Abstract
The activity of the kinase Aurora-A (Aur-A) peaks during mitosis and depends on phosphorylation by one or more unknown kinases. Mitotic phosphorylation sites were mapped by mass spec sequencing of recombinant Aur-A protein that had been activated by incubation in extracts of metaphase-arrested Xenopus eggs. Three sites were identified: serine 53 (Ser-53), threonine 295 (Thr-295), and serine 349 (Ser-349), which are equivalent to Ser-51, Thr-288, and Ser-342, respectively, in human Aur-A. To ask how phosphorylation of these residues might affect kinase activity, each was mutated to either alanine or aspartic acid, and the recombinant proteins were then tested for their ability to be activated by M phase extract. Mutation of Thr-295, which resides in the activation loop of the kinase, to either alanine or aspartic acid abolished activity. The S349A mutant had slightly reduced activity, indicating that phosphorylation is not required for activity. The S349D mutation completely blocked activation, suggesting that Ser-349 is important for either the structure or regulation of Aur-A. Finally, like human Aur-A, overexpression of Xenopus Aur-A transformed NIH 3T3 cells and led to tumors in nude mice. These results provide further evidence that Xenopus Aur-A is a functional ortholog of human Aur-A and, along with the recently described crystal structure of human Aur-A, should help in future studies of the mechanisms that regulate Aur-A activity during mitotic progression.
The serine/threonine kinase Aurora-A (Aur-A) is required for centrosome maturation and formation of a bipolar mitotic spindle, and for accurate segregation of both centrosomes and chromosomes into daughter cells during mitotic exit (1–3). In somatic cells, both the amount of Aur-A protein and its kinase activity peak during mitosis and then drop. Recent work has led to several insights about its regulated degradation. Aur-A is ubiquitinated by the Cdh1-activated form of the anaphase-promoting complex/cyclosome (APC/C; refs. 4 and 5), a multisubunit ubiquitin ligase that targets several proteins for proteasome-mediated proteolysis during the latter stages of mitotic exit. Recognition of Aur-A by APC/CCdh1 requires two domains, a C-terminal destruction box (D box) shared by many APC/C targets (4–6) and an N-terminal region, the A box, that is conserved in Aur-A family members but is not found in Aur-B or Aur-C (5). The A box contains Ser-53, which is phosphorylated during M phase and may control the timing of Aur-A destruction during mitotic exit (5).
By contrast, much less is known about how the kinase activity of Aur-A is regulated during cell-cycle progression. Phosphorylation is required for its activity (7, 8). Thr-295 resides in the predicted activation loop and is thus well situated to affect activity (9). This residue is part of a PKA consensus motif, and, in vitro, phosphorylation of recombinant Aur-A by PKA increases its activity (10), but endogenous activating kinase(s) have not been identified. Inactivation probably involves dephosphorylation by phosphatase 1 (PP1), as high concentrations of okadaic acid (OA) maintain high Aur-A kinase activity in cells and in extracts (8, 10), and Aur-A contains a functional PP1-binding site near the C terminus (11).
In somatic cells, kinase activity increases roughly in parallel with rising protein levels, but the lack of methods for achieving highly synchronous progression during G2, mitosis, and mitotic exit has made it difficult to study the temporal control of Aur-A activity in those cells. Because of this problem, answers to even the most basic questions of whether Aur-A activity is regulated simply by synthesis and destruction, or is also regulated by posttranslational modifications at specific times during cell-cycle progression, are not known. By contrast, the Xenopus early embryonic cell cycles provide good opportunities to investigate these questions. These cycles are rapid and naturally synchronous, and many events regulating mitotic progression can be reproduced in vitro by using concentrated extracts of these cells (12). Finally, unlike somatic cells, Aur-A protein levels remain constant during the early cell cycles, and Aur-A kinase activity is regulated solely by cycles of activation and inactivation (5).
Here, we show that extracts of unfertilized eggs, which are arrested naturally at metaphase of meiosis II, can activate recombinant Aur-A. We have used this assay to identify three residues that become phosphorylated during M phase (Ser-53, Thr-295, and Ser-349, which are equivalent to Ser-51, Thr-288, and S342 in human Aur-A) and to ask which of these are required for activation by M phase extracts. S53A and S53D mutations had no significant effect on kinase activity. T295A and T295D mutations abolished activity, confirming the importance of this residue. Mutation of Ser-349 to alanine slightly reduced activity, indicating that phosphorylation of Ser-349 is not required for activity. By contrast, mutation of Ser-349 to aspartic acid completely blocked activation, suggesting that this residue is important for the structure or regulation of Aur-A. Ser-349 is immediately adjacent to a PP1 binding site (11) and, when mapped onto the recent crystal structure of human Aur-A (9), is located in a region that could affect conformation of the protein.
Methods
Expression and Purification of Recombinant Aur-A.
Aur-A mutants were created by standard PCR methods and QuickChange site-directed mutagenesis (Stratagene). All constructs were sequenced in full. For expression in Sf9 cells, N-terminally histidine-tagged constructs were cloned into the pFastBacHT vectors (Invitrogen). Baculovirus was isolated by using the BAC-TO-BAC baculovirus expression system (Invitrogen) and was amplified in Sf9 cells. His-tagged Aur-A protein was prepared from Sf9 cells by three different methods.
To obtain active Aur-A (Aur-AOA), cells were incubated in the presence of 0.5 μM okadaic acid for the final 4 h of expression. Cells were solubilized in pulldown buffer (20 mM β-glycerophosphate/10 mM Hepes-KOH, pH 7.7/5 mM EGTA/5 mM β-mercaptoethanol/150 mM NaCl/1% Chaps/1 mM PMSF/protease inhibitors, Roche protease inhibitor tablets, EDTA-free). Samples were centrifuged for 30 min at 8,000 rpm in an SS-34 rotor (Sorvall). Ni-NTA agarose beads (Qiagen, Valencia, CA) were added to the supernatant, and samples were rocked at 4°C for 2 h. After a brief centrifugation (1 min at 250 × g), beads were collected and washed three times in a 10-ml wash buffer (20 mM β-glycerophosphate/10 mM Hepes-KOH, pH 7.7/5 mM EGTA/5 mM β-mercaptoethanol/500 mM NaCl/0.1% Chaps/1 mM PMSF/protease inhibitors, Roche protease inhibitor tablets, EDTA-free), eluted in elution buffer (20 mM β-glycerophosphate/10 mM Hepes-KOH, pH 7.7/5 mM EGTA/5 mM β-mercaptoethanol/25 mM NaCl/200 mM imidazole/1 mM DTT/1 mM EDTA/1 mM PMSF and protease inhibitors), dialyzed in dialysis buffer (10 mM Hepes-KOH, pH 7.7/100 mM NaCl/1 mM DTT/0.1 mM EDTA/10% glycerol), aliquoted, and stored at −80°C as described (8). Protein preparations routinely were about 0.2 mg/ml.
Cells were incubated with a serine/threonine kinase inhibitor mix (Calbiochem) for 4 h before harvesting to obtain inactive Aur-A protein (Aur-AKI). Inhibitors and their final concentrations were bisindolylmaleimide I (PKC inhibitor, 420 nM), H-89 dihydrochloride (PKA inhibitor, 100 nM), KN-93 (CaM kinase II inhibitor, 740 nM), ML-7 (myosin light chain kinase inhibitor, 600 nM), and staurosporine (PKC and broad range inhibitor, 40 nM).
Aur-A was expressed in the absence of inhibitors, purified, and then phosphatase-treated during the first step of protein purification. Bead-bound Aur-A was washed three times with 10 ml of PPase buffer (50 mM Tris⋅HCl/0.1 mM EDTA/5 mM DTT/2 mM MnCl2/0.01% Brij35, pH 7.5). One milliliter of PPase buffer containing 4,000 units of λ-phosphatase (New England Biolabs) was added, and samples were incubated at 30°C for 30 min. For dephosphorylation by phosphatase PP1, bead-bound Aur-A was washed with PP1 buffer (50 mM Tris⋅HCl/0.1 mM EDTA/5 mM DTT/0.01% Brij 35/1 mM MnCl2, pH 7.0), and 25 units of PP1 (New England Biolabs) was used in the dephosphorylation reaction.
Identification of Mitotic Phosphorylation Sites.
Concentrated extracts were prepared from G2-arrested Xenopus oocytes (8) and M phase-arrested Xenopus eggs (12). His-tagged inactive Aur-AKI was incubated with extracts, repurified, and assayed for kinase activity. For each reaction, 10 μl (2 μg) of Aur-AKI was incubated with 10 μl of G2 or M phase extracts plus 20 μl of 2× kinase buffer (1× kinase buffer: 20 mM Hepes, pH 7.7/10 mM MgCl2/0.1 mM EGTA/0.5 mM DTT/50 μM ATP) containing 1 μM okadaic acid for 30 min at 30°C. Aur-A was recovered by binding to Ni-NTA agarose beads in pulldown buffer for 1–2 h at 4°C. Samples were washed four times in pulldown buffer, eluted by boiling in SDS gel sample buffer, and separated by SDS/PAGE (13). Gels were stained with Coomassie blue, and the slower mobility, phosphorylated Aur-A bands were excised and sequenced at the Harvard Microchemistry Facility, by using microcapillary reverse-phase HPLC nanoelectrospray tandem mass spectrometry on a Finnigan LCQ DECA quadrupole ion trap mass spectrometer.
For in vitro kinase assays of recombinant protein, equimolar amounts of Aur-A (0.011 nM) were incubated in 1× kinase buffer, 0.5 μl of [γ-32P]ATP and 2.5 μg of myelin basic protein (MBP). For in vitro kinase assays of recombinant protein activated by extract, 2 μg of Aur-A protein were used in Fig. 1, and 0.5 μg was used in Fig. 2. Bead-bound Aur-A samples were washed twice with kinase buffer and then incubated in 20 μl of kinase buffer, 1 μl of [γ-32P]ATP (Perkin–Elmer, 3,000 Ci/mmol; 1 Ci = 37 GBq), and 2.5 μg MBP (Invitrogen) for 20 min at 30°C. Fifteen microliters of sample buffer was added to each reaction, and samples were separated on SDS/15% PAGE (13) and visualized by autoradiography. Most results were quantified by using a Molecular Imager FX PhosphorImager with QUANTITY ONE software (Bio-Rad).
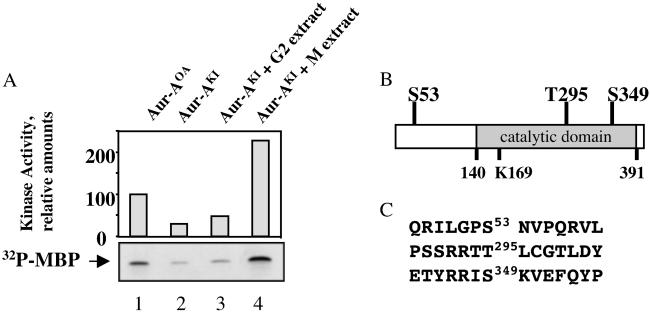
Fig 1.
Identification of sites phosphorylated on Aur-A by M phase extracts. (A) Activation of recombinant Aur-A. His-tagged recombinant Aur-A protein was expressed in Sf9 cells in the presence of okadaic acid (Aur-AOA, lane 1) or Ser/Thr kinase inhibitors (Aur-AKI, lanes 2–4) and purified. Aur-AKI samples were incubated with either G2-arrested oocyte extract (lane 3) or M phase extract (lane 4), repurified on Ni-NTA beads, and assayed for in vitro kinase activity toward MBP. Samples were separated by SDS/PAGE, quantified (Upper), and visualized by autoradiography (Lower). (B) Location of phosphorylated residues. (C) Sequences surrounding phosphorylated sites.
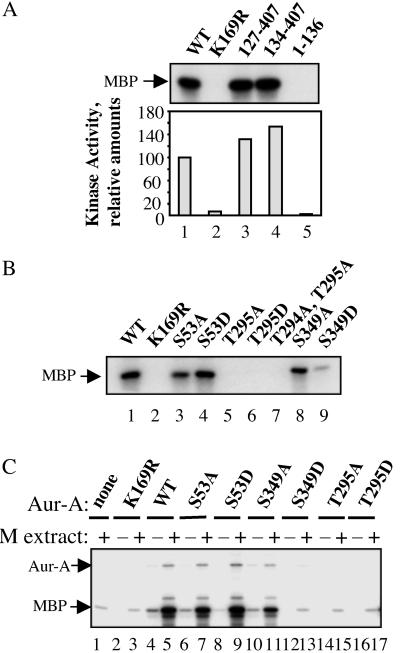
Fig 2.
Effects of N-terminal truncation and of Ser-53, Thr-295, and Ser-349 mutations on Aur-A kinase activity. (A and B) Equimolar amounts of wild-type Aur-A and the indicated mutants were expressed in Sf9 cells in the presence of okadaic acid, purified, and assayed for in vitro kinase activity toward MBP. Autoradiograms of the 32P-labeled MBP products are shown. (C) Activity of Aur-A mutants after incubation in M phase extract. His-tagged Aur-A proteins, either wild type or mutant, were expressed in Sf9 cells, dephosphorylated with λ-phosphatase, incubated with or without M phase extracts, repurified, and assayed for kinase activity toward MBP, as above.
Kinase Inhibitors.
M phase extracts (one volume extract to two volumes kinase buffer, final volume 29 μl) were incubated for 15 min at 4°C with one of the following inhibitors at the indicated concentrations: apigenin (Sigma), roscovitine (Calbiochem), bisindolylmaleimide I (Calbiochem), PKI (New England Biolabs), KN-93 and negative control KN-92 (Calbiochem), LY294002 (Calbiochem), ML-7 (Calbiochem), staurosporine (Calbiochem), and U0126 (Promega). One microliter (0.5 μg) of wild-type Aur-APpase was added and incubated at room temperature for 15 min. Aur-A was repurified on Ni-NTA beads and assayed for kinase activity.
Cell Growth and Transformation Assays.
Anchorage-independent growth assays were performed by suspending 104 cells in 0.36% Bactoagar (Difco) over a 0.6% agar base layer in DMEM plus 10% FCS in 35-mm dishes. Every 4 days, ≈300 μl of media was added to each plate. After 2–3 weeks, colonies were stained overnight with 0.5 mg/ml nitroblue tetrazolium (Sigma) in PBS and counted. Each experiment was performed in triplicate and repeated at least two times. In vivo transformation was measured by injecting 106 cells s.c. behind both front legs of two nude mice for each cell line (vector only, Aur-A, and Aur-A-K169R). Additionally, two mice were injected at three locations (behind both front legs and one hind leg) with the three constructs per mouse.
Results
Metaphase Extracts Phosphorylate Xenopus Aur-A on Ser-53, Thr-295, and Ser-349.
When His-tagged Xenopus Aur-A was produced in Sf9 cells, addition of the phosphatase inhibitor OA during the last 4 h of protein expression enhanced its final activity (Aur-AOA, Fig. 1), as described (8). When produced in the presence of a mix of kinase inhibitors (see Methods), Aur-A had much lower activity (Aur-AKI). When Aur-AKI was incubated with extracts prepared from G2-arrested Xenopus oocytes, where the activity of endogenous Aur-A is low (8, 14), and then repurified, its activity was essentially unchanged. By contrast, Aur-AKI was activated 8-fold by incubation with extracts prepared from metaphase II-arrested Xenopus eggs. These results strongly suggest that phosphorylation at one or more sites is required for Aur-A activity.
To determine which sites were phosphorylated by M phase extracts, Aur-AKI was activated by incubation with M phase extract, repurified, and separated by SDS/PAGE. The most electrophoretically retarded Aur-A bands were excised and sequenced by mass spectrometry. Three phosphorylated residues were identified: Ser-53, Thr-295, and Ser-349 (Fig. 1B). Ser-53 is part of the highly conserved A box in the N-terminal noncatalytic domain that is required for mitotic destruction (5). Thr-295 resides in the predicted activation loop (9). Ser-349 is immediately adjacent to K350VEF, a binding site for the phosphatase PP1 (11).
Full Activation of Aur-A Requires Thr-295 and Ser-349.
To investigate how phosphorylation of these residues might regulate Aur-A activity, wild-type and mutated versions of His-tagged Aur-A were assayed in two different ways. In the first, Aur-A proteins were produced in Sf9 cells in the presence of okadaic acid, purified, and assayed directly for kinase activity (Fig. 2 A and B). In the second, more physiologically relevant approach, the requirement of these residues for activation by M phase extracts was tested. Briefly, Aur-A was inactivated by phosphatase treatment, incubated with M phase extract, repurified, and assayed for kinase activity (Fig. 2C). Both approaches gave similar results.
In agreement with previous studies (9, 15), the N terminus was not required for activity, either when expressed directly in Sf9 cells (Fig. 2A) or when activated by M phase extracts (data not shown). However, Aur-A lacking the N terminus consistently showed an elevation in kinase activity when assayed by either method. Neither the S53A nor the S53D mutation significantly affected activity, although we routinely noted that the S53A mutant had slightly decreased activity.
Both the T295A and T295D mutations almost completely eliminated kinase activity, which was as low as that seen when the catalytic lysine was mutated to arginine (K169R). These results strongly argue that phosphorylation of Thr-295 is required for activity.
Mutation of Ser-349 to alanine only slightly reduced activity, indicating that phosphorylation of this residue is not required for activity. By contrast, mutation of Ser-349 to aspartic acid completely abolished activity. This result could indicate that phosphorylation of Ser-349 is inhibitory or that Ser-349 is critical for the conformation of Aur-A in some other way. These possibilities are considered further in Discussion.
Aur-A Activation by M Phase Extracts Is Not Blocked by Inhibitors of PKA, PKC, or cdc2 but Is Inhibited by High Concentration of Apigenin.
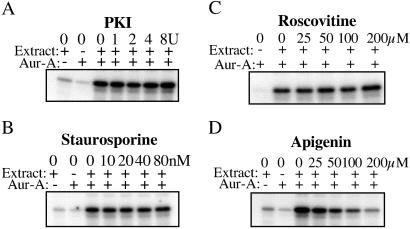
Both Thr-295 and Ser-349 are predicted to be good sites (RRTT295L and RRIS349K, respectively) for phosphorylation by PKA. In vitro, PKA can phosphorylate and increase the activity of Aur-A, both human (10) and Xenopus (data not shown). To ask whether PKA is required for activation of Aur-A, M phase extract was first incubated with PKI, the inhibitory subunit of PKA, under conditions known to reduce activity by more than 95% and then tested for its ability to reactivate recombinant Aur-A. As shown in Fig. 3, inhibition of PKA in the M phase extract did not interfere with activation of Aur-A. M phase extracts contain high levels of mitogen-activated protein kinase and cyclin B/cdc2 kinase activity; inhibition of cdc2 by preincubation with roscovitine failed to block activation of Aur-A. Preincubation with the MEK inhibitor U0126 also failed to block activation (data not shown). These results suggests that neither cdc2 nor mitogen-activated protein kinase is required for activation of Aur-A, nor are they required to maintain activity of an activator. We then tested other inhibitors, including those in the mix of kinase inhibitors used to produce low activity Aur-A (Aur-AKI) in Sf9 cells: bisindolylmaleimide I (PKC inhibitor), H-89 dihydrochloride (PKA inhibitor), KN-93 (CaM kinase II inhibitor), ML-7 (myosin light chain kinase inhibitor), staurosporine (PKC and broad range inhibitor), and apigenin (CK2 and PKC inhibitor). The only inhibitor that significantly blocked activation of Aur-A was apigenin. In vitro, CK2 neither phosphorylated nor activated recombinant Aur-A (data not shown), arguing that CK2 is not a direct activator of Aur-A.
Fig 3.
Effect of kinase inhibitors on the ability of M phase extracts to activate Aur-A. M phase extracts were first incubated in the absence or presence of individual kinase inhibitors, PKI (PKA inhibitor), roscovitine (cyclin B/cdc2 inhibitor), staurosporine (broad range Ser/Thr kinase inhibitor), and apigenin (CK2 inhibitor), and then tested for their ability to activate His-tagged Aur-A as in Fig. 2C.
Mutations of Thr-295 and Ser-349 Do Not Interfere with Cdh1-Induced Destruction of Aur-A During Mitotic Exit in Egg Extracts.
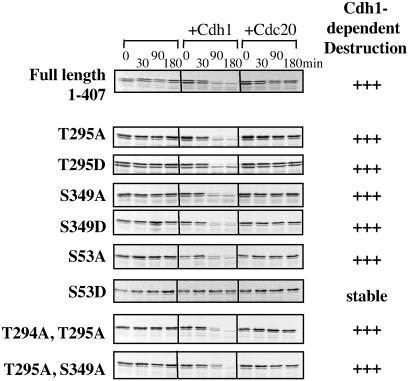
Mutation of Ser-53 to aspartic acid blocks the Cdh1-dependent destruction of Aur-A during mitotic exit, suggesting that phosphorylation of Ser-53 might block destruction during mitosis (5). To ask how mutations in Thr-295 and Ser-349 affected Aur-A destruction, radiolabeled in vitro translation products encoding mutant Aur-As were added to M phase extracts, Cdh1 was added to program the APC/C for recognition and ubiquitination of Aur-A, and calcium was added to induce the metaphase/anaphase transition and mitotic exit (Fig. 4). As seen previously, wild-type Aur-A was degraded between 30 and 60 min after release from metaphase arrest, whereas the S53D mutant was resistant to destruction. By contrast, mutation of either Thr-295 or Ser-349 to either alanine or aspartic acid did not significantly affect either the rate or extent of destruction. Thus, of the three M phase phosphorylation sites identified, only Ser-53 seems to affect destruction of Aur-A.
Fig 4.
Thr-295 and Ser-349 mutations do not interfere with Cdh1-dependent destruction of Aur-A. M phase extracts containing cycloheximide were incubated with Cdh1 or Cdc20, as indicated, and mixed with [35S]methionine-labeled Aur-A in vitro translation products. Calcium was added to initiate M phase exit, and samples were taken at the indicated times. Samples were analyzed by SDS/PAGE followed by autoradiography.
Overexpression of Xenopus Oocyte Aur-A Transforms NIH 3T3 Cells and Induces Tumors in Nude Mice.
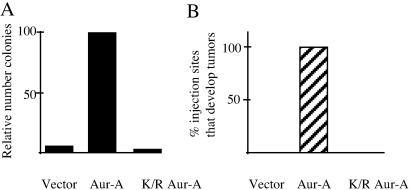
The properties and behavior of Xenopus Aur-A, which was originally isolated from oocytes where it was called Eg2 (8, 16, 17), resembles human Aur-A in many aspects. Their sequences are very similar, and the kinase activities of both peak during mitosis. Ectopic expression of both generates aneuploid cells containing multiple centrosomes (5, 18, 19). Both undergo APC/CCdh1-mediated destruction during mitotic exit (ref. 5, and R. F. Crane and L.E.L., unpublished data). Cells overexpressing human Aur-A are transformed and are highly tumorigenic (7). However, the oncogenic potential of Xenopus Aur-A had not been tested. We thus established NIH 3T3 cell lines stably transfected with empty vector, AU1-tagged wild-type Aur-A, or AU1-tagged kinase-dead Aur-A (K169R) and tested their oncogenic potential by using the approaches described for human Aur-A (7). Expression of Aur-A was confirmed by collecting nocadozole-arrested cells and immunoblotting for the AU1 tag (data not shown). These cells were first tested for their ability to form colonies in soft agar. At 20 days after plating, 1.7% of the Aur-A transfected cells formed colonies (Fig. 5). When the transformation efficiency of wild-type Aur-A was set as 100%, the relative transformation efficiency of cells transfected with empty vector was 6%. That of cells carrying kinase-dead Aur-A was even less (2%), suggesting that the kinase-dead mutant may behave like a dominant negative in this setting. Similar results were obtained in two additional experiments.
Fig 5.
Xenopus Aur-A is oncogenic. (A) Aur-A transforms NIH 3T3 cells. NIH 3T3 cells were transfected with empty vector, Aur-A, or catalytically inactive K169R Aur-A, and stably transfected cell lines were established. Cells from each line were plated on soft agar and assayed for growth 20 days later. Colonies were counted and compared in the bar graph (A), where efficiency of colony formation by Aur-A was normalized to 100%. (B) Cells overexpressing wild-type Aur-A are tumorigenic in nude mice. Cells (106) from each of the cell lines described above were injected s.c. in nude mice. Two mice were injected with each cell line at two locations right behind the front leg, and two additional mice were injected at three locations with the three constructs per mouse.
To test the oncogenic potential of Aur-A in whole animals, 106 cells stably expressing wild-type Aur-A, Aur-A K169R, or empty vector were injected s.c. into nude mice. Each of the three cell lines was injected individually into nude mice as described in Methods. By 5 weeks, all of the sites injected with Aur-A expressing cells had developed large tumors; no tumors were seen at the sites of injection of cells carrying kinase-dead Aur-A or empty vector, or at other sites in the any of the mice (Fig. 5). These results demonstrate that Xenopus Aur-A, like human Aur-A, is a potent protooncogene. Thus, Xenopus Aur-A and human somatic cell Aur-A are functionally equivalent by all criteria tested and seem to be functional orthologs.
Discussion
Progression through mitosis is regulated mainly by reversible phosphorylation and irreversible proteolysis (20, 21). The best understood examples include (i) activation of cyclin B/cdc2 by the phosphatase cdc25, which removes inhibitory phosphorylations from cdc2 and thus catalyzes the G2/M transition, (ii) the APC/C-mediated ubiquitin-dependent proteolysis of securin, the inhibitory subunit of the protease that cleaves proteins that hold the sister chromatids together and thus catalyzes anaphase onset, and (iii) the APC/C-mediated destruction of cyclin B, which results in inactivation of cdc2 and thus catalyzes exit from M phase into G1 of the next cell cycle. As outlined earlier, several studies show that misregulation of the mitotic kinase Aur-A has profound effects on mitotic progression, and overexpression of Aur-A is oncogenic. Although we have some insight into what is required for the regulated destruction of Aur-A during mitotic exit, much less is known about how phosphorylation contributes to its activation and inactivation during mitosis.
The identification of three sites in Xenopus Aur-A that are phosphorylated during M phase (Ser-53, Thr-295, and Ser-349) should help in understanding how phosphorylation contributes to regulation of Aur-A. This is especially true in view of the recently solved crystal structure of the catalytic domain of human Aur-A (9). Thr-295 (Thr-288 in human Aur-A), which is conserved in all Aurora family members, is part of the activation loop. However, it is in a disordered region, making it impossible to predict at this time the structural consequences of Thr-295 phosphorylation or mutations in this residue. Biochemically, we find that the single point mutations T295A and T295D each completely eliminate the kinase activity of Aur-A, both when expressed in Sf9 cells and after incubation in Xenopus M phase egg extracts. These results, which were obtained consistently, were done with constructs that had all been sequenced in full to confirm the desired mutation and to ensure that no other mutations had been introduced inadvertently. In contrast, an earlier report found that a T288D mutation in human Aur-A did not abolish activity and that mutation of both Thr-287 and Thr-288 was required for inactivation of Sf9 cell-expressed Aur-A (10). The different results could be caused by differences in assay conditions, or they could reflect a real difference in the regulation of the human and Xenopus enzymes. Introduction of a T288D mutant, which was reported to enhance kinase activity, into murine cells led to transformation and tumor formation (7). It is now known that even kinase-dead Aur-A constructs can induce aneuploidy (5, 19), leaving open the possibility that overexpression of the T288D mutant induces tumor formation by promoting genomic instability rather than through elevated kinase activity.
Mutating Ser-349 to alanine slightly reduced activity, indicating that phosphorylation is not essential for activity. The S349D mutant was completely inactive when expressed in Sf9 cells and unable to be activated by M phase extract. These results could indicate that phosphorylation of Ser-349 negatively regulates activity. Although this interpretation might seem flawed because Aur-A is phosphorylated on Ser-349 during M phase, when it is active, it is worth pointing out that the gel-purified Aur-A bands sequenced could have contained a mix of active and inactive Aur-A. Thus, we cannot eliminate the possibility that Ser-349 phosphorylation negatively regulates activity. Ser-349 is immediately adjacent to the PP1-binding sequence, K350VEF (11), and phosphorylation of Ser-349 could affect the recruitment or strength of PP1 binding.
Although the N terminus of Aur-A is not required for kinase activity, its removal elevates activity, and it clearly plays an important role in vivo. In addition to being required for destruction during mitotic exit (5), it is required for localization of Aur-A at the centrosomes during mitosis (15) and for the ability of the tumor suppressor gene product p53 to bind and inactivate Aur-A (22). Overexpression of Aur-A induces defects in cytokinesis that result in the formation of aneuploid cells containing multiple centrosomes (5, 19); this effect is greatly enhanced in cells lacking p53 and can be reversed by the reintroduction of p53 (19, 22). From these results, it might be concluded that p53 suppresses the oncogenic potential of Aur-A simply by inactivating Aur-A. This explanation cannot be the only one, however, as overexpression of inactive mutants of both human and Xenopus Aur-A induce aneuploidy (5, 19). These findings thus argue strongly that p53 must also control a second function of Aur-A. It would not be surprising if future studies revealed an effect of p53 on the ability of Aur-A to associate with or regulate other interacting proteins, including the APC/C activator cdc20 (23), the translational regulator CPEB (24), the kinesin-like motor Eg5 (25), the transforming protein TACC that can regulate spindle structure (26), TPX2, which is required for targeting Aur-A to spindles (27), a newly identified protein called AIP that can promote Aur-A destruction (28), or others yet to be discovered.
Acknowledgments
We thank all members of the Ruderman laboratory for discussions and especially Dawn Farruggio, Holger Bastians, Brian Duckworth, Sammy Kim, and Richard Crane for helpful input. We thank Joan Brugge for helpful suggestions and interest. This work was supported by a National Institutes of Health grant (to J.V.R.). L.E.L. was supported by a National Science Foundation predoctoral fellowship.
Abbreviations
Aur-A, Aurora-A
APC/C, anaphase-promoting complex/cyclosome
PP1, phosphatase 1
MBP, myelin basic protein
OA, okadaic acid
References
- 1.Bischoff J. R. & Plowman, G. D. (1999) Trends Cell Biol. 9 454-459. [DOI] [PubMed] [Google Scholar]
- 2.Nigg E. A. (2001) Mol. Cell. Biol. 2 21-32. [DOI] [PubMed] [Google Scholar]
- 3.Dutertre S., Descamps, S. & Prigent, C. (2002) Oncogene 21 6175-6183. [DOI] [PubMed] [Google Scholar]
- 4.Castro A., Arlot-Bonnemains, Y., Vigneron, S., Labbe, J. C., Prigent, C. & Lorca, T. (2002) EMBO Rep. 3 457-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Littlepage L. E. & Ruderman, J. V. (2002) Genes Dev. 16 2274-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arlot-Bonnemains Y., Klotzbucher, A., Giet, R., Uzbekov, R., Bihan, R. & Prigent, C. (2001) FEBS Lett. 508 149-152. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff J. R., Anderson, L., Zhu, Y., Mossie, K., Ng, L., Souza, B., Schryver, B., Flanagan, P., Clairvoyant, F., Ginther, C., et al. (1998) EMBO J. 17 3052-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andresson T. & Ruderman, J. V. (1998) EMBO J. 17 5627-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheetham, G. M., Knegtel, R. M., Coll, J. T., Renwick, S. B., Swenson, L., Weber, P., Lippke, J. A. & Austen, D. A. (2002) J. Biol. Chem., in press. [DOI] [PubMed]
- 10.Walter A. O., Seghezzi, W., Korver, W., Sheung, J. & Lees, E. (2000) Oncogene 19 4906-4916. [DOI] [PubMed] [Google Scholar]
- 11.Katayama H., Zhou, H., Li, Q., Tatsuka, M. & Sen, S. (2001) J. Biol. Chem. 276 46219-46224. [DOI] [PubMed] [Google Scholar]
- 12.Murray A. (1991) Methods Cell Biol. 36 581-605. [PubMed] [Google Scholar]
- 13.Anderson C. W., Baum, P. R. & Gesteland, R. F. (1973) J. Virol. 12 241-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank-Vaillant M., Haccard, O., Thibier, C., Ozon, R., Arlot-Bonnemains, Y., Prigent, C. & Jessus, C. (2000) J. Cell Sci. 113 1127-1138. [DOI] [PubMed] [Google Scholar]
- 15.Giet R. & Prigent, C. (2001) J. Cell Sci. 114 2095-2104. [DOI] [PubMed] [Google Scholar]
- 16.Paris J., Osborne, H. B., Couturier, A., LeGuellec, R. & Philippe, M. (1988) Gene 72 169-176. [DOI] [PubMed] [Google Scholar]
- 17.Roghi C., Giet, R., Uzbekov, R., Morin, N., Chartrain, I., Le Guellec, R., Couturier, A., Doree, M., Philippe, M. & Prigent, C. (1998) J. Cell Sci. 111 557-572. [DOI] [PubMed] [Google Scholar]
- 18.Zhou H., Kuang, J., Kuo, W., Gray, J., Sahin, A., Brinkley, B. R. & Sen, S. (1998) Nat. Genet. 20 189-193. [DOI] [PubMed] [Google Scholar]
- 19.Meraldi P., Honda, R. & Nigg, E. A. (2002) EMBO J. 21 483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Farrell P. H. (2001) Trends Cell Biol. 11 512-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nasmyth K. (2002) Science 297 559-565. [DOI] [PubMed] [Google Scholar]
- 22.Chen S. S., Chang, P. C., Cheng, Y. W., Tang, F. M. & Lin, Y. S. (2002) EMBO J. 21 4491-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farruggio D. C., Townsley, F. M. & Ruderman, J. V. (1999) Proc. Natl. Acad. Sci. USA 96 7306-7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendez R., Hake, L. E., Andresson, T., Littlepage, L. E., Ruderman, J. V. & Richter, J. D. (2000) Nature 404 302-307. [DOI] [PubMed] [Google Scholar]
- 25.Giet R., Uzbekov, R., Cubizolles, F., Le Guellec, K. & Prigent, C. (1999) J. Biol. Chem. 274 15005-15013. [DOI] [PubMed] [Google Scholar]
- 26.Giet R., McLean, D., Descamps, S., Lee, M. J., Raff, J. W., Prigent, C. & Glover, D. M. (2002) J. Cell Biol. 156 437-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kufer T. A., Sillje, H. H., Korner, R., Gruss, O. J., Meraldi, P. & Nigg, E. A. (2002) J. Cell Biol. 158 617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiat, L., Hui, K. M. & Gopalan, G. (2002) J. Biol. Chem., in press. [DOI] [PubMed]