared with a 5′-bridging phosphorothiolate linkage (12) at the preferred site of topo I cleavage (13). The oligonucleotide sequence was 5′-AAAAAGACTTsTGAAAAATTTTT-3′in the binary topo70-DNA crystal form and 5′-AAAAAGACTTsGGAAAAATTTTT-3′ in the ternary topo70-DNA–Topotecan crystal form, where “s” represents the 5′ bridging phosphorothiolate of the cleaved strand. Crystals of the ternary complex were grown in sitting drops at 16°C by vapor equilibration against a precipitant of 10% (wt/vol) polyethylene glycol (PEG) 8000, 200 mM lithium sulfate, and 100 mM Mes, pH 6.5, at 16°C (15). Crystallization drops were prepared by mixing in order 2 μl of precipitant, 1.5 μl of 0.05 mM suicide duplex substrate, 0.3 μl of 10 mM Topotecan (dissolved in water), and 1.5 μl of 4 mg/ml topo70. Crystals of the binary complex were grown in sitting drops at 16°C by vapor equilibration against a precipitant of 10% wt/vol PEG 8000, 100 mM Tris⋅HCl, pH 8.0, 100 mM Na/K phosphate, pH 6.2, 100 mM KCl, and 10 mM DTT. Crystallization drops were prepared by mixing in order 2 μl of precipitant, 2 μl of 0.1 mM suicide duplex substrate, and 2 μl of 4 mg/ml topo70.
For structure determination and refinement, crystals were cryoprotected by soaking them in precipitant plus 30% vol/vol PEG 400 for 30 s. Data were collected at 100 K at the National Synchrotron Light Source, Brookhaven National Laboratory beamline X25, and COM-CAT, Sector 32, Advanced Photon Source, Argonne National Laboratory. The structures were solved by molecular replacement with AMORE (16) by using the structure of the noncovalent topo70–DNA complex (17). The DNAs were placed into the electron density of SigmaA-weighted phase-combined maps and rigid body refined using CNX. The structures were rebuilt and refined by CNX with simulated annealing (18) and iterative model adjustments with XTALVIEW (19). Topotecan was placed unambiguously into the SigmaA-weighted |Fobs|–|Fcalc| maps of the ternary complex. The final ternary complex model contains residues 201–765 of topo70, Topotecan, and the 22-bp DNA duplex oligonucleotide, with good geometry and no Ramachandran outliers (Table 1). Crystals of the binary topo70–DNA covalent complex displayed merohedral twinning. The twin fraction was estimated using CNX, and the final binary complex model was refined using torsion angle-simulated annealing and restrained B-factor refinement against the twinned data using CNX. All DNA calculations were made with the program X3DNA (20) using the standard reference frame for calculations recommended by the Tsukuba workshop on nucleic acid structure and interactions (21).
Table 1.
Refinement statistics
| Crystallographic data | Ternary complex | Binary complex |
|---|---|---|
| Inhibitor bound | Topotecan | — |
| Resolution, Å | 2.1 | 3.2 |
| No. of reflections | 46,866 | 17,874 |
| Rsym | 6.2 | 13.0 |
| Completeness | 81.1 | 85.9 |
| Space group | P21 | P3(2) |
| a | 57.1 Å | 73.2 |
| b | 116.6 Å | 73.2 |
| c | 75.2 Å | 186.6 |
| β | 94.2 | — |
| Reflections used in Rfree | 10% | 10% |
| No. of protein atoms | 4685 | 3498 |
| No. of DNA atoms | 892 | 892 |
| No. of Topotecan atoms | 31 | — |
| No. of solvent atoms | 227 | — |
| No. of PEG atoms | 13 | — |
| Rfactor | 23.2 | 21.7 |
| Rfree | 27.1 | 28.4 |
| rms deviations from ideal stereochemistry | ||
| Bond lengths, Å | 0.0068 | 0.0088 |
| Bond angles, ° | 1.17 | 1.36 |
| Dihedrals, ° | 21.3 | 22.16 |
| Impropers, ° | 3.6 | 3.35 |
| Mean B factor—all atoms, Å3 | 39.3 | 41.8 |
Rsym = Σ|li − lm|/Σlm where Ii is the intensity of the measured reflection and Im is the mean intensity of all symmetry-related reflections.
Nondrug-bound crystals were merohedrally twinned (twin law h, -h-k, -l; twin fraction = 0.151 estimated using cnx). R factors were calculated using twinned data.
Results
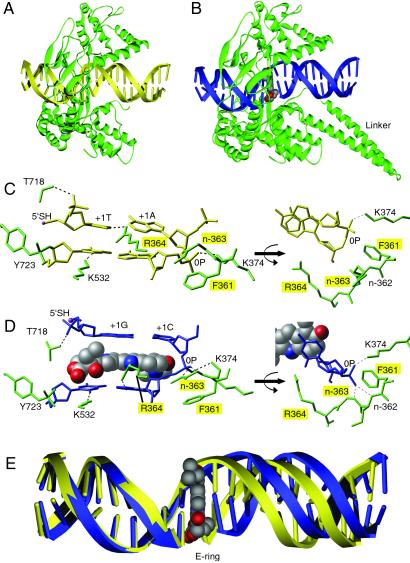
To define the binding mode of Topotecan, we have solved the x-ray crystal structures (Table 1) of a 70-kDa form of human topo I (topo70) in covalent complex with duplex DNA in the absence (binary complex; Fig. 1A) and presence of Topotecan (ternary complex; Fig. 1B). Topo70 is a fully active enzyme that displays in vitro activity and sensitivity to Topotecan that is indistinguishable from the native full-length enzyme (14). Both crystal forms have one unit cell dimension formed by pseudocontinuous end-to-end packing of the 22-mer duplex DNAs. This unit cell edge is 73.2 Å in the binary complex (the a edge) and 75.2 Å in the ternary complex (the c edge). A comparison of the Topotecan ternary complex structure to the non-Topotecan binary structure reveals minor differences in the overall C-α backbone trace with an rms deviation of 1.33 Å, excluding linker domain residues Gln-633–Gln-697, which were not visible in the electron density of the nondrug-bound protein. The linker domain residues are typically disordered in topo70 crystal structures (22).
Fig 1.
Structure of topo I–DNA complex without (A) and with (B) bound Topotecan. The binary complex is diagrammed with protein (green) and DNA (yellow). The ternary complex is diagrammed with protein (green), DNA (blue), and Topotecan (CPK gray as carbon, red as oxygen, blue as nitrogen). The 2.1-Å drug-bound structure represents the most complete topo I structure reported to date, providing visible electron density from Gln-201 to the COOH-terminal Phe-765. The linker domain (Pro-636 to Lys-712) could not be visualized in the binary complex (22) but was visible in the ternary complex. Previously reported crystal structures of human topo I include the inactive Y723F version of topo70 and topo58/6.3 (a reconstituted linkerless enzyme) in noncovalent complex with DNA and topo58/6.3 in covalent complex with DNA (13, 17). Unlike topo70, the reconstituted enzyme has altered kinetics and is not sensitive to CPT in a plasmid relaxation assay (17). (C) Molecular diagram showing the nondrug-bound topo I–DNA complex. The +1 and −1 bases of the duplex DNA (yellow, stick) are shown making four contacts to the surrounding protein (green, stick) to stabilize the protein–DNA complex. (D) Topotecan (CPK) intercalates between the +1 and −1 bases of the duplex DNA (blue, stick). Six protein (green, stick) contacts stabilize the open form of the DNA. A contact with the main chain nitrogen of Arg-362 is not labeled. Topo70 residues whose mutation leads to drug resistance are highlighted with yellow boxes. The 5′-SH of the +1 G is highlighted in pink. The covalent phosphotyrosine attachment to DNA is shown between Tyr-723 (green, stick) and the −1 T (blue, stick) of the cleaved strand. The mobile phosphodiester of the intact DNA strand is labeled 0P. (E) Comparison of the 22-mer duplex oligonucleotides of the drug-bound (blue) and nondrug-bound (yellow) complexes reveals that Topotecan (CPK) binds to the enzyme–substrate complex by intercalating in the DNA and shifting the downstream bases by ≈3.6 Å, equivalent to the rise of one base pair.
A comparative analysis of these structures demonstrates that Topotecan intercalates at the site of DNA cleavage, forming base-stacking interactions with both the −1 (upstream) and +1 (downstream) base pairs (Fig. 1). The planar 5-membered polycyclic ring system of Topotecan mimics a DNA base pair in the DNA duplex and occupies the same space as the +1 base pair in the structure without drug bound (Fig. 1E); ≈60% of solvent accessible surface of Topotecan (380 Å2 of the total 630 Å2) is covered by base-stacking interactions. Intercalation by Topotecan extends the rise between the −1 and +1 base pairs from 3.6 Å in the binary complex to 7.2 Å in the ternary complex. In addition, the twist between the −1 and +1 base pairs in the ternary complex is only ≈10°, as compared with ≈36° twist in the binary complex. As a result, the distance from the 5′-SH to the phosphorous of the 3′-phosphotyrosine increases from 3.5 Å in the binary complex to 11.5 Å in the ternary complex.
The intercalation binding site is created by conformational changes of the phosphodiester bond between the +1 and −1 base pairs of the uncleaved strand (0P, Fig. 1D), which effectively “open” the DNA duplex. Positioning of the 0P phosphodiester in the ternary complex is stabilized through hydrogen bond contacts to the main chain nitrogens of residues Arg-362 and Gly-363 (Fig. 1D). A hydrogen bond contact to Lys-374 is present in both structures (Fig. 1 C and D). The intercalation binding site is also stabilized by several protein–DNA interactions (Fig. 1D). The hydroxyl of Thr-718 makes a hydrogen bond with the nonbridging phosphodiester oxygen of guanosine at position +1 of the cleaved strand (+1G). Arg-364 makes a hydrogen bond with N3 of adenosine at position −1 of the uncleaved strand (−1A), and Lys-532, a known catalytic residue (23), makes a hydrogen bond with the O2 oxygen of thymidine at position −1 (−1T).
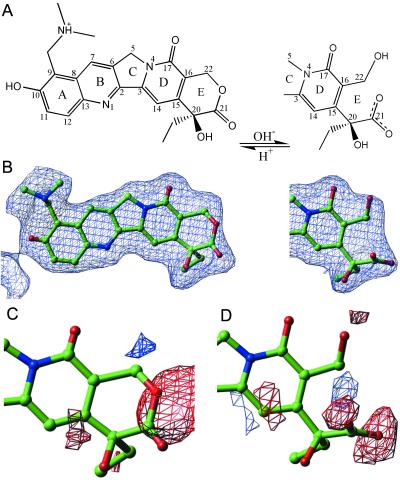
The E-ring of Topotecan is known to be in equilibrium between the closed lactone form and a hydrolyzed open carboxylate form (4) (Fig. 2A). Close inspection of the Topotecan electron density allowed positioning of both the open and closed E-ring conformers (Fig. 2B). An unrestrained full matrix refinement of occupancy factors (24) (with all positional and thermal parameters fixed) for the closed lactone and open carboxylate versions of Topotecan converged to an occupancy of 63% (standard uncertainty 7%) closed lactone and 37% (standard uncertainty 7%) open carboxylate conformers. When each conformer is placed into the structure and independently refined, difference Fourier maps demonstrate the presence of both conformations of Topotecan (Fig. 2 C and D). Fluorescence spectrophotometric measurements of Topotecan at 1 mM concentration in precipitant solution were determined as a function of pH (without added topo70 or duplex oligonucleotide) (25). This analysis revealed a similar ratio of closed (75%) and open (25%) forms of the drug dissolved in the precipitant solution that was used to grow the ternary complex crystals, 10% wt/vol PEG 8000, 200 mM lithium sulfate, 100 mM Mes, pH 6.5 (data not shown).
Fig 2.
Topotecan electron density. (A) Topotecan with reversible hydrolysis of the E-ring from the closed lactone form to the open carboxylate form is diagrammed. (B) A 3.0 σ |Fo|–|Fc| omit map of electron density for Topotecan is illustrated. (C) A 3.0 σ |Fo|–|Fc| electron density map calculated with the lactone form of Topotecan (100% closed E-ring) is diagrammed. Negative electron density (red) is seen in the vicinity of the lactone oxygen, and positive (blue) electron density peaks are located nearby. (D) A 3.0 σ |Fo|–|Fc| electron density calculated with the carboxylate form of Topotecan (100% open E-ring) is diagrammed. Negative electron density (red) surrounds the terminal hydroxyl and carboxylic acid moieties, whereas a positive (blue) electron density peak is in the location of what would be the lactone oxygen in the closed E-ring conformation.
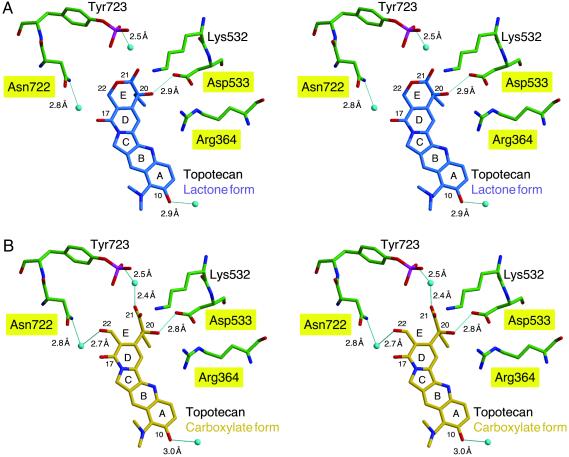
There is only one direct hydrogen bond between the enzyme and Topotecan; Asp-533 hydrogen bonds to the 20(S)-hydroxyl in both the carboxylate and lactone models (Fig. 3). Asp-533 is also coordinated by the ɛ-nitrogen of Arg-364. In the carboxylate model (Fig. 3B), there are two water-mediated hydrogen bonds that assist in coordinating the Topotecan into the cleaved DNA intermediate. The 22-hydroxyl is bridged through a water to the R-group of Asn-722, and one of the 21-carboxylate oxygens is bridged to the phosphotyrosine (P-Tyr-723). Finally, in both the lactone and carboxylate models, the 10-hydroxyl of Topotecan is 2.8 Å from a water molecule.
Fig 3.
Stereoview of Topotecan binding. The Topotecan interactions with protein side chains for the lactone (A, blue) and carboxylate (B, gold) forms of the drug are depicted. Hydrogen bonds (predicted by contact distance and geometry of the refined atomic positions) are shown as solid blue lines, and the calculated interatomic distances are indicated. Labels for residues that result in resistance to CPT when mutated are highlighted in yellow. The oxygen atoms of water molecules are depicted as light blue spheres. Protein side chains are green, and noncarbon atoms are colored red for oxygen, blue for nitrogen, and magenta for phosphorus. The DNA and protein side chains that do not interact with Topotecan have been omitted from the figure.
Discussion
The ternary topo I–DNA–Topotecan complex demonstrates that Topotecan intercalates at the site of DNA cleavage and is stabilized by base-stacking interactions with both the −1 (upstream) and +1 (downstream) base pairs. The planar ring system of Topotecan mimics a DNA base pair and occupies the same space as the +1 base pair in the structure without drug bound. Specifically, the long axis of the Topotecan molecule is parallel with the axis of base pairing, which differs from published computer models of CPT binding, which invoke only partial intercalative binding perpendicular to the long axes of the −1 and +1 bases (13, 26, 27).
The intercalation binding pocket is created by simple rotation of the phosphodiester bond (0P) of the uncleaved strand at the site of topo I-mediated cleavage and does not require rotation of a DNA base out of the helix or breakage of any hydrogen bonds between DNA base pairs as previously hypothesized (13, 26). The observed rotation does require unstacking of the +1 and −1 base pairs, which may be energetically promoted by the concomitant movement of the 0P phosphodiester into a binding pocket, where it is stabilized by two additional hydrogen bond interactions with the main-chain nitrogens of Arg-362 and Gly-363.
The intercalation binding mode explains how CPTs specifically block DNA religation, as the binding/intercalation results in a 3.6 Å shift of the downstream base pair and displaces the reactive 5′-OH (or 5′SH in the structures presented) of the cleaved strand 8 Å further away from the phosphotyrosine. Because the Topotecan binding pocket is located within the DNA substrate and can form only after the first transesterification, the structures explain why CPTs target the enzyme–substrate complex and not the enzyme alone (7). It should be noted that at concentrations of 10–20 μM, Topotecan has been shown to bind directly to duplex DNA (28, 29), and this binding is likely to be of an intercalative nature (30). Our crystal structure of the ternary complex demonstrates that Topotecan can mimic a DNA base pair, and this property may partially explain why at relatively high concentrations the drug can intercalate into duplex DNA in the absence of enzyme. It has also been shown that CPT can preferentially stabilize a covalent complex that has a G:C base pair at the +1 position (31). Our ternary complex structure was prepared with a G:C base pair in the +1 position, but we do not see any specific interactions that explain the reported preference for this base pair. Additional ternary complex structures with other base pairs in the +1 position are needed to address this issue in a comparative manner.
Previous functional analyses have demonstrated that a large number of modifications can be placed at positions 7, 9, and 10 of CPT (32), and in some cases these modifications can increase in vivo activity without changing in vitro activity. The structural model demonstrates that these positions face into the major groove of the DNA, and modifications that improve solubility, stability, etc. would not sterically interfere with drug binding. The structure model also predicts that there is only one direct hydrogen bond between the enzyme and Topotecan; Asp-533 hydrogen bonds to the 20(S)-hydroxyl. The importance of the 20(S)-hydroxyl in stabilizing Topotecan is supported by a number of structure/activity studies. For example, the 20(R) stereoisomer of CPT is inactive in vitro (33, 34). According to the model, the 20(R)-hydroxyl would not be able to contact Asp-533, and the 20(R)-ethyl group would create a steric clash with Asp-533 and Lys-532. In addition, substitution of the 20(S)-hydroxyl for hydrogen eliminates activity (35, 36); however, substitution of chlorine or bromine for the 20(S)-hydroxyl only partially eliminates activity (36). The structure model explains these results because the halogen substitutions, unlike the 20(S)-deoxy substitution, would eliminate the 20(S)-hydroxyl:Asp-533 interaction but still allow E-ring opening and would therefore allow water-bridged contacts of the 22-hydroxyl to Asn-722 and a 21-carboxylate oxygen to the P-Tyr-723 in the carboxylate model (Fig. 3B). In addition, the 21-carboxylate oxygens would be positioned for ionic interactions with Lys-532.
Although the ternary crystal structure cannot be used to calculate the relative affinities of open carboxylate vs. closed lactone forms of Topotecan, the observed ternary complex structure demonstrates that both the closed lactone ring and open carboxylate conformers can bind within the same intercalation pocket and presumably prevent religation. This conclusion is surprising, because it is generally believed that only the lactone form is active. We cannot rule out the possibility that the high concentrations of Topotecan that are present in the crystallization drops (500 μM) allow one form of the drug to bind that would not bind under conditions normally used to measure inhibition of DNA relaxation (1–200 μM) or DNA religation (0.1–10 μM) in vitro. However, a similar ratio of carboxylate:lactone was observed in the precipitant solution used to grow the ternary complex crystals, suggesting that at high concentrations of drug there is no preferential binding of either form to the covalent complex.
It should also be emphasized that the drug is always in equilibrium between open carboxylate and closed lactone forms. If one form is present in the solution, the other form will also be present. In fact, Hertzberg et al. (35) have demonstrated that addition of either the sodium salt carboxylate form or the lactone form of CPT induced accumulation of topo I–DNA covalent adducts in vitro (see, however, refs. 34 and 37). Because drug binding can only be assayed indirectly by assaying topo I-mediated cleavage and/or relaxation, the assays can only be done in a limited pH range. This limitation also makes it difficult to assay only one form of the drug.
Chemical modifications to the drug can shift the carboxylate/lactone equilibrium; however, these modifications may disrupt other interactions or introduce new interactions. For example, CPT analogs containing a lactam E-ring shift the equilibrium in favor of the closed E-ring form and are not active in vitro (35). The simplest interpretation of this result is that the lactone form is not active in vitro, but the inactivity of the lactam modification may also result from the fact that this modification converts a hydrogen bond acceptor (lactone, O) to a hydrogen bond donor (lactam, NH). We observe no potential contacts between the DNA or enzyme to this atom in the lactone form of our model, which argues that E-ring opening is important for CPT poisoning. CPT analogs containing a seven-membered β-hydroxylactone ring (homo-CPTs), instead of the naturally occurring six-membered α-hydroxylactone E-ring, are potent topo I poisons (38). Examination of the ternary complex demonstrates that there is adequate space within the binding pocket to accommodate the slightly larger E-ring and the critical 20(S)-hydroxyl–Asp-533 interaction could be maintained. Although the rate of hydrolysis of the seven-membered E-ring is slower (38), a fact that is often taken to suggest that the carboxylate form of CPT is inactive, the overall equilibrium of homo-CPT is actually shifted in favor of the open carboxylate form (39). Clearly, additional crystal structures of homo-CPT bound to the enzyme–substrate complex are necessary to directly address the binding mode of this CPT analog.
Several CPT resistance (CPTr) point mutations in human topo I have been identified, including Asp-533 (40, 41), Arg-364 (42, 43), Asn-722 (44), Phe-361 (45, 46), Gly-363 (47, 48), Ala-653 (48), Glu-418 (49), and Thr-729 (50). Our ternary complex crystal structure model helps to clarify the structural role of amino acid side chains that, if mutated, produce a CPT-resistant enzyme. For example, our model predicts that mutations at Asp-533, Arg-364, and Asn-722 will result in CPT resistance by removing important interactions that contribute directly to drug binding. The model also predicts that the CPTr amino acids Phe-361 and Gly-363 do not contact the drug directly but are important for creating/stabilizing the intercalation binding pocket. Comparison of the binary and ternary complexes demonstrates that the 0P phosphodiester must rotate into a binding pocket to allow opening of the DNA duplex. In support of the proposed model, substitutions at Phe-361 and Gly-363 would be expected to destabilize the 0P phosphodiester in the ternary complex and result in CPT resistance (42). The proposed structure model does not explain all mutations that affect CPT sensitivity. For example, mutations at positions Glu-418 (49), Ala-653 (48), and Thr-729 (50) in human topo I also lead to CPT resistance; however, these residues are too distant to contact the drug directly, and it is not obvious how they affect the structure or stability of the intercalation binding pocket. Clearly, the structure determination of mutant topoisomerases in the presence and absence of CPT(s) is necessary to understand how these mutations confer drug resistance. We cannot rule out the possibility that the drug may also bind at other sites in the complex; however, careful analysis of the electron density map of the ternary complex allowed visualization of a single PEG molecule (Mr = 194) that was introduced during cryoprotection of the crystals (Table 1). This result argues that if Topotecan (Mr = 422) was stably bound at another site, it would be apparent.
Mutations at Thr-718 act as CPT mimics and result in an increase in the steady-state concentration of covalent topo I–DNA adducts even in the absence of drug (48). The structure model suggests that this phenotype may result from greater mobility of the reactive 5′-OH (5′-SH in presented structures). The hydroxyl of Thr-718 makes a hydrogen bond contact to the nonbridging oxygen of the +1 phosphodiester in both the binary and ternary complexes, and this interaction may help position the 5′-OH for nucleophilic attack at the 3′-phosphotyrosine bond.
In addition to preventing DNA religation, high concentrations of CPT and Topotecan have been shown to inhibit DNA relaxation in vitro (51). It has previously been hypothesized that CPT inhibits DNA relaxation, not by prolonging the lifetime of the covalent complex but by somehow hindering DNA rotation (51). One interpretation of the differences observed between our binary (no drug bound) and ternary (drug bound) structures is that Topotecan binding stabilizes the covalent complex and prevents conformational changes that would normally allow relaxation. A related possibility may be that drug binding specifically interferes with rotation around the 0P phosphodiester bond during the relaxation process (17). The hydrogen bond contacts between the nonbridging oxygens of 0P and the main chain nitrogens of Gly-363 and Arg-362 and Lys-374 would restrain three (α, β, γ) of the five potentially rotatable backbone bonds (52). This tight packing arrangement could hinder the downstream DNA from rotating about 0P. It is important to note that the packing arrangement of 0P would not eliminate all possible DNA rotation, and DNA relaxation may proceed by other mechanisms. For example, rotation could still occur at the +2 (or +3, etc.) phosphodiester. However, additional base-pair hydrogen bond interactions would have to be broken to allow this rotation. Finally, it is important to note that higher concentrations of drug are required to inhibit DNA relaxation than to inhibit DNA religation. It is therefore possible that Topotecan is inhibiting DNA relaxation by binding to different and/or additional sites on the protein or DNA.
A caveat to the interpretation of all crystal structures is that the high concentrations of protein and ligand, packing interactions, etc. may lead to complexes that are not biologically relevant. We believe the validity of our model can be supported by its congruence with many of the known structure–activity relationships of Topotecan and related compounds. However, there are several points that must be considered in this analysis. First, the model may not explain the activity of certain CPT analogs in vivo. For example, modifications that change features of the drug (solubility, stability, nonspecific binding to serum, etc.) may result in more potent drugs, but these same modifications may not alter binding to the topo I–DNA complex. The structure model is therefore most useful for addressing structure–activity relationship data obtained in vitro. These data determined in vitro can also be hard to compare because it has not been possible to assign an equilibrium dissociation binding constant to any topoisomerase poison; in vitro assays rely upon catalytic events (DNA cleavage or DNA relaxation) that may happen before or after drug binding. Finally, it is important to remember that a single ternary complex structure can only represent one step in the reaction cycle that probably involves several different conformational intermediates.
In conclusion, the 2.1-Å structure of Topotecan bound to the human topo I–DNA covalent complex demonstrates that Topotecan is an uncompetitive inhibitor and binds to the complex by intercalating between DNA bases of both strands at the enzyme-induced nick. Intercalation at the enzyme-induced nick explains why the drug binds the enzyme–substrate complex more efficiently than enzyme or DNA alone, and provides an explanation for how these drugs cause the accumulation of cytotoxic topoisomerase–DNA adducts. Topotecan binding is stabilized by stacking interactions with the DNA, a hydrogen bond contact with Asp-533, and water-bridged contacts to the active site phosphotyrosine and Asn-722. Residues that contribute to direct contacts with the drug (Asp-533, Asn-722, and Arg-364) result in CPT resistance when mutated. In addition, the model reveals that another class of CPT-resistant mutants results from disruption of interactions that are important for stabilizing the intercalation by contributing to a binding pocket for the 0P of the intact strand (Phe-361, Arg-362, Gly-363). Close examination of the ternary complex also indicates that the bound drug exists in both the closed lactone and open carboxylate forms. Clearly, additional structures will be needed to understand how other CPT analogs and other non-CPT topo I poisons mediate their inhibitory effects.
Acknowledgments
We thank G. Ireton, E. Wallace, D. Connor, and A. Raymond for technical assistance; H. Kim and M. A. Bjornsti for intellectual support and review of this manuscript; S. Wasserman at COM-CAT (Sector 32) Advanced Photon Source, Argonne National Laboratory for assistance in data collection; and L. Berman and the staff of X25 at the National Synchrotron Light Source, Brookhaven National Laboratory, for assistance in data collection and helpful support. This work was supported by National Cancer Institute Grants R43 CA82964 and R43 CA79439; by the Pacific West Cancer Fund; and by National Institute for General Medicine Grant GM58598.
Abbreviations
PEG, polyethylene glycol
CPT, camptothecin
topo I, DNA topoisomerase I
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes and ).
References
- 1.Wang J. C. (1996) Annu. Rev. Biochem. 65 635-692. [DOI] [PubMed] [Google Scholar]
- 2.Champoux J. J. (2001) Annu. Rev. Biochem. 70 369-413. [DOI] [PubMed] [Google Scholar]
- 3.Pommier Y. (1998) Biochimie 80 255-270. [DOI] [PubMed] [Google Scholar]
- 4.Wall M. E., Wani, M. C., Cook, C. E., Palmer, K. H., McPhail, A. T. & Sim, G. A. (1966) J. Am. Chem. Soc. 88 3888-3890. [Google Scholar]
- 5.Hsiang Y. H., Hertzberg, R., Hecht, S. & Liu, L. F. (1985) J. Biol. Chem. 260 14873-14878. [PubMed] [Google Scholar]
- 6.Nitiss J. L. & Wang, J. C. (1988) Proc. Natl. Acad. Sci. USA 85 7501-7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hertzberg R. P., Caranfa, M. J. & Hecht, S. M. (1989) Biochemistry 28 4629-4638. [DOI] [PubMed] [Google Scholar]
- 8.Chen A. Y. & Liu, L. F. (1994) Rev. Pharmacol. Toxicol. 34 191-218. [DOI] [PubMed] [Google Scholar]
- 9.Bjornsti M. A., Benedetti, P., Viglianti, G. A. & Wang, J. C. (1989) Cancer Res. 49 6318-6323. [PubMed] [Google Scholar]
- 10.Li T. K. & Liu, L. F. (2001) Annu. Rev. Pharmacol. Toxicol. 41 53-77. [DOI] [PubMed] [Google Scholar]
- 11.Horwitz S. B., Chang, C. K. & Grollman, A. P. (1971) Mol. Pharmacol. 7 632-644. [PubMed] [Google Scholar]
- 12.Burgin A. B., Huizenga, B. N. & Nash, H. A. (1995) Nucleic Acids Res. 23 2973-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redinbo M. R., Stewart, L., Kuhn, P., Champoux, J. J. & Hol, W. G. J. (1998) Science 279 1504-1513. [DOI] [PubMed] [Google Scholar]
- 14.Stewart L., Ireton, G. C., Parker, L. H., Madden, K. R. & Champoux, J. J. (1996) J. Biol. Chem. 271 7593-7601. [DOI] [PubMed] [Google Scholar]
- 15.Stewart L., Clark, R. & Behnke, C. (2002) Drug Discovery Today 7 187-196. [DOI] [PubMed] [Google Scholar]
- 16.Navaza J. (1994) Acta Crystallogr. A 50 157-163. [Google Scholar]
- 17.Stewart L., Redinbo, M. R., Qiu, X., Hol, W. G. J. & Champoux, J. J. (1998) Science 729 1534-1541. [DOI] [PubMed] [Google Scholar]
- 18.Brünger A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54 905-921. [DOI] [PubMed] [Google Scholar]
- 19.McRee D. E. (1999) J. Struct. Biol. 125 156-165. [DOI] [PubMed] [Google Scholar]
- 20.Lu X.-J., Shakked, Z. & Olson, W. K. (2000) J. Mol. Biol. 300 819-840. [DOI] [PubMed] [Google Scholar]
- 21.Olson W. K., Bansal, M., Burley, S. K., Dickerson, R. E., Gerstein, M., Harvey, S. C., Heinemann, U., Lu, X. J., Neidle, S., Shakked, Z., et al. (2001) J. Mol. Biol. 313 229-237. [DOI] [PubMed] [Google Scholar]
- 22.Redinbo M. R., Stewart, L., Champoux, J. J. & Hol, W. G. (1999) J. Mol. Biol. 292 685-696. [DOI] [PubMed] [Google Scholar]
- 23.Krogh B. O. & Shuman, S. (2000) Mol. Cell 5 1034-1041. [DOI] [PubMed] [Google Scholar]
- 24.Sheldrick G. M. & Schneider, T. R. (1997) Methods Enzymol. 277 319-343. [PubMed] [Google Scholar]
- 25.Chourpa I., Millot, J.-M., Sockalingum, G. D., Riou, J.-F. & Manfait, M. (1998) Biochim. Biophys. Acta 1379 353-366. [DOI] [PubMed] [Google Scholar]
- 26.Laco G. S., Collins, J. R., Luke, B. T., Kroth, H., Sayer, J. M., Jerina, D. M. & Pommier, Y. (2002) Biochemistry 41 1428-1435. [DOI] [PubMed] [Google Scholar]
- 27.Kerrigan J. E. & Pilch, D. S. (2001) Biochemistry 40 9792-9798. [DOI] [PubMed] [Google Scholar]
- 28.Yang D., Strode, J. T., Spielmann, H. P., Wang, A. H.-J. & Burke, T. G. (1998) J. Am. Chem. Soc. 120 2979-2980. [Google Scholar]
- 29.Yao S., Murali, D., Seetharamulu, P., Haridas, K., Petluru, P. N., Reddy, D. G. & Hausheer, F. H. (1998) Cancer Res. 58 3782-3786. [PubMed] [Google Scholar]
- 30.Miller S. E. & Pilch, D. S. (2000) Ann. N.Y. Acad. Sci. 922 309-313. [DOI] [PubMed] [Google Scholar]
- 31.Champoux J. J. & Aronoff, R. (1989) J. Biol. Chem. 264 1010-1015. [PubMed] [Google Scholar]
- 32.Kehrer D. F. S., Soepenberg, O., Loows, W. J., Verweij, J. & Sparreboom, A. (2001) Anti-Cancer Drugs 12 89-105. [DOI] [PubMed] [Google Scholar]
- 33.Jaxel C., Kohn, K. W., Wani, M. C., Wall, M. E. & Pommier, Y. (1989) Cancer Res. 49 1465-1469. [PubMed] [Google Scholar]
- 34.Hsiang Y. H., Liu, L. F., Wall, M. E., Wani, M. C., Nicholas, A. W., Manikumar, G., Kirschenbaum, S., Silber, R. & Potmesil, M. (1989) Cancer Res. 49 4385-4389. [PubMed] [Google Scholar]
- 35.Hertzberg R. P., Caranfa, M. J., Holden, K. G., Jakas, D. R., Gallagher, G., Mattern, M. R., Mong, S. M., Bartus, J. O., Johnson, R. K. & Kingsbury, W. D. (1989) J. Med. Chem. 32 715-720. [DOI] [PubMed] [Google Scholar]
- 36.Wang X., Zhou, X. & Hecht, S. M. (1999) Biochemistry 38 4374-4381. [DOI] [PubMed] [Google Scholar]
- 37.Potmesil M. (1994) Cancer Res. 54 1431-1439. [PubMed] [Google Scholar]
- 38.Lesuer-Ginot L., Demarquay, D., Kiss, R., Kasprzyk, P. G., Dassonneville, L., Bailly, C., Camara, J., Lavergne, O. & Bigg, C. H. (1999) Cancer Res. 59 2939-2943. [PubMed] [Google Scholar]
- 39.Chauvier D., Chourpa, I., Bigg, D. C. H. & Manfait, M. (2000) Ann. N.Y. Acad. Sci. 922 314-316. [DOI] [PubMed] [Google Scholar]
- 40.Saleem A., Ibrahim, N., Patel, M., Li, X. G., Gupta, E., Mendoza, J., Pantazis, P. & Rubin, E. H. (1997) Cancer Res. 57 5100-5106. [PubMed] [Google Scholar]
- 41.Tamura H., Kohchi, C., Yamada, R., Ikeda, T., Koiwai, O., Patterson, E., Keene, J. D., Okada, K., Kjeldsen, E., Nishikawa, K., et al. (1991) Nucleic Acids Res. 19 69-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X. G., Haluska, P., Jr., Hsiang, Y. H., Bharti, A. K., Kufe, D. W., Liu, L. F. & Rubin, E. H. (1997) Biochem. Pharmacol. 53 1019-1027. [DOI] [PubMed] [Google Scholar]
- 43.Urasaki Y., Laco, G. S., Pourquier, P., Takebayashi, Y., Kohlhagen, G., Gioffre, C., Zhang, H., Chatterjee, D., Pantazis, P. & Pommier, Y. (2001) Cancer Res. 61 1964-1969. [PubMed] [Google Scholar]
- 44.Fujimori A., Harker, W. G., Kohlhagen, G., Hoki, Y. & Pommier, Y. (1995) Cancer Res. 55 1339-1346. [PubMed] [Google Scholar]
- 45.Bailly C., Carrasco, C., Hamy, F., Vezin, H., Prudhomme, M., Saleem, A. & Rubin, E. (1999) Biochemistry 38 8605-8611. [DOI] [PubMed] [Google Scholar]
- 46.Rubin E., Pantazis, P., Bharti, A., Toppmeyer, D., Giovanella, B. & Kufe, D. (1994) J. Biol. Chem. 269 2433-2439. [PubMed] [Google Scholar]
- 47.Benedetti P., Fiorani, L., Capuani, L. & Wang, J. C. (1993) Cancer Res. 53 4343-4348. [PubMed] [Google Scholar]
- 48.Fiorani P., Amatruda, J. F., Silvestri, A., Butler, R. H., Bjornsti, M. A. & Benedetti, P. (1999) Mol. Pharmacol. 56 1105-1115. [DOI] [PubMed] [Google Scholar]
- 49.Chang J. Y., Liu, J. F., Juang, S. H., Liu, T. W. & Chen, L. T. (2002) Cancer Res. 62 3716-3721. [PubMed] [Google Scholar]
- 50.Kubota N., Kanzawa, F., Nishio, K., Takeda, Y., Ohmori, T., Fujiwara, Y., Terashima, Y. & Saijo, N. (1992) Biochem. Biophys. Res. Commun. 188 571-577. [DOI] [PubMed] [Google Scholar]
- 51.Champoux J. J. (2000) Ann. N.Y. Acad. Sci. 922 56-64. [DOI] [PubMed] [Google Scholar]
- 52.Saenger W., (1983) Principles of Nucleic Acid Structure (Springer, New York).