Abstract
Leukemic disease can be linked to aberrant gene expression. This often is the result of molecular alterations in transcription factors that lead to their misrouting within the nucleus. The acute myelogenous leukemia-related fusion protein AML1/ETO is a striking example. It originates from a gene rearrangement t(8;21) that fuses the N-terminal part of the key hematopoietic regulatory factor AML1 (RUNX1) to the ETO (MTG8) repressor protein. AML1/ETO lacks the intranuclear targeting signal of the wild-type AML1 and is directed by the ETO component to alternate nuclear matrix-associated sites. To understand this aberrant subnuclear trafficking of AML1/ETO, we created a series of mutations in the ETO protein. These were characterized biochemically by immunoblotting and in situ by immunofluorescence microscopy. We identified two independent subnuclear targeting signals in the N- and C-terminal regions of ETO that together direct ETO to the same binding sites occupied by AML1/ETO. However, each segment alone is targeted to a different intranuclear location. The N-terminal segment contains a nuclear localization signal and the conserved hydrophobic heptad repeat domain responsible for protein dimerization and interaction with the mSin3A transcriptional repressor. The C-terminal segment spans the nervy domain and the zinc finger region, which together support interactions with the corepressors N-CoR and HDACs. Our findings provide a molecular basis for aberrant subnuclear targeting of the AML1/ETO protein, which is a principal defect in t(8;21)-related acute myelogenous leukemia.
Acute myelogenous leukemia (AML) is a prevalent hematopoietic malignancy, characterized by abnormal proliferation and differentiation of myeloid progenitor cells (1–3). The gene encoding the hematopoietic transcription factor AML1 (RUNX1, CBFA2, PEBP2αB) is a primary target of leukemia-associated chromosomal translocations (for review, see ref. 4). ETO (MTG8) originally was identified as a component of the AML1/ETO fusion protein resulting from the t(8;21) gene rearrangement (5–7). AML1/ETO encompasses the N terminus of AML1 including the runt homology DNA-binding domain and a nearly intact ETO protein lacking only the first 30 aa (7). There is limited information about the normal gene regulatory mechanisms in which ETO participates. Most studies have focused on the function of ETO in the context of the AML1/ETO fusion protein that causes aberrant transcriptional regulation of genes usually activated by AML1. One general model proposes that AML1/ETO antagonizes AML1 function by binding to AML1-responsive promoters (8) but, instead of supporting transcription, recruits repressor proteins that include N-CoR, mSin3A, SMRT, and the histone deacetylase HDAC1, HDAC2, or HDAC3 (9–12). The AML1/ETO fusion protein now represses rather than activates target genes.
ETO normally is not highly expressed in some hematopoietic lineages, and targeted mutation of the mouse ETO (MTG8) locus at exon 2 has revealed that the ETO protein has a crucial role in gut development (13). The role of ETO in leukemia results from fusion with AML1. The AML1/ETO fusion protein is a chimeric inhibitor that interferes with gene regulatory mechanisms controlling hematopoiesis (14). Ectopic expression of AML1/ETO prevents granulocytic differentiation of 32Dcl3 and L-G myeloid cell lines, monocytic differentiation of U937 cells, and erythroid differentiation of K562 and TF-1 cells (15–17). Thus, ETO-mediated deregulation of AML1-dependent genes has substantial physiological consequences. In addition, there are indications that AML1 and AML1/ETO might be involved in distinct gene regulatory pathways and may activate or inhibit different subsets of genes (18, 19).
We have proposed that targeting of AML1 to specific intranuclear sites is critical for accurate control of hematopoietic gene expression and that molecular alterations that cause misrouting of transcription factors may result in aberrant gene expression and development of disease (20, 21). Our studies have shown that AML1 contains a unique intranuclear trafficking sequence, referred to as the nuclear matrix targeting signal (NMTS), that localizes the protein to transcriptionally active subnuclear domains (22, 23). The AML1/ETO fusion protein lacks the NMTS that directs wild-type AML1 to appropriate gene regulatory sites within the nucleus. Instead, the AML1/ETO fusion protein is redirected by the ETO component to alternate nuclear matrix-associated foci (24). In the present study, we have addressed the mechanisms by which intranuclear trafficking of AML1/ETO is compromised. We establish that ETO contains two independent N- and C-terminal regions that mediate targeting of this repressor to nuclear matrix-associated sites that are transcriptionally inactive. Our findings provide a molecular basis for aberrant subnuclear targeting of the AML1/ETO protein, which is a principal defect in t(8;21)-related acute myelogenous leukemia.
Materials and Methods
Cell Culture and Transient Transfections.
Saos-2 cells were grown in McCoy's 5A Medium (GIBCO) supplemented with 15% FBS. HeLa cells were maintained in DMEM (GIBCO) with 10% FBS. Cells were plated on 0.5% gelatin-coated coverslips (Fisher) in six-well tissue culture trays at a density of 0.3 × 106 cells per well. Transfections were performed at 20–24 h after plating, when cells reach 70% confluency. HeLa cells were transfected with Superfect (Qiagen, Chatsworth, CA), and Saos-2 cells were transfected with Effectene (Qiagen). The transfection procedures were optimized to achieve low but detectable expression levels. We used 1 μg of expression vector per well for HeLa cells and 0.5 μg for Saos-2 cells. Cells were processed for immunofluorescence microscopy 18 h after transfection as described below.
Immunofluorescence Microscopy.
Cells were processed for whole-cell (WC) or nuclear matrix intermediate filament (NMIF) preparations, as described (25). Cell preparations were incubated with appropriate primary and secondary antibodies as described (24). The following primary antibodies were used at the indicated dilutions: rabbit polyclonal antiserum to the HA epitope (1:1,000; SC-805; Santa Cruz Biotechnology); mouse mAb to the Flag epitope (1:1,000; F3165; Sigma); mouse mAb to the GAL-4 DBD (1:1,000; SC-510; Santa Cruz Biotechnology); and rabbit polyclonal to ETO (1:750). Secondary antibodies (1:500 dilution) were goat anti-rabbit IgG conjugated to Alexa 488 (Molecular Probes), donkey anti-mouse IgG conjugated to fluorescein (Jackson ImmunoResearch), and donkey anti-mouse IgG conjugated to Alexa 594 (Molecular Probes).
Digital imaging of cells was performed by using a Zeiss Axioplan 2 microscope equipped with epifluorescence filters and a charge-coupled device camera (Hamamatsu, Middlesex, NJ) interfaced with the MetaMorph Imaging System (Universal Imaging, Media, PA). Cells also were visualized by using a Leica SP2 confocal microscope (Leica Microsystems; www.leica-microsystems.com/).
Plasmids.
Constructs pCMV5-HA-AML1/ETO, pCMV5-HA-ETO, and pCMV5-ETO as well as pCMV5-based vectors encoding ETO proteins in which different functional domains are deleted (Fig. 1A) were described (10, 26). Additional Flag-tagged deletion mutants of ETO and Flag-ETO were prepared as follows. Inserts were generated by PCR amplification of segments of the ETO coding sequences by using primers containing HindIII and XhoI restriction sites. After restriction enzyme digestion, the PCR products were cloned into the pCMV-Flag 2 mammalian expression vector (Stratagene). For the construction of vectors expressing fusion proteins of GAL4 DNA-binding domain (GAL4-DBD), we used the pCMX-GAL4-DBD vector (27). ETO encoding segments were generated by PCR and inserted by using SalI (or BamHI) and NheI restriction sites that were present in the PCR primers.
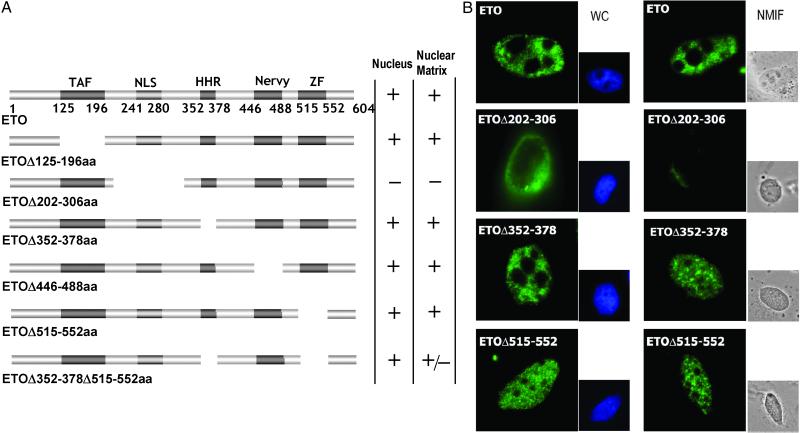
Fig 1.
Subnuclear targeting of wild-type ETO and internal deletion mutants. (A) Schematic representations of ETO mutants with internal deletions. The top line shows the structure of wild-type ETO. The four evolutionarily conserved functional domains are depicted: TBP-associated factor homology domain (TAF), HHR, the region homologous with the Nervy protein of Drosophila (nervy), and the domain corresponding to the two ZF motifs. The columns to the right indicate whether the protein is localized in the nucleus and whether it is retained in the NMIF. The results were obtained from four independent experiments in HeLa and Saos-2 cells. No significant difference was observed between these cell lines. Plus sign (+) indicates that >90% of cells score positively for nuclear matrix association in NMIF preparations relative to the number of cells that exhibit nuclear staining in WC preparations; plus and minus sign (+/−) indicates 40–60% positive cells. (B) In situ analysis of nuclear distribution of ETO mutants. Immunofluorescence microscopy images of representative cells transfected with vectors expressing the indicated ETO mutant proteins are shown. Proteins were visualized by using antibodies against the ETO protein and a fluorescein-conjugated secondary antibody. The 4′,6-diamidino-2-phenylindole-stained nuclei for WC preparations and differential interference contrast images for NMIF preparations also are shown.
Subcellular Fractionation and Western Blotting.
Cells were fractionated 18–20 h after transfection by using a standard protocol (28) we described recently in detail (29). The WC fraction was obtained by resuspending cells in RIPA buffer (150 mM NaCl/50 mM Tris, pH 7.5/1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS). The cytoskeletal fraction was obtained by extracting cells twice in CSK buffer (100 mM NaCl/0.3 M sucrose/10 mM Pipes/3 mM MgCl2/1 mM EGTA/0.5% Triton X-100, pH 6.8). Nuclei were resuspended in nuclease digestion buffer (50 mM NaCl/0.3 M sucrose/10 mM Pipes/3 mM MgCl2/1 mM EGTA/0.5% Triton X-100, pH 6.8) and incubated with DNase I. After nuclease digestion, ammonium sulfate was added to a final concentration of 250 mM. Extracted nuclei were subjected to centrifugation to separate soluble nuclear proteins (chromatin fraction) and the NMIF fraction. The same volume percentage (10%) of each fraction was separated electrophoretically in an 8% denaturing polyacrylamide gel, transferred to a poly(vinylidene difluoride) membrane, and processed for Western blotting. Proteins were detected by using antibodies with the indicated dilutions: mouse mAb against Flag tag (1:12,000; Sigma), goat polyclonal antibody against lamin B (1:2,000; Santa Cruz Biotechnology; SC-6217), and the corresponding horseradish peroxidase-conjugated secondary antibodies (1:3,000; Santa Cruz Biotechnology; SC-2042). Immunoreactive bands were visualized by using the enhanced chemiluminescence kit (Amersham Pharmacia).
Results
Delineation of Subnuclear Targeting Sequences in the ETO/MTG8 Protein.
The ETO protein contains four evolutionarily conserved domains (30–32), at least two of which [i.e., the hydrophobic heptad repeat (HHR) and zinc finger (ZF) region] contribute to transcriptional repression by AML1/ETO, a major protein in (8;21)-related leukemia (for review, see ref. 3). To assess whether specific, functional domains of ETO are important for the subnuclear compartmentalization of the protein, we tested a panel of ETO proteins with internal deletions that eliminate individual, conserved domains (Fig. 1A). Upon short-term expression of these proteins after transfection, we analyzed the subnuclear distribution of the ETO deletion mutants by in situ immunofluorescence microscopy of WC and NMIF preparations (Fig. 1B). The data show that the wild-type ETO protein, as well as four of six ETO mutants, is targeted to punctate subnuclear foci associated with the nuclear matrix (Fig. 1B and data not shown). The ETO mutant (ETOΔ202–306 aa) that lacks a known nuclear import signal (33) is not detected in the nucleus in either WC or NMIF preparations (Fig. 1B). Deletion of both the HHR and ZF domains decreases but does not abolish association with the NMIF (data not shown). Thus, none of the known conserved domains of ETO is individually necessary for nuclear matrix association within the context of the full-length ETO protein.
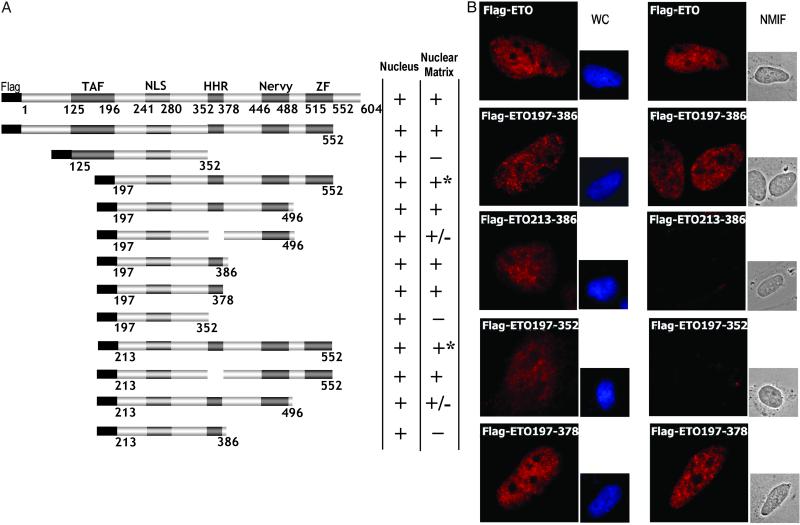
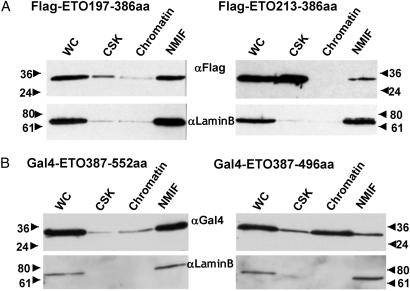
To identify protein regions responsible for nuclear matrix association of ETO, we systematically analyzed a series of ETO mutants with C- and/or N-terminal deletions (Fig. 2A). These mutant proteins are tagged with the Flag epitope and contain the native nuclear import signal of ETO (33). In situ immunofluorescence analysis shows that removal of sequences N-terminal of amino acid 196 and C-terminal of amino acid 378 does not influence targeting of ETO to the nuclear matrix (Fig. 2B). Therefore, the minimal NMTS in the N-terminal part of ETO is located between amino acids 197 and 378. Protein segments from amino acids 197–212 and 352–378 are critical within this region, because removal of either of these segments abolishes nuclear matrix targeting (Fig. 2B). Differences in nuclear matrix association between selected ETO constructs (e.g., Flag-ETO197–386 and Flag-ETO213–386) were confirmed by biochemical analysis of subcellular fractions (Fig. 3A). Thus, these data show that the region between amino acid 197 and 378 represents a minimal domain that supports subnuclear targeting of ETO.
Fig 2.
Delineation of an NMTS in the N terminus of ETO. (A) Schematic representation of Flag-tagged ETO deletion mutants lacking N- and/or C-terminal segments. Symbols are as described in Fig. 1. Plus sign with asterisk (+*) indicates that protein exhibits a distinct granular pattern in the nucleus. (B) Subnuclear targeting of Flag-tagged full-length and deletion mutants of ETO. Immunofluorescence microscopy images of representative cells transiently expressing the indicated ETO mutant proteins are shown. Proteins were visualized by using antibodies against the Flag epitope tag and Alexa 594-conjugated secondary antibody. 4′,6-Diamidino-2-phenylindole staining is shown for WC preparations and differential interference contrast for NMIF preparations.
Fig 3.
Western blot analysis of subcellular fractions from cells transiently expressing Flag-tagged (A) or GAL4DBD-fused (B) ETO mutants. WC lysate, a cytoplasmic fraction (CSK), a nuclease-digested and high-salt-extracted chromatin fraction (Chromatin), and the NMIF were prepared and analyzed by protein blotting with the indicated antibodies. Molecular mass markers are indicated at the left or right of each image.
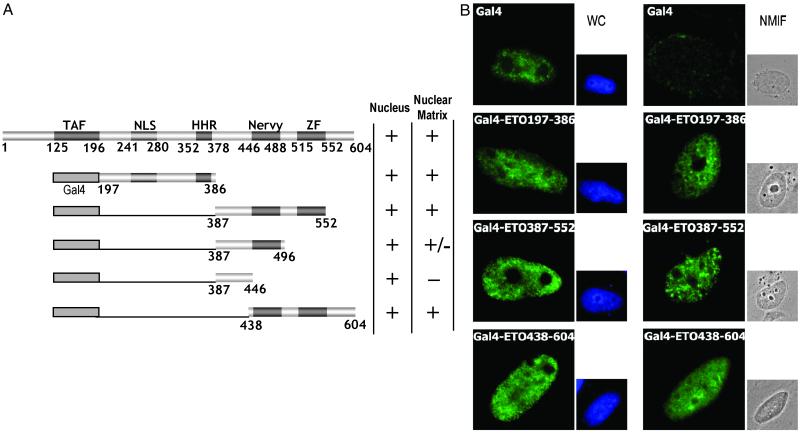
To test whether the N-terminal ETO targeting module is capable of directing a heterologous protein to the nuclear matrix, we tethered the 197–386 segment of ETO to the GAL4 (amino acid 1–147) DNA-binding domain (Fig. 4A). The GAL4-DBD protein alone does not associate with the NMIF (Fig. 4B). However, the GAL4/ETO197–386 fusion protein is targeted to the nuclear matrix (Fig. 4B). Thus, the 197–386 segment of ETO represents an autonomous subnuclear targeting module.
Fig 4.
Identification of a subnuclear targeting signal in the C terminus of ETO. (A) Schematic representation of GAL4-fused ETO deletion mutants. Symbols are as described in Fig. 1. (B) Subnuclear targeting of GAL4 and GAL4-fused deletion mutants of ETO. Immunofluorescence microscopy images of representative cells expressing the indicated GAL4-ETO fusion proteins. Proteins were visualized by using antibodies against the GAL4-DBD domain and Alexa 488-conjugated secondary antibodies. 4′,6-Diamidino-2-phenylindole-stained nuclei are shown for WC preparations, and differential interference contrast images are shown for NMIF preparations.
To assess whether the 197–378 domain represents the only region of ETO that supports nuclear matrix association, we examined several other constructs in which C-terminal segments of the ETO protein were fused to GAL4-DBD. Because C-terminal proteins lack a nuclear import signal, fusion to the GAL4-DBD, which contains an intrinsic signal for nuclear import, permits assessment of nuclear matrix association. We first tested two segments (respectively, amino acids 387–552 and 438–604) that together span the C terminus of ETO. Immunofluorescence microscopy reveals that both proteins are directed to the nuclear matrix (Fig. 4B). These results suggest that the region of overlap between GAL4/ETO387–552 and GAL4/ETO438–604 is important for nuclear matrix association of the C-terminal segment of ETO. Analysis of additional truncated ETO variants fused to GAL4 confirmed this finding. Our data indicate that the GAL4/ETO387–496 fusion protein, which lacks the ZF motifs, is impaired in its ability to associate with the nuclear matrix as reflected by a decrease in the percentage of cells scoring positive for fluorescent signals in NMIF preparations (Fig. 4A). Furthermore, the ETO387–446 segment, which lacks both the ZF and nervy regions, is completely defective for nuclear matrix association. The immunofluorescence microscopy results with the GAL4/ETO C-terminal fusion proteins were confirmed by biochemical methods (Fig. 3B). Based on these data, we propose that the ETO segment from 438–552 is a C-terminal subnuclear targeting signal that functions independently of the 197–378 module.
Two ETO NMTSs Specify Distinct Subnuclear Locations.
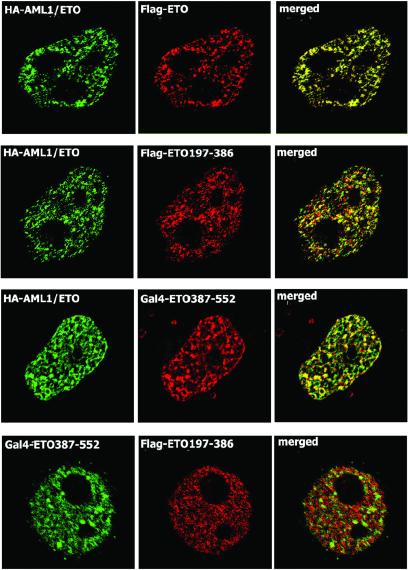
Previous results have shown that AML1/ETO and ETO proteins colocalize in the same subnuclear foci (ref. 24; see also Fig. 5). We analyzed the in situ function of these subnuclear sites in which the AML1/ETO and ETO proteins reside. Based on BrUTP labeling, ETO containing foci are not associated with nascent RNA transcripts, nor do they colocalize with RNA-processing domains containing mRNA splicing factors (i.e., SC-35; data not shown). To test whether the NMTS of ETO can direct proteins to the same foci in the nucleus in which the AML1/ETO protein localizes, we coexpressed Flag-tagged ETO mutant proteins or GAL4/ETO fusion proteins with HA-tagged, full-length ETO or AML1/ETO proteins and examined their subnuclear localization by using in situ immunofluorescence microscopy. ETO deletion mutants that are incapable of (or impaired in) nuclear matrix association do not colocalize with either ETO or AML1/ETO protein in WC preparations (data not shown). The Flag-ETO197–378 and GAL4/ETO387–552, which contain the N- and C-terminal targeting signals, respectively, each show only partial overlap of fluorescent signal with ETO or AML1/ETO in WC and NMIF preparations (Fig. 5 and data not shown). Thus, the individual NMTS segments are directed to only a subset of the sites in which the full-length ETO protein resides.
Fig 5.
The ETO NMTSs define different subnuclear locations. Subnuclear distributions of AML1/ETO and ETO mutant proteins were analyzed in NMIF preparations of HeLa cells by using laser-scanning confocal microscopy. Epitope-tagged proteins were detected with antibodies against the HA, Flag, or GAL4 tags and secondary antibodies conjugated to Alexa 488 (green) or Alexa 568 (red). Overlap of red and green immunofluorescence signals results in yellow staining and indicates colocalization of coexpressed proteins.
We directly compared the relative subnuclear locations specified by the N- or C-terminal NMTS sequences of ETO by coexpressing the Flag-tagged ETO197–386 protein encompassing the N-terminal NMTS and the GAL4/ETO387–552 fusion protein that contains the C-terminal NMTS. The data indicate that these two proteins have different subnuclear distributions (Fig. 5, bottom row). Hence, the two independent NMTS sequences of ETO are targeted to distinct subnuclear locations.
Discussion
In the present study, we have identified the determinants of subnuclear targeting of the AML1/ETO t(8;21) fusion protein, which is involved in AML. Our previous work has demonstrated that the subnuclear distribution of AML1/ETO is the same as that of wild-type ETO but different from that of wild-type AML1 (24). Thus, and AML1 are targeted to different subnuclear sites and presumably become components of different nuclear matrix-associated gene regulatory complexes, which may play a critical role in the transforming ability of the translocation-related AML1/ETO protein. Here, we demonstrate that there are two separate segments in the ETO protein that, together, direct ETO to nuclear matrix-associated foci. Our data indicate that, within these two subnuclear targeting regions of ETO, there are multiple determinants that influence nuclear matrix association and/or intranuclear distribution of this protein.
Specific mechanisms may coordinate the spatial organizations of genes, transcripts, and regulatory proteins within the nucleus (34). Consequently, analysis of targeting signals that direct regulatory factors to nuclear matrix-associated subnuclear sites may provide insight into structure–function interplay of those factors. Several regulatory proteins such as RUNX/CBFA/AML factors (22, 23, 35) and the Pit1 transcription factor (36) have a single nuclear matrix targeting determinant. In contrast, steroid hormone receptors (37–39), as well as the multifunctional regulator YY1 (25, 40) and the NP/NMP4 transcription factor (41, 42), have intricate bi- or multipartite signals that mediate targeting to the nuclear matrix. Based on the results presented here, the subnuclear localization of the ETO and AML1/ETO proteins is mediated similarly by a composite signal. Both the N- and C-terminal signals that are responsible for nuclear matrix association of ETO contain conserved functional domains including the HHR and ZF region (see Fig. 6). ZF motifs also have been shown to play crucial roles in mediating subnuclear targeting of YY1 and NP/NMP4 (25, 42).
Fig 6.
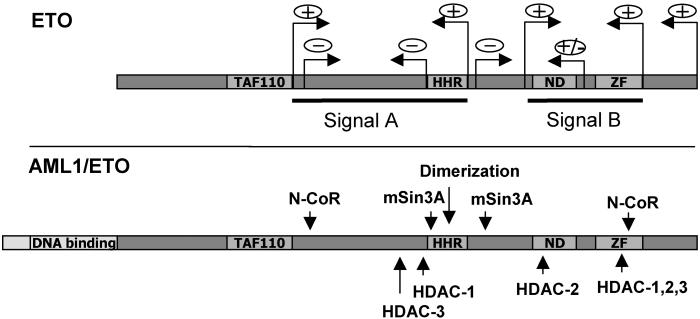
ETO subnuclear targeting signals overlap corepressor-interacting domains. Two independent NMTSs (signals A and B) are indicated (black lines) in the diagram representing full-length ETO. Arrows with plus signs reflect the minimal region of ETO required for nuclear matrix association in signals A and B. The lower diagram shows the AML1/ETO fusion protein; regions mediating corepressor interactions are indicated by vertical arrows (43).
The regions responsible for protein–protein interactions of ETO may play a major role in aberrant intranuclear trafficking of AML1/ETO. For example, the N-terminal subnuclear targeting signal (amino acid 197–386) and the ZF region within the C-terminal NMTS both are independently capable of interacting with N-CoR (Fig. 6). The 387–496 segment of the C-terminal targeting signal also encompasses the nervy domain and the proline-rich region, both of which are known to interact with mSin3A (9, 10, 12, 26). Because mSin3A represents an integral component of a histone deacetylase complex, and because histone deacetylases are intrinsic nuclear matrix components (reviewed in ref. 44), the possibility arises that mSin3A may play a role in nuclear matrix association of the ETO and AML1/ETO proteins.
Two distinct regions in the N- and C-terminal halves of the ETO protein independently support protein association with the nuclear matrix. However, colocalization studies with AML1/ETO, wild-type ETO, and shorter ETO proteins indicate that, individually, the N- and C-terminal targeting sequences direct proteins to different subnuclear addresses. Proteins containing only the N- or C-terminal signal are routed to a portion of the sites to which the full-length, wild-type ETO and AML1/ETO proteins are directed. Thus, our findings indicate that each targeting signal cannot fully recapitulate the subnuclear targeting behavior of ETO and AML1/ETO. Instead, subnuclear distribution of ETO and AML1/ETO proteins is defined by complementary contributions of independent NMTSs.
Acknowledgments
This study was supported by National Institutes of Health Grant CA82834.
Abbreviations
HHR, hydrophobic heptad repeat
AML, acute myelogenous leukemia
NMTS, nuclear matrix targeting signal
ZF, zinc finger
WC, whole cell
NMIF, nuclear matrix intermediate filament
References
- 1.Bloomfield C. D., Lawrence, D., Byrd, J. C., Carroll, A., Pettenati, M. J., Tantravahi, R., Patil, S. R., Davey, F. R., Berg, D. T., Schiffer, C. A., et al. (1998) Cancer Res. 58 4173-4179. [PubMed] [Google Scholar]
- 2.Grimwade D., Walker, H., Oliver, F., Wheatley, K., Harrison, C., Harrison, G., Rees, J., Hann, I., Stevens, R., Burnett, A., et al. (1998) Blood 92 2322-2333. [PubMed] [Google Scholar]
- 3.Downing J. R. (1999) Br. J. Haematol. 106 296-308. [DOI] [PubMed] [Google Scholar]
- 4.Downing J. R., Higuchi, M., Lenny, N. & Yeoh, A. E. (2000) Semin. Cell Dev. Biol. 11 347-360. [DOI] [PubMed] [Google Scholar]
- 5.Miyoshi H., Shimizu, K., Kozu, T., Maseki, N., Kaneko, Y. & Ohki, M. (1991) Proc. Natl. Acad. Sci. USA 88 10431-10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erickson P. F., Robinson, M., Owens, G. & Drabkin, H. A. (1994) Cancer Res. 54 1782-1786. [PubMed] [Google Scholar]
- 7.Miyoshi H., Kozu, T., Shimizu, K., Enomoto, K., Maseki, N., Kaneko, Y., Kamada, N. & Ohki, M. (1993) EMBO J. 12 2715-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyers S., Lenny, N. & Hiebert, S. W. (1995) Mol. Cell. Biol. 15 1974-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J., Hoshino, T., Redner, R. L., Kajigaya, S. & Liu, J. M. (1998) Proc. Natl. Acad. Sci. USA 95 10860-10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lutterbach B., Westendorf, J. J., Linggi, B., Patten, A., Moniwa, M., Davie, J. R., Huynh, K. D., Bardwell, V. J., Lavinsky, R. M., Rosenfeld, M. G., et al. (1998) Mol. Cell. Biol. 18 7176-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelmetti V., Zhang, J., Fanelli, M., Minucci, S., Pelicci, P. G. & Lazar, M. A. (1998) Mol. Cell. Biol. 18 7185-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amann J. M., Nip, J., Strom, D. K., Lutterbach, B., Harada, H., Lenny, N., Downing, J. R., Meyers, S. & Hiebert, S. W. (2001) Mol. Cell. Biol. 21 6470-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calabi F., Pannell, R. & Pavloska, G. (2001) Mol. Cell. Biol. 21 5658-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okuda T., Cai, Z., Yang, S., Lenny, N., Lyu, C. J., van Deursen, J. M., Harada, H. & Downing, J. R. (1998) Blood 91 3134-3143. [PubMed] [Google Scholar]
- 15.Kohzaki H., Ito, K., Huang, G., Wee, H. J., Murakami, Y. & Ito, Y. (1999) Oncogene 18 4055-4062. [DOI] [PubMed] [Google Scholar]
- 16.Le X. F., Claxton, D., Kornblau, S., Fan, Y. H., Mu, Z. M. & Chang, K. S. (1998) Eur. J. Haematol. 60 217-225. [DOI] [PubMed] [Google Scholar]
- 17.Westendorf J. J., Yamamoto, C. M., Lenny, N., Downing, J. R., Selsted, M. E. & Hiebert, S. W. (1998) Mol. Cell. Biol. 18 322-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L. Q., Ilaria, R., Jr., Kingsley, P. D., Iwama, A., van Etten, R. A., Palis, J. & Zhang, D. E. (1999) Mol. Cell. Biol. 19 3029-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhoades K. L., Hetherington, C. J., Harakawa, N., Yergeau, D. A., Zhou, L., Liu, L. Q., Little, M. T., Tenen, D. G. & Zhang, D. E. (2000) Blood 96 2108-2115. [PubMed] [Google Scholar]
- 20.Stein G. S., van Wijnen, A. J., Stein, J. L., Lian, J. B., Pockwinse, S. & McNeil, S. (1998) J. Cell Biochem. 70 200-212. [PubMed] [Google Scholar]
- 21.Stein G. S., van Wijnen, A. J., Stein, J. L., Lian, J. B., Javed, A., McNeil, S. & Pockwinse, S. M. (1999) Exp. Cell Res. 253 110-116. [DOI] [PubMed] [Google Scholar]
- 22.Zeng C., van Wijnen, A. J., Stein, J. L., Meyers, S., Sun, W., Shopland, L., Lawrence, J. B., Penman, S., Lian, J. B., Stein, G. S., et al. (1997) Proc. Natl. Acad. Sci. USA 94 6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng C., McNeil, S., Pockwinse, S., Nickerson, J. A., Shopland, L., Lawrence, J. B., Penman, S., Hiebert, S. W., Lian, J. B., van Wijnen, A. J., et al. (1998) Proc. Natl. Acad. Sci. USA 95 1585-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNeil S., Zeng, C., Harrington, K. S., Hiebert, S., Lian, J. B., Stein, J. L., van Wijnen, A. J. & Stein, G. S. (1999) Proc. Natl. Acad. Sci. USA 96 14882-14887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNeil S., Guo, B., Stein, J. L., Lian, J. B., Bushmeyer, S., Seto, E., Atchison, M. L., Penman, S., van Wijnen, A. J. & Stein, G. S. (1998) J. Cell. Biochem. 68 500-510. [PubMed] [Google Scholar]
- 26.Lutterbach B., Sun, D., Schuetz, J. & Hiebert, S. W. (1998) Mol. Cell. Biol. 18 3604-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J., Zamir, I. & Lazar, M. A. (1997) Mol. Cell. Biol. 17 6887-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fey E. G., Krochmalnic, G. & Penman, S. (1986) J. Cell Biol. 102 1654-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaidi S. K., Javed, A., Choi, J.-Y., van Wijnen, A. J., Stein, J. L., Lian, J. B. & Stein, G. S. (2001) J. Cell Sci. 114 3093-3102. [DOI] [PubMed] [Google Scholar]
- 30.Gross C. T. & McGinnis, W. (1996) EMBO J. 15 1961-1970. [PMC free article] [PubMed] [Google Scholar]
- 31.Kitabayashi I., Ida, K., Morohoshi, F., Yokoyama, A., Mitsuhashi, N., Shimizu, K., Nomura, N., Hayashi, Y. & Ohki, M. (1998) Mol. Cell. Biol. 18 846-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gamou T., Kitamura, E., Hosoda, F., Shimizu, K., Shinohara, K., Hayashi, Y., Nagase, T., Yokoyama, Y. & Ohki, M. (1998) Blood 91 4028-4037. [PubMed] [Google Scholar]
- 33.Odaka Y., Mally, A., Elliott, L. T. & Meyers, S. (2000) Oncogene 19 3584-3597. [DOI] [PubMed] [Google Scholar]
- 34.Stein G. S., van Wijnen, A. J., Stein, J. L., Lian, J. B., Montecino, M., Zaidi, S. K. & Javed, A. (2000) J. Cell. Biochem. 35 84-92. [PubMed] [Google Scholar]
- 35.Stein G. S., van Wijnen, A. J., Stein, J. L., Lian, J. B., Montecino, M., Choi, J.-Y., Zaidi, K. & Javed, A. (2000) J. Cell Sci. 113 2527-2533. [DOI] [PubMed] [Google Scholar]
- 36.Mancini M. G., Liu, B., Sharp, Z. D. & Mancini, M. A. (1999) J. Cell. Biochem. 72 322-338. [PubMed] [Google Scholar]
- 37.DeFranco D. B. & Guerrero, J. (2000) Crit. Rev. Eukaryotic Gene Expression 10 39-44. [PubMed] [Google Scholar]
- 38.Tang Y., Getzenberg, R. H., Vietmeier, B. N., Stallcup, M. R., Eggert, M., Renkawitz, R. & DeFranco, D. B. (1998) Mol. Endocrinol. 12 1420-1431. [DOI] [PubMed] [Google Scholar]
- 39.van Steensel B., van Binnendijk, E. P., Hornsby, C. D., van der Voort, H. T., Krozowski, Z. S., de Kloet, E. R. & van Driel, R. (1996) J. Cell Sci. 109 787-792. [DOI] [PubMed] [Google Scholar]
- 40.Bushmeyer S. M. & Atchison, M. L. (1998) J. Cell. Biochem. 68 484-499. [PubMed] [Google Scholar]
- 41.Alvarez M., Long, H., Onyia, J., Hock, J., Xu, W. & Bidwell, J. (1997) Endocrinology 138 482-489. [DOI] [PubMed] [Google Scholar]
- 42.Feister H. A., Torrungruang, K., Thunyakitpisal, P., Parker, G. E., Rhodes, S. J. & Bidwell, J. P. (2000) J. Cell. Biochem. 79 506-517. [PubMed] [Google Scholar]
- 43.Hiebert S. W., Lutterbach, B. & Amann, J. (2001) Curr. Opin. Hematol. 8 197-200. [DOI] [PubMed] [Google Scholar]
- 44.Davie J. R. (1997) Mol. Biol. Rep. 24 197-207. [DOI] [PubMed] [Google Scholar]