Abstract
Isolated newt (Notophthalmus viridescens) chromosomes were studied by using micromechanical force measurement during nuclease digestion. Micrococcal nuclease and short-recognition-sequence blunt-cutting restriction enzymes first remove the native elastic response of, and then to go on to completely disintegrate, single metaphase newt chromosomes. These experiments rule out the possibility that the mitotic chromosome is based on a mechanically contiguous internal non-DNA (e.g., protein) “scaffold”; instead, the mechanical integrity of the metaphase chromosome is due to chromatin itself. Blunt-cutting restriction enzymes with longer recognition sequences only partially disassemble mitotic chromosomes and indicate that chromatin in metaphase chromosomes is constrained by isolated chromatin-crosslinking elements spaced by ≈15 kb.
The question of how the millimeter-long chromatin fibers in eukaryote chromosomes are folded up into compacted metaphase chromatid remains open (1, 2). It has been proposed that metaphase chromatin is organized into loops attached to an underlying protein-rich structure called the metaphase scaffold (3–5). This scaffold has been visualized in the electron microscope by histone-depleting chromosomes; the resulting micrographs show ≈40-kb DNA loops dangling from a protein-rich aggregate, which is the shape of the original chromosome (3). Isolated scaffolds were shown to contain nonhistone proteins (5) including topoisomerase II (6) and structural maintenance of chromosomes (7). Further studies indicated that the metaphase scaffold is helically folded (8) and contains AT-rich DNA sequences (9, 10). The basic “scaffold-loop” model suggested by these studies is widely accepted (11–14).
A fundamental question is whether the nonhistone proteins of the scaffold are connected. Early studies stated that the scaffold was responsible for the basic shape of metaphase chromosomes and was essentially a fibrous network of nonhistone proteins (3), which could be isolated as a structurally independent stable entity (5). However, later discussions of this issue suggest that the question of whether the native scaffold is stabilized through protein–protein interactions is unresolved (6, 15). Furthermore, an old literature of whole-chromosome digestion experiments (16) appears inconsistent with a contiguous protein scaffold.
The alternative is that the proteins of the scaffold are not contiguously connected through the chromosome, and that DNA itself links the scaffold together. Here we report micromanipulation experiments that establish the non-DNA content of the mitotic chromosome to be mechanically disconnected, and therefore that mitotic chromosomes are best thought of as a chromatin network. Our experimental approach uses chromosome elastic response to report on structure and structural changes. We have recently developed techniques to carry out micromechanical experiments on isolated individual mitotic chromosomes (17, 18) using methods similar to those used in Nicklas' classic study of meiotic metaphase chromosome physical properties (19).
Observations that mitotic chromosomes can be stretched to five times their native length without permanent elongation (17–19) suggested to us that the folded chromatin must be organized into a crosslinked chromatin network. To directly test this hypothesis, we carried out mechanical measurements on chromosomes during in situ digestion using DNA-cutting nuclease and restriction nucleases. Our aim was to monitor how chromosome elasticity and connectivity change dynamically as DNA cuts are made.
Our result is that, as DNA cuts are made within a mitotic chromosome by either micrococcal nuclease (MN) or frequently cutting restriction enzymes (REs), first chromosome elasticity is lost, and then the chromosome is completely dissolved. REs with higher specificity only partially reduce chromosome elasticity, showing that DNA and therefore chromatin are the mechanically contiguous components of mitotic chromosome structure and ruling out the possibility that the chromatin is organized by being tethered to a mechanically contiguous internal protein scaffold. Instead, we conclude that a mitotic chromosome is essentially a “network” of chromatin, with crosslinks about every 15 kb.
Materials and Methods
Chromosome Micromanipulation.
Micromechanical experiments were done with an inverted microscope (IX-70, 60X, 1.4 NA, Olympus, New Hyde Park, NY) equipped with two micromanipulators (MP-285, Sutter Instruments, Novato, CA) and a charge-coupled device camera controlled by a computer with labview and imaq (National Instruments, Austin, TX). Newt cells were grown in laboratory-made dishes designed to allow micropipettes to extract mitotic chromosomes [TVI cell line (20)].
After extraction, chromosomes were suspended between two micropipettes, one of which was used as a force transducer (17–19). This force-measuring micropipette was pulled and cut to have a bending force constant of ≈0.1 nN/μm (1 nN = 10−9 N) and then mounted directly to the microscope stage to minimize mechanical noise. Force-extension experiments were done by moving the stiffer pipette at rates of ≈0.01 μm/sec, while observing the bending of the force-measuring pipette. After each experiment, the force-measuring pipette was calibrated as described in ref. 18.
Previous experiments have shown that forces in the nanonewton range generate reversible stretching of mitotic chromosomes (17–19, 21). The newt mitotic spindle is capable of generating nanonewton forces, and chromatids are often observed to be appreciably stretched during mitosis, particularly during anaphase. Therefore, the nanonewton forces used are comparable to forces applied to chromosomes in vivo.
All experiments were performed in the extracellular buffer, ≈50 μm above the cover glass on which the cells were cultured. The conditions for our experiments are therefore those of the extracellular medium (pH 7.5, ≈100 mM net univalent salt, mainly Na+ and K+). A number of results indicate there is no appreciable restructuring of chromatin during or shortly after chromosome extraction into extracellular buffer. First, elasticity of chromosomes in the extracellular medium (17, 18) is close to that measured in vivo (19, 22, 23). Second, we observe no morphological change of chromosomes during extraction from cytoplasm to extracellular buffer (17–18). Third, we observe no gradual changes of physical properties of mitotic chromosomes from ≈20-sec to 6-h exposure to extracellular buffer (likely due to the similarity of our extracellular buffer to solution conditions widely used to study chromatin structure). We therefore consider the extracted chromosomes to have essentially the same internal chromatin structure at the beginning of our experiments as they had inside the cell.
Combined Enzymatic–Micromechanical Measurements.
Once a single mitotic chromosome was isolated and held between two pipettes, the native elastic response of a chromosome was measured by observation of deflection of the force-measuring pipette while the other pipette is moved. Then, one of two types of experiments was done with an enzyme that was microinjected from a third 3-μm-diameter micropipette ≈10 μm from the chromosome (Fig. 1a). In the first type of experiment, some tension is applied to the chromosome before exposing it to a DNA-cutting enzyme. Then, the enzyme is sprayed onto the chromosome while monitoring the tension (Fig. 1). Digital phase-contrast images were acquired at 10 frames/sec before, during, and after each enzyme exposure, recording pipette positions (therefore force and extension) and chromosome morphology. In the second type of experiment, the chromosome is sprayed with an enzyme with no applied tension. Then the force response of the chromosome is measured in the absence of any enzyme (Fig. 2).
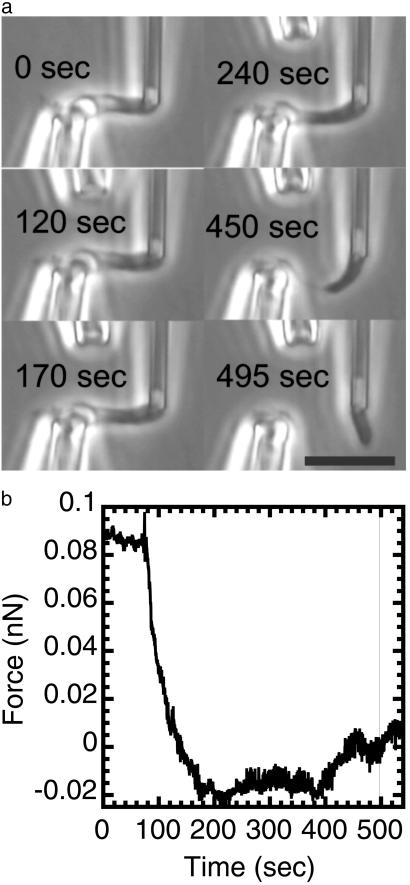
Fig 1.
Mechanical response of a newt mitotic chromosome microdigested with 1 nM MN in 60% PBS with 1 mM CaCl2. The chromosome was put under 0.1 nN of force before microdigestion. MN causes a relaxation of the initially applied force before any apparent change in chromosome morphology in phase contrast microscopy. Longer exposures lead to dissolution of the chromosome into unobservably small fragments, showing that a large-scale protein scaffold does not exist within mitotic chromosomes. (a) Phase images of the chromosome being digested by MN. The time in each image corresponds to the time axis of b. (Bar = 10 μm.) A video is available at www.uic.edu/∼jmarko/published/enz. (b) Time series of the force supported by the chromosome during the nuclease digestion. The thin vertical line indicates the time at which the chromosome was severed. Digestion was initiated at t = 60 sec.
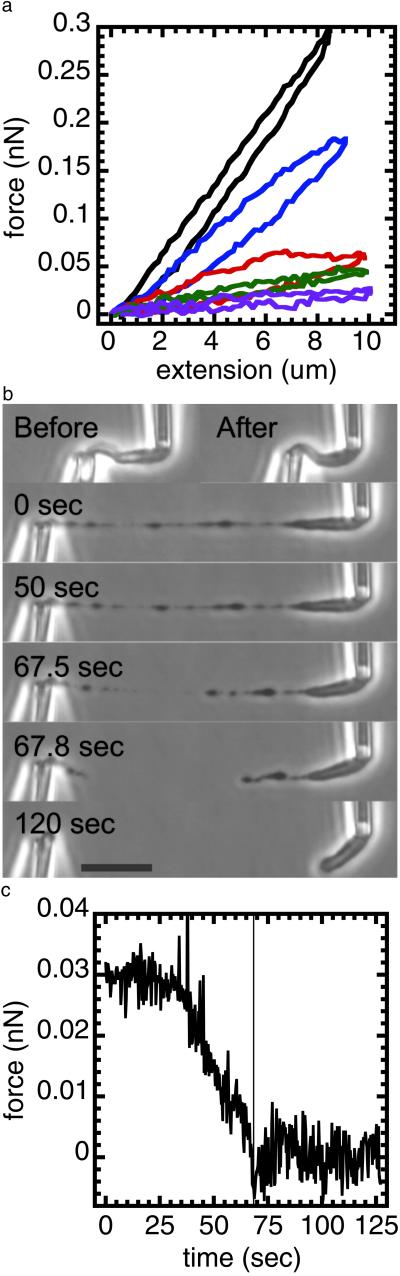
Fig 2.
Partial digestion of a mitotic chromosome by MN shows it to behave as a chromatin network. A mitotic newt chromosome was digested with 10 nM MN for 90 sec without an applied tension. It was then subjected to four extension–retraction cycles in the absence of MN. The chromosome force constant was reduced after each extension–retraction cycle. After this, the mildly digested chromosome was extended to 40 μm. The chromosome does not elongate homogeneously; instead, there are blobs connected by thin fibers. To test whether the thin fibers contain DNA, we exposed the extended blob-thin fiber structure to 10 nM MN while monitoring the force. The force relaxes in response to the exposure and the blob-thin fiber structure is cut through, indicating that the thin fibers contain DNA and that it is required to support the applied tension. (a) Force–extension response of a chromosome before (black) and after a 90-sec chromosome digestion with 10 nM MN. After digestion (without an applied tension), successive extension–relaxation cycles (blue, red, green, and purple) progressively reduce the force response; there is no longer reversible elasticity. (b) Top images show the chromosome unextended before (Left) and after (Right) 90-sec MN microdigestion; there is no obvious change in morphology under zero force. Lower images labeled with a time show the final cutting of the chromosome extended to 40 μm, after the microdigestion and the extension–relaxation cycles of a. The t = 0 image shows the blob-link structure produced by microdigestion and stretching, whereas the t > 0 images show that the chromosome is completely cut by spraying with MN. (Bar = 10 μm.) (c) Time series of the digestion experiment of b. The chromosome was extended by ≈40 μm and supported a force of ≈30 pN. The microdigestion began at ≈40 sec. The force relaxes to zero as the chromosome is cut. The low levels of force in this experiment are insufficient to break a single DNA molecule. A video is available at www.uic.edu/∼jmarko/published/enz.
Spraying was controlled with a microinjection controller (World Precision Instruments, Sarasota, FL); all experiments used between 1- and 20-min sprays driven by 1,000-Pa pressure. When spraying is stopped, diffusion rapidly dissipates (≈0.1 sec; see ref. 24) the sprayed reagent, abruptly stopping the reaction. In separate experiments with fluorescent dyes, we checked that the enzyme concentration at the chromosomes was within a factor of two of that inside the pipette (data not shown). Enzyme experiments were done with either small forces <1 nN or with zero force initially applied to the chromosome.
DNA-Cutting Enzymes.
MN and type II REs were used to induce cuts in double-stranded DNA. MN was prepared at 1–10 nM in 60% PBS with 1 mM CaCl2. The REs used were AluI (AG↓CT, Promega); HaeIII (GG↓CC, Roche); Cac8I (GCN↓NGC, New England Biolabs; N denotes “any base”); HincII [GT(T/C)↓ (A/G)AC, Promega; T/C denotes either T or C]; HindII [GT (T/C)↓(A/G)AC, Roche Diagnostics, Indianapolis, IN], DraI (TTT↓AAA, Promega), StuI (AGG↓CCT, New England Biolabs); and PvuII (CAG↓CTG, Promega), all of which produce blunt DNA cuts. REs were prepared at a concentration of 0.4–1.2 units/μl in appropriate reaction buffers (in each case, Tris⋅HCl, pH 7.5–8/50–100 mM NaCl/5–10 mM MgCl2). Because a relatively high RE concentration at room temperature in the chromosome experiments was used, each preparation of enzyme was tested by digestion of either 20 ng/μl pBR322 (Promega) or 10 ng/μl λ-DNA (Promega) at 25°C for 15, 30, and 60 min. Electrophoresis gel analysis showed that in each case, digestion was complete after 30 min, with no excess cutting or “star activity” (data not shown).
Results
MN Digestion of Mitotic Chromosomes.
To investigate how MN affects chromosome elasticity and therefore structure, an extracted chromosome held by two micropipettes was elongated and retracted three times to determine its native elastic response. The chromosome was then extended so that that it supported ≈0.1 nN. Before the spray of MN is initiated, the chromosome continuously supports this tension. After 60 sec (t = 60 sec; Fig. 1b), the MN spray (1 nM) begins; the force supported by an elongated mitotic chromosome drops and is reduced to zero by MN after ≈60 sec of spraying (t = 120 sec; Fig. 1b), indicating reduction of elastic modulus to nearly zero. This softening occurs before any apparent morphological change (Fig. 1a). After another 60 sec of exposure (t = 180 sec), the chromosome “thins,” and after 500 sec, it is severed and subsequently completely dissolved. Similar results were found for four separate chromosomes with initial tensions ranging between 0.1 and 1 nN, and MN concentrations varied between 1 and 10 nM.
To further probe the structural change during the initial digestion by MN, a second type of experiment was done where an isolated chromosome with no applied tension was exposed to 10 nM MN for 90 sec. After this amount of digestion, no morphological change was observed. Then, the chromosome was repeatedly extended and retracted with no further MN spraying. Before digestion, a chromosome can be repeatedly extended and retracted without any change in its elastic response (18). By contrast, after this 90-sec MN digestion, repeated extension–retraction cycles are no longer reversible, with the force needed to double chromosome length dropping with each successive extension–retraction cycle (Fig. 2a). Also, the chromosome no longer extends homogeneously: instead, relatively dense domains connected by thin fibers appear for >40-μm extensions (Fig. 2b; t = 0 sec). To test whether the thin fibers contain DNA, they were extended and then sprayed with 10 nM MN. Immediately, the force relaxes and the thin fiber is cut (Fig. 2 b and c), indicating that its contiguous structural element is DNA and not a network of protein.
RE Digestion of Mitotic Chromosomes.
Experiments with blunt-cutting REs were done to estimate how often double-stranded cuts need to be made to disconnect the mitotic chromosome (REs cleave DNA at specific sequence and therefore with a given statistical frequency). Chromosomes were extended to ≈1.5 times native length (force ≈0.5 nN) and then were sprayed with a RE in appropriate reaction buffer. REs with either four- or six-base recognition sequences were used. The six-base REs varied in the number of six-base sequences recognized, which in turn varied the cut frequency.
Experiments with AluI produce results similar to MN, with force dropping to zero after 30 sec of spraying, and chromosome disintegration occurring after 200 sec of spraying (Fig. 3). HaeIII gives the same result (data not shown). These enzymes cut bare random-sequence DNA on average once every 44 = 256 bases or with a frequency 1/256 that of MN and still lead to complete disintegration of the chromosome. During AluI digestion, we do not observe any chromatin fragments.
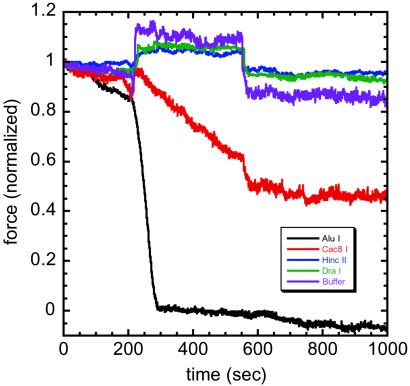
Fig 3.
REs with increasing specificity show decreasing effects on chromosome elastic response. Force data are before, during, and after 350-sec exposures to various REs; force is normalized to units of initial applied force, which ranged between 0.2 and 0.8 nN in the five separate experiments shown. AluI AG↓CT (black) relaxes the force in ≈30 sec; Cac8I GCN↓NGC (red) only partially reduces the force. HincII GT(T/C)↓(A/G)AC (blue) and DraI TTT↓AAA (green) induce an increase in force during spraying, with a return to the original force when spraying stops (≈600 sec), similar to spraying with reaction buffer and no enzyme (violet). These results indicate that chromatin–chromatin crosslinks occur roughly every 15 kb (see text). A video of AluI digestion is available at www.uic.edu/∼jmarko/published/enz.
Experiments with Cac8I were done to examine the effect of size of the recognition sequence length on enzyme activity. Cac8I's recognition sequence has the same statistical frequency on random-sequence DNA (1/44 = 1/256) as those of AluI and HaeIII but is spread over a six-base footprint. Cac8I exposure only partially reduces the applied force (Fig. 3). After spraying for 60 min, the force constant converges to 40% of its native value (data not shown). The rate of force reduction by Cac8I is 1/10 that of AluI and HaeIII, indicating that the increase in recognition sequence size has reduced accessibility by ≈10 times.
REs with high specificity do not reduce chromosome elasticity. HincII (Fig. 3) and HindII (data not shown), which cut random DNA once every 45 = 1,024 bases and one-fourth that of Cac8I, do not reduce chromosome elasticity. Experiments with DraI (TTT↓AAA, Promega, Fig. 3), StuI (AGG↓CCT, New England Biolabs; data not shown), and PvuII (CAG↓CTG, Promega; data not shown), which cut random DNA once every 46 = 4,096 bases, also do not produce observable force reduction.
The force increase observed during spraying with the less effective cutters (“step” response; Fig. 3) is due to the reaction buffers' divalent ions (6–10 mM Mg2+), which weakly and reversibly hypercondense mitotic chromosomes (24). Fig. 3 includes a force trace for reaction buffer with no enzyme, indicating that after spraying is complete, chromosome elastic response returns to its native value. The irreversible changes are therefore due to enzyme and not a buffer effect.
Discussion
There Is No Contiguous Protein Scaffold Within Mitotic Chromosomes.
MN and 4-bp blunt REs eliminate a chromosome's ability to support a force. The reduction in elastic modulus occurs before any apparent morphological change occurs; long exposures “dissolve” the mitotic chromosome into unobservably small fragments. This result indicates that the mechanical integrity of the mitotic chromosome comes from DNA itself and shows there is no contiguous protein structure to which chromatin “loops” are merely tethered. Any protein “scaffold” should be made of relatively small isolated elements, connected together by chromatin (Fig. 4).
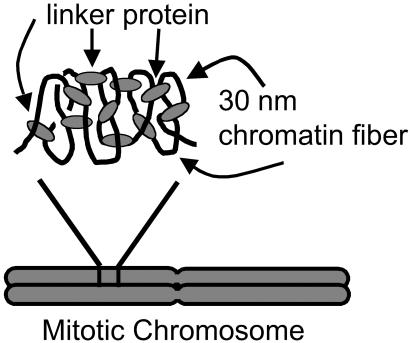
Fig 4.
Proposed “network” model of higher mitotic chromosome structure. The black lines represent 30-nm chromatin fiber, and the gray ovals represent proteins connecting the chromatin fiber to form a network-type structure. This model has a structure where the proteins crosslink chromatin, which maintains higher chromosome structure. When this structure is exposed to MN, cuts in the DNA (chromatin) between the crosslinks will be induced. The chromosome will no longer support an applied tension, which is what is observed experimentally. In addition, we estimate the average number of base pairs between crosslinks to be ≈15 kb, based on the results from digesting the mitotic chromosome with various REs. Note that the crosslinks need not be homogeneously distributed through the chromatids.
This “network” picture is supported by experiments where short exposures to MN are made that do not disrupt chromosome morphology. Extension then leads the chromosome to break up into chromatin islands, attached by thin fibers (Fig. 2b). These thin fibers can be cut by MN. This result, plus the greatly reduced elastic response (Fig. 2a) and the weaker effects of more specific REs (Fig. 3), are all consistent with a network whose interconnects are randomly cut (25). In this proposed native state, chromosome elastic response is due to the self-adhesion and elastic response of the crosslinked folded chromatin [note that destabilizing chromatin–chromatin interactions reversibly decondenses whole chromosomes (16, 24, 26)], combined with the elastic response of the protein crosslinks themselves.
One might imagine that part of the internal protein structure of the chromosome is lost as a result of extraction, either as a result of dissociation under dilute conditions or as a consequence of a lack of energy sources or other cofactors not present in the extracellular medium. However, such “lost” protein structures do not contribute significantly to the mechanical properties of mitotic chromosomes, because their bending and stretching properties in vivo are similar to those measured in the extracellular medium (23). Repetition of the experiments of this paper in mitotic cell extracts may be able to further address this question.
Chromatin in Mitotic Chromosomes Is Crosslinked Roughly Every 15 kb.
MN and REs with a 4-bp recognition sequence lead to compete disintegration of the chromosome. Four-base-pair REs cut bare random-sequence DNA on average once every 44 = 256 bases or with a frequency 1/256 that of MN. Because the access of REs to DNA is reduced in chromatin by ≈10-fold (27, 28), we conclude that roughly one cut every 2.5 kb (12 nucleosomes) is sufficient to completely disassemble a mitotic chromosome.
REs with higher specificities were used to reduce the number of DNA cuts made in a mitotic chromosome, using gradually rarer recognition sequences. A complication is that longer recognition sequences will be less accessible along the short internucleosomal linker DNAs in chromatin. To control for this effect, Cac8I was used. This enzyme recognizes a 6-bp recognition sequence but has 16-fold degeneracy and therefore cuts with the same statistical frequency as AluI and HaeIII. The rate of force reduction by Cac8I is 1/10 that of AluI and HaeIII, indicating that the 2-bp increase in recognition sequence size reduces accessibility by ≈10 times. We therefore estimate that Cac8I can cut metaphase chromatin about once every 25 kb. For a network architecture, reduction in force constant to 40% of native requires cutting of 60% of the links. On the basis of the estimate that Cac8I cuts once per 25 kb, we conclude that the length of chromatin between crosslinks in the mitotic chromosome is ≈15 kb. We emphasize that this estimate is approximate and that it may be affected by a number of factors, including kinetics of RE access to DNA.
We therefore expect that six-base blunt-cutting enzymes (i.e., with the same DNA access as Cac8I) with lower cutting frequencies will produce proportionally smaller force relaxations. Given our force resolution of ≈0.05 nN, no observable change is expected. This is the experimental result observed for all of the higher specificity enzymes we used: HincII (Fig. 3), HindII (data not shown), DraI (Fig. 3), StuI (data not shown), and PvuII (data not shown).
The large difference in activity of Cac8I and AluI is somewhat surprising, because the recognition sequences differ by only 2 bp in length, a small fraction of the 20–40 bp between nucleosomes. It is possible that shape and size of these enzymes play significant roles. Alternately, only subregions of internucleosomal linker DNA may be accessible between the footprints of other DNA-bound proteins such as topoisomerases and SMCs (structural maintenance of chromosome proteins).
Thus, sufficiently frequent DNA cuts in a mitotic chromosome completely disassemble it, proving that a contiguous protein scaffold does not exist within mitotic chromosomes. Instead, our results indicate that DNA itself provides the mechanical integrity of chromosomes, and that mitotic chromosomes can be considered to be crosslinked networks of chromatin (Fig. 4). Our result that chromatin is crosslinked every ≈15 kb is comparable to the sizes of DNA fragments obtained from classical chromatin “loop size” analyses (6). Our experiments reveal that the classical loops are better thought of as network links, because in the mitotic state, the non-DNA portions of the loop “bases” are not solidly connected to one another. It should be noted that if the construct of Fig. 4 were treated so that the network nodes adhered to one another, it is natural to expect that apparent loops emanating from an apparent protein-rich condensate would be observed after histone removal, explaining the classical loop visualization experiments. Finally, we emphasize that a network as in Fig. 4 can result from a hierarchical folding process (29). Further insight may come from analysis of the chromatin fragment size distributions produced by digestion by various REs.
Acknowledgments
We are indebted to D. Chatenay, E. L. Zecheidrich, and J. Widom for advice regarding our experiments. We also acknowledge the help of L. Miller, L. Kaufman, and H. Buhse. The TVI cells were a generous gift from D. Reese, U.S. Environmental Protection Agency. This research was supported by a Biomedical Research Engineering Grant from the Whitaker Foundation; by National Science Foundation Grants DMR-9734178 and DMR-0203963; by a Focused Giving Award from Johnson and Johnson Corporation; by a Research Innovation Award from Research Corporation; and by the Petroleum Research Foundation of the American Chemical Society.
Abbreviations
MN, micrococcal nuclease
RE, restriction enzyme
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hirano T. (2000) Annu. Rev. Biochem. 69 115-144. [DOI] [PubMed] [Google Scholar]
- 2.Dietzel S. & Belmont, A. S. (2001) Nat. Cell Biol. 3 767-770. [DOI] [PubMed] [Google Scholar]
- 3.Paulson J. R. & Laemmli, U. K. (1977) Cell 12 817-828. [DOI] [PubMed] [Google Scholar]
- 4.Marsden M. P. F. & Laemmli, U. K. (1979) Cell 17 849-858. [DOI] [PubMed] [Google Scholar]
- 5.Laemmli U. K., Cheng, S. M., Adolph, K. W., Paulson, J. R., Brown, J. A. & Baumbach, W. R. (1978) Cold Spring Harbor Symp. Quant. Biol. 42 351-360. [DOI] [PubMed] [Google Scholar]
- 6.Gasser S. M., Laroche, T., Falquet, J., Boy de la Tour, E. & Laemmli, U. K. (1986) J. Mol. Biol. 188 613-629. [DOI] [PubMed] [Google Scholar]
- 7.Saitoh N., Goldberg, I. G., Wood, E. R. & Earnshaw, W. C. (1994) J. Cell Biol. 127 303-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boy de la Tour E. & Laemmli, U. K. (1988) Cell 55 937-944. [DOI] [PubMed] [Google Scholar]
- 9.Saitoh Y. & Laemmli, U. K. (1994) Cell 76 609-622. [DOI] [PubMed] [Google Scholar]
- 10.Hart C. M. & Laemmli, U. K. (1998) Curr. Opin. Genet. Dev. 8 519-525. [DOI] [PubMed] [Google Scholar]
- 11.Stack S. M. & Anderson, L. K. (2001) Chromosome Res. 9 175-198. [DOI] [PubMed] [Google Scholar]
- 12.Lodish H., Baltimore, D., Berk, A., Zipursky, S. L., Matsudaria, P. & Darnell, J., (1995) Molecular Cell Biology (Scientific American, New York), pp. 349–353.
- 13.Wolffe A. P., (1995) Chromatin: Structure and Function (Academic, New York), Sec. 2.4.1.
- 14.Lewin B., (2000) Genes VII (Oxford, New York), pp. 551–552.
- 15.van Holde K. E., (1989) Chromatin (Springer, New York), pp. 346–350.
- 16.Cole A. (1967) Theor. Exp. Biol. 1 305-375. [Google Scholar]
- 17.Houchmandzadeh B., Marko, J. F., Chatenay, D. & Libchaber, A. (1997) J. Cell Biol. 139 1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirier M., Eroglu, S., Chatenay, D. & Marko, J. F. (2000) Mol. Biol. Cell 11 269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicklas R. B. (1983) J. Cell Biol. 97 542-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reese D. H., Yamada, T. & Moret, R. (1976) Differentiation (Berlin) 6 75-81. [DOI] [PubMed] [Google Scholar]
- 21.Houchmandzadeh B. & Dimitrov, S. (1999) J. Cell Biol. 145 215-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall W. F., Marko, J. F., Agard, D. A. & Sedat, J. W. (2001) Curr. Biol. 11 569-578. [DOI] [PubMed] [Google Scholar]
- 23.Poirier M., Eroglu, S. & Marko, J. F. (2002) Mol. Biol. Cell 13 2170-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poirier M. G., Monhait, T. & Marko, J. F. (2002) J. Cell. Biochem. 85 422-434. [DOI] [PubMed] [Google Scholar]
- 25.Plischke M. & Barsky, S. J. (1998) Phys. Rev. E 58 3347-3352. [Google Scholar]
- 26.Maniotis A. J., Bojanowski, K. & Ingber, D. E. (1997) J. Cell. Biochem. 65 114-130. [PubMed] [Google Scholar]
- 27.Polach K. J. & Widom, J. (1995) J. Mol. Biol. 254 130-149. [DOI] [PubMed] [Google Scholar]
- 28.Anderson J. D. & Widom, J. (2000) J. Mol. Biol. 296 979-987. [DOI] [PubMed] [Google Scholar]
- 29.Belmont A. S., Sedat, J. W. & Agard, D. A. (1987) J. Cell Biol. 105 77-92. [DOI] [PMC free article] [PubMed] [Google Scholar]