Abstract
The adult mouse intestine contains an intricate vascular network. The factors that control development of this network are poorly understood. Quantitative three-dimensional imaging studies revealed that a plexus of branched interconnected vessels developed in small intestinal villi during the period of postnatal development that coincides with assembly of a complex society of indigenous gut microorganisms (microbiota). To investigate the impact of this environmental transition on vascular development, we compared the capillary networks of germ-free mice with those of ex-germ-free animals colonized during or after completion of postnatal gut development. Adult germ-free mice had arrested capillary network formation. The developmental program can be restarted and completed within 10 days after colonization with a complete microbiota harvested from conventionally raised mice, or with Bacteroides thetaiotaomicron, a prominent inhabitant of the normal mouse/human gut. Paneth cells in the intestinal epithelium secrete antibacterial peptides that affect luminal microbial ecology. Comparisons of germ-free and B. thetaiotaomicron-colonized transgenic mice lacking Paneth cells established that microbial regulation of angiogenesis depends on this lineage. These findings reveal a previously unappreciated mechanism of postnatal animal development, where microbes colonizing a mucosal surface are assigned responsibility for regulating elaboration of the underlying microvasculature by signaling through a bacteria-sensing epithelial cell.
Keywords: small intestine, gnotobiotics, ecology, symbiosis
The mucosal surface of the adult mouse and human small intestine consists of invaginations (crypts of Lieberkühn) and finger-shaped projections (villi). The gut epithelium undergoes constant renewal throughout postnatal life. This process is fueled by one or more long-lived active multipotent stem cells located at or near the base of each crypt (1, 2). The stem cell gives rise to four lineages, three of which (enterocytic, goblet, and enteroendocrine) complete their differentiation as they migrate out of crypts up onto adjacent villi. In contrast, members of the fourth lineage (Paneth cell) complete their differentiation at the crypt base (3, 4). Paneth cells are critical components of the intestine's innate immune system; they secrete a wide variety of antimicrobial peptides and proteins into the gut lumen (5, 6).
The intestinal epithelium is uniquely positioned to receive and transduce signals from two vastly different cellular populations. It sits at the interface between a subepithelial ensemble of mesenchymal fibroblasts, immune and endothelial cells, and a vast community of indigenous microorganisms (the microbiota) that reside in the gut lumen (7). Endothelial cells in the villus core form a dense and intricate capillary network (8) for efficient distribution of absorbed nutrients.
Collectively, the microbiota can be viewed as a metabolically active “organ” that provides functions beneficial for both microbes and host. For example, by supporting assembly of a society of resident microbes with the capacity to break down carbohydrate polymers in the distal intestine (e.g., refs. 9–16), mice (and humans) did not have to evolve their own repertoire of glycosylhydrolases to cleave the wide variety of linkages that exist in dietary polysaccharides. In this arrangement, the host is able to extract nutrient value from an otherwise poorly used dietary substrate, while microbes are provided with a niche where they can gain access to abundant, readily fermentable carbon sources.
The idea that our resident microbial communities are important contributors to normal mammalian physiology and health is not new: the concept dates back to Pasteur (17). However, we are now developing a broader appreciation of the wide range of host functions that are regulated by symbiotic microorganisms (reviewed in refs. 18–21). In the present study, we examine the interactions between the gut microbiota, the small intestinal epithelium, and the villus' mesenchymal microvascular network. We show that the microbiota plays a key role in constructing this microvascular network, and that this regulation depends on a central component of the gut's innate immune system: the Paneth cell. These findings reveal a previously unappreciated pathway for control of angiogenesis, and emphasize the importance of considering features of postnatal mammalian development as manifestations of mutually beneficial partnerships with microbes.
Materials and Methods
Mice.
All experiments with mice were performed using protocols approved by our institutional animal studies committee. Germ-free mice belonging to the NMRI inbred strain were maintained in plastic gnotobiotic isolators (22) under a 12-h light cycle, and given free access to autoclaved water and chow (B & K Universal, East Yorkshire, U.K.). We re-derived FVB/N CR2-tox176 transgenic mice (23) as germ-free using protocols described in ref. 22, and maintained the pedigree by crosses to germ-free nontransgenic littermates. Animals were genotyped by PCR assays of tail DNA (23). Genotypes were confirmed at the time of sacrifice by documenting Paneth cell loss through staining small intestinal sections with indocarbocyanine (Cy3)-labeled Ulex europeaus agglutinin-1 (ref. 24; Sigma).
Colonization of Germ-Free Animals.
Mice were inoculated with either mid-log phase cultures of Bacteroides thetaiotaomicron (type strain VPI 5482), or an intestinal microbiota harvested from the distal third of the small intestines and cecums of age-matched, conventionally raised, specified pathogen-free animals (22). Mice were killed 7, 10, or 30 days after inoculation. Animals with ≥107 colony forming units per ml luminal contents in their distal small intestines were used to evaluate the villus microvasculature.
Quantitative Analysis of the Villus Microvasculature.
All mice (conventionally raised, germ-free, and ex-germ-free) were killed at the same time of day (12:00 p.m.). Animals were anesthetized by metofane inhalation for 5 min, and then injected with 100 μl of a 2 mg/ml solution of high-molecular-weight (2,000 kDa) fluorescein isothiocyanate (FITC)-labeled dextran (Sigma). A 30-gauge needle, attached to a 1-ml syringe, was used to infuse this material over a 10-s period into the retro-orbital plexus. Animals were killed 3 min later. The small intestine was removed and measured without stretching. An 8-cm segment, spanning the junction of the middle and distal thirds of the small bowel, was excised, perfused with fixation solution containing 0.5% paraformaldehyde, 15% picric acid, and 0.1 M sodium phosphate buffer (pH 7.0), opened with an incision along its cephalocaudal axis, pinned on black wax, and shaken gently at 4°C for 12 h in fixation solution. Specimens were rinsed in ice-cold PBS (three washes, 5 min each), followed by a 3-h incubation in 10% sucrose/PBS (4°C) and an overnight incubation in 20% sucrose/10% glycerol/PBS (4°C).
Villi were examined using two different methods. In the first method, 2.5-cm-long portions of the intestinal segment were frozen in OCT compound, and 120-μm-thick sections were cut along the cephalocaudal axis. Sections were then air-dried for 2 h at room temperature in the dark, followed by rehydration in ice-cold PBS (1 min) and an overnight incubation at 4°C in 3% deoxycholic acid (Sigma). Sections were rinsed two times in water (5 min per cycle, room temperature), and one time in PBS (5 min, room temperature) to remove residual deoxycholic acid. Sections were stained with Syto61 (Molecular Probes; 1:1,000 dilution in PBS) for 1 h at room temperature, followed by three PBS washes (5 min per cycle, room temperature). Sections were mounted in 50% glycerol/PBS and stored at 4°C before viewing. In the second method, whole-mount preparations are used rather than cryosections. A 1-cm portion of the fixed intestinal segment was washed in PBS and stained with Syto61 exactly as in the first method. The stained whole-mount preparation was placed on a glass slide (villus side up), and 100 μl of a solution of 50% glycerol/PBS was added, followed by a coverslip.
Cryosections and whole mounts were viewed with an LSM 510 confocal microscope (Zeiss) and scanned at 5-μm-thick intervals. Scans were projected in three dimensions by taking 12–16 serial images, aligning them at 7–10° intervals, and compiling/rotating them about the y axis by using LSM 510 software (Zeiss).
The capillary network in the upper third of villi was quantified by counting the number of Syto61-positive nuclei per window as visualized from a fixed perspective of the three-dimensional reconstruction (a window was defined as an open area bounded by capillaries; n = 20–25 villi per mouse; n = 3 animals per group). Mean values ± SD of nuclei per window were calculated for each mouse and mean values ± SEM computed for each group. The statistical significance of differences observed between groups was assessed using Student's t test.
photoshop (Version 5.0, Adobe Systems, Mountain View, CA) was used to create two-dimensional images from rotating three-dimensional images. These two-dimensional images are shown in Figs. 1–3.
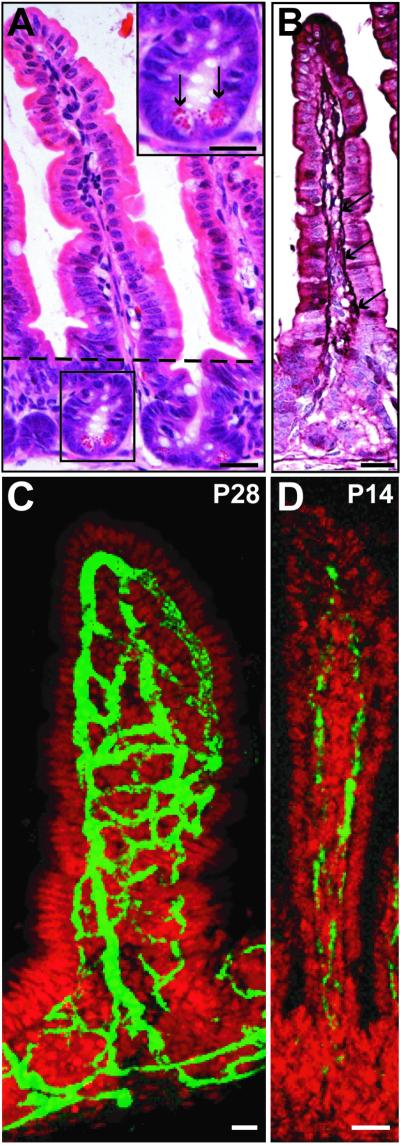
Fig 1.
Villus capillary networks in conventionally raised mice. (A–C) Sections prepared from the junction between the middle and distal thirds of the small intestine of a normal P28 mouse. (A) Hematoxylin/eosin (H&E)-stained section of a crypt–villus unit. The villus is located above, and the crypt below the dashed line. (Inset) A higher power view of the crypt showing Paneth cells (arrows). (B) Section from the same animal as in A, but stained with H&E and antibodies to the endothelial marker, von Willebrand's factor (purple; arrows). (C) Single confocal microscopic scan of a 120-μm-thick cryosection. The capillary network is stained with FITC-tagged high-molecular-weight dextran (green), and epithelial nuclei with Syto61 (red). To view a three-dimensional rotating image of a compiled set of serial scans of this type, see Movies 1 and 2, which are published as supporting information on the PNAS web site, www.pnas.org, or go to http://gordonlab.wustl.edu/vasculature. (D) Confocal view of FITC-dextran-labeled vessels in a villus positioned at the junction of the middle and distal thirds of the small intestine of a normal, conventionally raised P14 mouse. (Bars, 25 μm.)
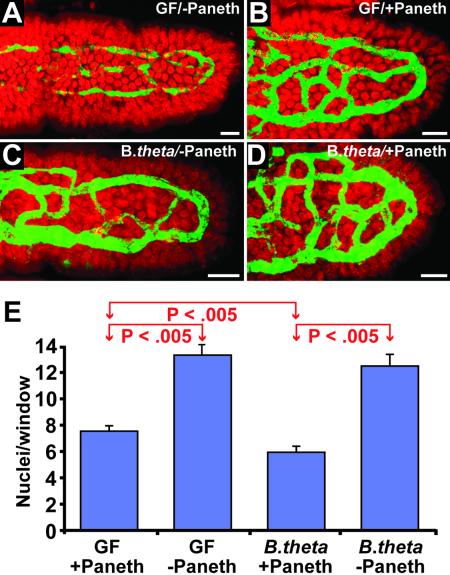
Fig 3.
Paneth cell and microbial regulation of angiogenesis. (A–D) Confocal scans of 120-μm-thick cryosections showing the upper thirds of villi. (A) Germ-free, Paneth cell-deficient P28 male CR2-tox176 mouse. (B) Age- and gender-matched, germ-free, Paneth-cell-containing normal littermate. (C) Ex-germ-free P28 CR2-tox176 mouse examined 7 days after colonization with B. thetaiotaomicron. (D) P28 nontransgenic mouse killed 7 days after mono-association with B. thetaiotaomicron. To view three-dimensional rotating images of the capillary networks shown in A–D, see Movies 6–9, which are published as supporting information on the PNAS web site, or go to http://gordonlab.wustl.edu/vasculature. (E) Quantitation of capillary network density. Mean values ± SEM for each condition are plotted. The statistical significance of differences between various groups is noted (Student's t test). (Bars in A–D, 25 μm.)
Results
A Quantitative Assay for Visualizing Development of the Microvasculature in Three Dimensions Within the Mouse Small Intestine.
Two-dimensional views are not adequate for characterizing the organization of the villus microvasculature (Fig. 1 A and B). Therefore, we developed a quantitative method for visualizing blood vessels in three dimensions. This method employs retro-orbital injection of a solution containing high-molecular-weight FITC-conjugated dextran, followed by confocal microscopy of villi present in either whole-mount preparations of small intestine or 120-μm-thick cryosections. Epithelial nuclei were labeled with Syto61, and the density of the microvascular network was quantified by counting the number of stained nuclei overlying the open spaces or “windows” framed by the underlying interconnected capillaries. The higher the density of capillary branches and connections, the smaller the average number of nuclei contained per window. By assembling serial confocal images, taken at 5-μm intervals, a rotating three-dimensional “movie” was constructed for viewing the microvascular network from multiple perspectives (Fig. 1C).
Vascular network development was first characterized in conventionally raised mice (i.e., mice with a microbiota). We focused on the distal half of the intestine, where the density of microbial colonization in the adult is several orders of magnitude greater than in the proximal half (25, 26). Three time points were selected: postnatal day 7 (P7), P14, and P28. During the suckling period (P1–P14), the crypt stem cell hierarchy is established (27, 28), the preweaning microbiota is predominated by facultative anaerobes (26), and mature Paneth cells have yet to emerge (24). At P7 and P14, the villus microvascular pattern consists of a simple arch with few or no cross-linking branches (Fig. 1D; n = 3 mice per time point). The suckling–weaning transition and crypt–villus morphogenesis are both completed by P28, at which time the villus capillary network has evolved to a complex latticework of branched interconnecting vessels (Fig. 1C).
The interval between P14 and P28 corresponds to the transition between the preweaning microbiota and assembly of a climax microbial community predominated by obligate anaerobes (26). In addition, a full census of differentiated Paneth cells appears at the base of now fully formed crypts. Paneth cells are able to sense, respond to, and subsequently shape the intestinal microbiota through their production and export of antimicrobial peptides (6). Based on these considerations, we proceeded to test the contributions of the microbiota and Paneth cells to angiogenesis.
The Adult Mouse Germ-Free Small Intestine Has an Arrested Angiogenic Program That Can Be Rapidly Restarted and Completed After Bacterial Colonization.
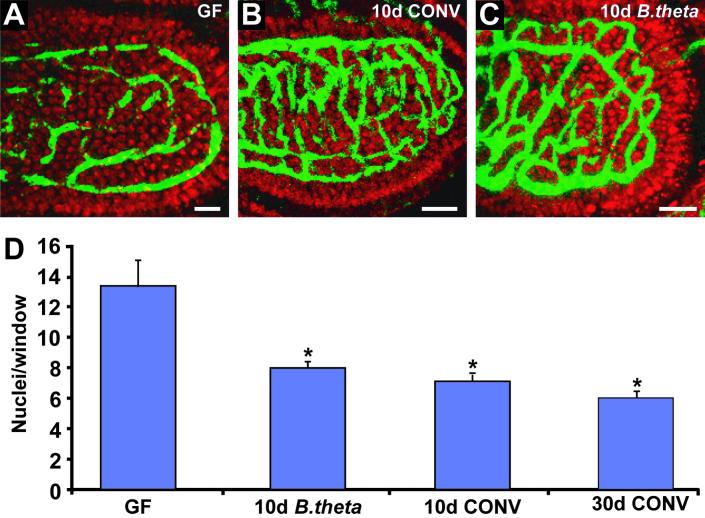
Recent DNA microarray-based profiling of small intestinal gene expression in adult NMRI mice raised without indigenous microbes (germ-free) suggested that a state of functional immaturity persists through adulthood when a microbiota is absent (ref. 29; see Table 1, which is published as supporting information on the PNAS web site, or http://gordonlab.wustl.edu/vasculature). Therefore, we compared the structures of villus capillary networks in 6-week-old male NMRI germ-free mice, and in age- and gender-matched ex-germ-free animals colonized for 10 or 30 days with an unfractionated distal small intestinal microbiota harvested from age- and gender-matched, conventionally raised adult mice. “Conventionalization” resulted in a marked increase in the density of the capillary network, as evidenced by a statistically significant 2-fold reduction in the average number of Syto61-labeled nuclei per “window” (P < 0.05; Fig. 2 A, B, and D). Microbial induction of angiogenesis was completed over a short time interval (10 days): colonization for 30 days produced no statistically significant changes in network density compared with a 10-day colonization [Fig. 2D; note that the density of epithelial nuclei per unit of villus surface area does not change during colonization (data not shown)].
Fig 2.
Rapid microbial induction of angiogenesis in small intestinal villi of adult ex-germ-free mice. (A–C) Confocal scans of the capillary network present in the upper third of small intestinal villi. Whole mounts are from the junction between the middle and distal thirds of the small intestines of 6-week-old NMRI mice (capillaries, green; nuclei, red). (A) Germ-free (GF) mouse. (B) Age-matched ex-germ-free conventionalized (CONV) mouse killed 10 days after colonization with an unfractionated microbiota harvested from a conventionally raised “donor.” (C) Ex-germ-free mouse 10 days after colonization with B. thetaiotaomicron (B. theta) alone. To view three-dimensional rotating images of the capillary networks shown in A–C, see Movies 3–5, which are published as supporting information on the PNAS web site, or go to http://gordonlab.wustl.edu/vasculature. (D) Quantitation of villus capillary network density. Mean values ± SEM are plotted for each condition. The asterisk indicates that the difference relative to germ-free mice is statistically significant (P < 0.005 according to Student's t test). (Bars in A–C, 25 μm.)
B. thetaiotaomicron is a genetically manipulatable, abundant component of the normal human and mouse distal gut microbial society (30, 31). It plays an important role in breaking down otherwise undigestable dietary polysaccharides (9–16, 32). A 10-day colonization of adult germ-free NMRI mice with this Gram-negative anaerobe recapitulates many of the changes in gene expression prompted by colonization with an unfractionated “conventional” distal small intestinal microbiota (ref. 29; see Table 1 or http://gordonlab.wustl.edu/vasculature). It does so without detectable binding to the epithelium (33). B. thetaiotaomicron colonization of 6-week-old germ-free NMRI mice for just 10 days is sufficient to induce a statistically significant (P < 0.005) 2-fold increase in the density of the villus capillary network. The resulting decrease in window size was indistinguishable from that produced by colonization with an unfractionated distal small intestinal microbiota (Fig. 2 B–D).
Paneth Cells Produce Factors That Play a Key Role in the Development of the Villus Microvasculature.
We next assessed the effects of Paneth cells, and of B. thetaiotaomicron, on development of the capillary network by using a pedigree of germ-free FVB/N transgenic mice that lacks this cell type. Lineage ablation was achieved by expressing an attenuated diphtheria toxin A fragment (tox176) under the control of nucleotides −6,500 to +34 of the Paneth-cell-specific mouse cryptdin-2 (CR2) gene (23). tox176 expression reduced the average Paneth cell census per crypt by >95% (data not shown). Electron microscopic studies disclosed that undifferentiated columnar cells (34) replace Paneth cells. Loss of Paneth cells did not affect cell division rates in the crypt, as judged by the number S-phase cells/5-μm-thick sections prepared from normal and transgenic littermates treated with 5-bromo-2′deoxyuridine (BrdUrd) 90 min before sacrifice (100 sectioned crypts scored per mouse; n = 3 mice per group). Similarly, we observed no detectable effects on crypt and villus architecture, or on the differentiation programs of the other three small intestinal epithelial cell lineages, as defined by histochemical stains and immunohistochemical surveys with a panel of previously characterized (35) antibodies and lectins (n = 3 mice studied per group).
The capillary networks of germ-free P28 normal mice and Paneth cell-deficient CR2-tox176 littermates were compared. Without Paneth cells, the network was significantly less complex (2-fold larger average window size in distal small intestinal villi; n = 3 mice per group; P < 0.005; Fig. 3 A, B, and E). This finding indicates that even in the absence of a microbiota, Paneth cells produce factors that play a key role in the development of the villus microvasculature.
B. thetaiotaomicron Stimulates Angiogenesis in the Developing Small Intestine via Paneth Cells.
We then colonized weaning P21 male germ-free normal and CR2-tox176 mice for 7d with B. thetaiotaomicron so that animals could be killed at the conclusion of crypt–villus morphogenesis. Comparisons of the resulting P28 germ-free and B. thetaiotaomicron-mono-associated CR2-tox176 mice revealed that in the absence of Paneth cells, the bacterium was unable to exert an effect on the density of the distal small intestinal villus microvasculature (Fig. 3 A, C, and E). In contrast, in the presence of Paneth cells a 7-day colonization produced a statistically significant 25% increase in capillary network density (P < 0.005; Fig. 3 B, D, and E). The effects of Paneth cells and B. thetaiotaomicron were additive, resulting in a >2-fold difference (P < 0.005) in network density between colonized normal and CR2-tox176 littermates (Fig. 3 C, D, and E).
Discussion
This study indicates that indigenous microbes act as environmental agents that shape development of the intestinal villus microvasculature through Paneth cells. By showing that indigenous microbes play a key role in the postnatal development of host niches they occupy, our findings illustrate of the importance of the coevolution of animals and their microbial partners, as well as the concept of “ecological developmental biology” (20, 21).
The notion of indigenous gut bacteria contributing to vascular development is appealing from the perspective of what constitutes the underpinnings of symbiotic host–microbial relationships. Microbes facilitate luminal breakdown of dietary macromolecules (to the benefit of both partners). By stimulating expression of host genes involved in the uptake of the resulting digestion products (9–16, 32), and by increasing the intestine's absorptive capacity through promotion of angiogenesis, the microbiota provides benefit to the host. Microbial regulation of angiogenesis also allows the marked increase in bacterial density and species complexity that occurs during the postnatal period to be coordinated with an accompanying increase in crypt–villus units. The net result of such coordination is creation of an intestinal “bioreactor” capable of fulfilling the growing nutrient and energetic needs of both partners.
The elegance of this regulatory system is emphasized by the fact that a key cellular component of the innate immune system (Paneth cells) is strategically positioned to coordinate development of both the microbiota and the microvasculature. The appearance of Paneth cells coincides with initial colonization of the gut. Their subsequent differentiation is influenced by the microbiota, while at the same time their secreted antimicrobial peptides/proteins effects microbial ecology (6, 24). Moreover, their placement at the crypt base may allow them to further influence gut development by affecting the properties of the stem cell niche.
Paneth expressed factor(s) may act directly on the microvasculature, or indirectly by influencing expression of regulators produced by other epithelial or mesenchymal cell populations. DNA microarray analysis of laser-capture microdissected epithelial cells harvested from the crypt bases of P28 germ-free normal and CR2-tox176 mice yielded a list of 63 genes whose expression is enriched in Paneth cells (T.S. and J.G., unpublished work). No known regulators of angiogenesis are included in this list, although one gene encodes a newly described (36), microbially regulated (29) member of the angiogenin family of RNases. Further dissection of the microbial, cellular, and molecular components of this angiogenesis pathway may provide important new therapeutic principles for preventing or minimizing ischemic insults to the gut, facilitating villus regeneration after injury, improving absorptive function, and/or treating neoplastic processes. The gnotobiotic normal and transgenic mice described in this report provide a starting point for this pathway dissection in a simplified, genetically and environmentally defined system.
Supplementary Material
Acknowledgments
We thank David O'Donnell, Maria Karlsson, and Sabrina Wagoner for superb technical assistance. We are grateful to Richard Hotchkiss and Mitchell Grayson for suggestions concerning labeling of capillary networks. This work was funded in part by grants from the National Institutes of Health (DK30292 and DK52574) and AstraZeneca. T.S.S. is the recipient of a National Institutes of Health K08 Career Development Grant. L.V.H. is supported by a Burroughs Wellcome Foundation Career Award in the Biomedical Sciences.
Abbreviations
Pn, postnatal day n
References
- 1.Wong M. H., Saam, J. R., Stappenbeck, T. S., Rexer, C. H. & Gordon, J. I. (2000) Proc. Natl. Acad. Sci. USA 97 12601-12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Booth C. & Potten, C. S. (2000) J. Clin. Invest. 105 1493-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng H. (1974) Am. J. Anat. 141 521-536. [DOI] [PubMed] [Google Scholar]
- 4.Bjerknes M. & Cheng, H. (1981) Am. J. Anat. 160 51-63. [DOI] [PubMed] [Google Scholar]
- 5.Ouellette A. J. & Selsted, M. E. (1996) FASEB J. 10 1280-1289. [DOI] [PubMed] [Google Scholar]
- 6.Ayabe T., Satchell, D. P., Wilson, C. L., Parks, W. C., Selsted, M. E. & Ouellette, A. J. (2000) Nat. Immunol. 1 113-118. [DOI] [PubMed] [Google Scholar]
- 7.Vaughan E. E., Schut, F., Heilig, H. G., Zoetendal, E. G., de Vos, W. M. & Akkermans, A. D. (2000) Curr. Issues Intest. Microbiol. 1 1-12. [PubMed] [Google Scholar]
- 8.Hashimoto H., Ishikawa, H. & Kusakabe, M. (1998) Anat. Rec. 250 488-492. [DOI] [PubMed] [Google Scholar]
- 9.Salyers A. A., Vercellotti, J. R., West, S. E. & Wilkins, T. D. (1977) Appl. Environ. Microbiol. 33 319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salyers A. A., West, S. E., Vercellotti, J. R. & Wilkins, T. D. (1977) Appl. Environ. Microbiol. 34 529-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salyers A. A., Harris, C. J. & Wilkins, T. D. (1978) Can. J. Microbiol. 24 336-338. [DOI] [PubMed] [Google Scholar]
- 12.Salyers A. A., Gherardini, F. & O'Brien, M. (1981) Appl. Environ. Microbiol. 41 1065-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Elia J. N. & Salyers, A. A. (1996) J. Bacteriol. 178 7173-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Elia J. N. & Salyers, A. A. (1996) J. Bacteriol. 178 7180-7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reeves A. R., Wang, G. R. & Salyers, A. A. (1997) J. Bacteriol. 179 643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho K. H. & Salyers, A. A. (2001) J. Bacteriol. 183 7198-7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schottelius M. (1902) Arch. Hyg. Bakteriol. 42 48-70. [Google Scholar]
- 18.Steinert M., Hentschel, U. & Hacker, J. (2000) Naturwissenschaften 87 1-11. [DOI] [PubMed] [Google Scholar]
- 19.Hooper L. V. & Gordon, J. I. (2001) Science 292 1115-1118. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert S. F. (2001) Dev. Biol. 233 1-12. [DOI] [PubMed] [Google Scholar]
- 21.McFall-Ngai M. J. (2002) Dev. Biol. 242 1-14. [DOI] [PubMed] [Google Scholar]
- 22.Hooper L. V., Mills, J. C., Roth, K. A., Stappenbeck, T. S., Wong, M. H. & Gordon, J. I. (2002) Methods Microbiol. 31 559-589. [Google Scholar]
- 23.Garabedian E. M., Roberts, L. J. J., McNevin, M. S. & Gordon, J. I. (1997) J. Biol. Chem. 272 23729-23740. [DOI] [PubMed] [Google Scholar]
- 24.Bry L., Falk, P., Huttner, K., Ouellette, A., Midvedt, T. & Gordon, J. I. (1994) Proc. Natl. Acad. Sci. USA 91 10335-10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ushijima T., Takahashi, M., Tatewaki, K. & Ozaki, Y. (1983) Microbiol. Immunol. 27 985-993. [DOI] [PubMed] [Google Scholar]
- 26.Savage D. C. (1977) Annu. Rev. Microbiol. 31 107-133. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt G. H., Winton, D. J. & Ponder, B. A. (1988) Development (Cambridge, U.K.) 103 785-790. [DOI] [PubMed] [Google Scholar]
- 28.Wong M. H., Huelsken, J., Birchmeier, W. & Gordon, J. I. (2002) J. Biol. Chem. 277 15843-15850. [DOI] [PubMed] [Google Scholar]
- 29.Hooper L. V., Wong, M. H., Thelin, A., Hansson, L., Falk, P. G. & Gordon, J. I. (2001) Science 291 881-884. [DOI] [PubMed] [Google Scholar]
- 30.Moore W. E. C. & Holdeman, L. V. (1974) Appl. Microbiol. 27 961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salyers A. A., Bonheyo, G. & Shoemaker, N. B. (2000) Methods 20 35-46. [DOI] [PubMed] [Google Scholar]
- 32.Hooper L. V., Midtvedt, T. & Gordon, J. I. (2002) Annu. Rev. Nutr. 22 283-307. [DOI] [PubMed] [Google Scholar]
- 33.Bry L., Falk, P. G., Midtvedt, T. & Gordon, J. I. (1996) Science 273 1380-1383. [DOI] [PubMed] [Google Scholar]
- 34.Cheng H. & Leblond, C. P. (1974) Am. J. Anat. 141 461-480. [DOI] [PubMed] [Google Scholar]
- 35.Hermiston M. L. & Gordon, J. I. (1995) J. Cell Biol. 129 489-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holloway D. E., Hares, M. C., Shapiro, R., Subramanian, V. & Acharya, K. R. (2001) Protein Expression Purif. 22 307-317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.