Abstract
We describe a strategy to obtain highly enriched long-term repopulating (LTR) hematopoietic stem cells (HSCs) from bone marrow side-population (SP) cells by using a transgenic reporter gene driven by a stem cell enhancer. To analyze the gene-expression profile of the rare HSC population, we developed an amplification protocol termed “constant-ratio PCR,” in which sample and control cDNAs are amplified in the same PCR. This protocol allowed us to identify genes differentially expressed in the enriched LTR-HSC population by oligonucleotide microarray analysis using as little as 1 ng of total RNA. Endoglin, an ancillary transforming growth factor β receptor, was differentially expressed by the enriched HSCs. Importantly, endoglin-positive cells, which account for 20% of total SP cells, contain all the LTR-HSC activity within bone marrow SP. Our results demonstrate that endoglin, which plays important roles in angiogenesis and hematopoiesis, is a functional marker that defines LTR HSCs. Our overall strategy may be applicable for the identification of markers for other tissue-specific stem cells.
In mammals, hematopoietic stem cells (HSCs) are responsible for the daily production of billions of mature cells of all blood lineages throughout adult life. HSCs are defined by their ability to self-renew and differentiate into all blood cell types (1–3). Their extraordinary ability to self-renew and differentiate was demonstrated by repopulation of the entire blood system by a single stem cell (3, 4). To better understand the mechanisms that control HSC self-renewal and differentiation, it is essential to identify and purify these cells unambiguously and identify genes that are involved specifically in regulating HSC activity. This is a difficult task because of the extremely low frequency of HSCs in normal bone marrow, ≈1 in 105 (5). A variety of methods have been developed to enrich this rare pluripotent HSC population. For example, mouse bone marrow HSCs can be enriched significantly as c-Kit, Sca-1, Thy-1-positive, lineage marker low or negative (c-Kitpos, Thy-1pos, Sca-1pos, Linneg/low) cells (6). HSCs can be enriched also as side-population (SP) cells based on the differential efflux of the fluorescent dye Hoechst 33342 (7). Despite significant enrichment for HSC activity, the c-Kitpos, Thy-1pos, Sca-1pos, Linneg/low and SP cells remain heterogeneous (7, 8), and additional surface markers are required to further enrich HSCs to homogeneity. Moreover, markers that are functionally important for HSC activity may shed light on the molecular mechanisms that control HSC self-renewal and differentiation.
The paucity and heterogeneity of the HSC population has greatly hindered systematic efforts to identify HSC-specific genes. A number of laboratories have used large-scale cDNA subtraction and microarray hybridization approaches to address this question (9, 10). These technologies are extremely laborious and time-consuming. Thus far, no applicable markers that can be used to purify long-term repopulating (LTR) HSCs have resulted from these screens. The application of cDNA/oligonucleotide microarray technology to identification of HSC-specific genes has been prohibited by the amount of RNA required: 50–100 μg of total RNA and 2–5 μg of poly(A) RNA (11). A number of amplification protocols, particularly antisense RNA (aRNA) amplification, were used for amplifying limiting amount of RNA for microarray analysis (12, 13). However, application of these protocols to amplify extremely low amounts of RNA (<10 ng) for oligonucleotide microarray analysis was not assessed (14). Recently two groups independently carried out comprehensive expression profile analyses on enriched HSC populations and other stem cells (15, 16). Although they generated a number of genes that appear to be differentially expressed in HSCs or other stem cell types, their differential expression on actual stem cells was not validated (15, 16).
Here we describe an approach to enrich HSCs from the SP cells by using a transgenic stem cell marker and a method for identifying stem cell-specific genes by oligonucleotide microarray analysis. The strategy was validated by the identification and demonstration of endoglin as a functional surface marker that defines LTR HSCs. These results suggest that the overall strategy can be applied for the identification of markers for other tissue-specific stem cells.
Materials and Methods
Mouse Strains.
Nonobese diabetic/severe combined immunodeficient (NOD-SCID) mice, FVB, and C57BL/6J (ly5.2),B6/SJL (Ly5.1) congenic mice, 6–8 weeks of age, were purchased from The Jackson Laboratory. The transgenic construct 6E5-βgeo-3′En was generated by replacing the lacZ gene in the pGL2–6E5/lacZ/3′En vector (17) with the βgeo (lacZneo) reporter gene. Transgenic mice were analyzed by whole-mount 5-bromo-4-chloro-3-indolyl β-D-galactoside staining and fluorescence-activated cell sorter (FACS) using the β-galactosidase substrate fluorescein di-β-galactopyranoside (FDG) as described (17). Irradiated animals were provided with antibiotic and acidified water for the first 2 weeks posttransplantation. All animals were maintained at the Whitehead Institute animal facility according to institutional guidelines.
FACS Analysis.
Murine bone marrow was obtained from the femurs and tibias. For detection of SP cells, bone marrow cells were stained with Hoechst 33342 dye (Sigma) as described (7). For antibody staining, primary antibodies were typically added at 1/50 to 1/100 dilutions. Secondary staining reagents including fluorescent conjugated secondary antibodies and streptavidin were titrated individually for minimal nonspecific signal. Antibody staining was carried out for 15 min on ice (for endoglin staining) or at 4–8°C. When multiple antibody staining was required, primary and secondary staining for endoglin was carried out first. R-phycoerythrin- or allophycocyanin-conjugated goat anti-rat antibodies were used as the secondary staining reagent for the antiendoglin antibody (rat MJ7/18). In case of multiple staining, the staining order was Hoechst 33342, FDG, antiendoglin/secondary antibodies, lineage mixture, and other antibodies. Lineage antibodies included anti-CD11b (M1/70), anti-Gr-1 (RB6-8C5), anti-B220 (RA3-6B2), anti-CD3ɛ (145-2C11), and anti-Ter119 (Ter119). Other antibodies used in this study included anti-CD4 (GK1.5), anti-CD8a (53-6.7), anti-Cd45.1 (A20), anti-CD45.2 (104), anti-Sca-1 (E13–161.7), and anti-c-Kit.
Constant Ratio (CR)-PCR and Chip Hybridization.
For details see Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Competitive Repopulation Assay.
The competitive repopulation assay was performed by using the congenic Ly5.1/Ly5.2 system as described (18). Total bone marrow cells (2 × 105 cells) from female C57/BL6 (Ly5.2) mice were used as competitors. NOD-SCID mice ≈8 weeks old were sublethally irradiated (350–400 Gy) and used to measure HSC activity of cells from stem cell leukemia (SCL)-βgeo-3′En transgenic mice (FVB). Transplanted mice were analyzed for donor cell contribution by retroorbital bleeding at various times (4, 16, or 28 weeks) posttransplantation. Nucleated cells were stained with appropriate conjugated Ly5.1 and Ly5.2 antibodies to determine the percentage of donor contribution in the recipient mice. The cells were stained simultaneously with R-phycoerythrin-conjugated anti-B220, a mixture of R-phycoerythrin-conjugated anti-Mac-1 and anti-Gr-1 antibodies, or a mixture of R-phycoerythrin-conjugated anti-CD4 and anti-CD8 antibodies to determine multilineage repopulation. Competitive repopulation units (CRU) per 105 were calculated by the formula [%Ly5.1/(100 − %Ly5.1)] × (2 × 105 cells per number of cells injected).
Results
Enrichment of HSCs from Bone Marrow SP Cells with a Transgenic Stem Cell Marker.
Although a combination of surface markers can be used to enrich LTR HSCs, such markers are rarely available for the purification of other adult stem cells. Nevertheless, many types of adult stem cells from a variety of tissues such as bone marrow (7), muscle (19), neurosphere (20), and testis can be enriched as SP cells based on the differential efflux of the fluorescent dye Hoechst 33342. This suggests that the conserved SP phenotype may represent a functional feature shared by stem cells of different tissue origin (21). However, SP cells purified from bone marrow and other tissues are heterogeneous and may contain progenitors or differentiated cells or adult stem cells of different tissue origins (22). We hypothesized that a transgenic stem cell marker, a reporter transgene that recapitulates the in vivo expression of a functionally defined critical regulator of stem cell formation and maintenance, may be used to enrich correspondent tissue-specific stem cells from SP cells.
To test this hypothesis we selected the SCL gene as a functional genetic marker to enrich HSCs from bone marrow SP cells. SCL, a basic helix–loop–helix transcription factor, is essential for mouse primitive and definitive hematopoiesis and also plays an important role in endothelial development (23). Detailed analysis of the regulation of the mouse SCL gene identified one 3′ enhancer (3′En) that directed expression of a linked transgene specifically to HSCs/progenitor cells (17, 24). Therefore, we generated transgenic mice using the reporter construct 6E5-βgeo-3′En (SCLβgeo) in which βgeo expression is controlled by the SCL promoter (6E5) and the 3′En. As shown by whole-mount 5-bromo-4-chloro-3-indolyl β-D-galactoside embryo staining (17), the selected founder line no. 336 showed predominant expression of the SCLβgeo transgene in embryonic day 11.5–12.5 fetal liver (Fig. 1 A and B). Further analysis by FDG staining (17) showed ≈1–2% of bone marrow cells from the transgenic mice was positive for the SCLβgeo transgene (Fig. 1C). More than 30% of the FDG-positive bone marrow cells were lineage-negative or -low (data not shown), suggesting that SCLβgeo-positive cells were enriched for hematopoietic progenitor or stem cells.
Fig 1.
Use of the SCLβgeo-3′En transgene as a functional genetic marker to enrich HSCs from mouse bone marrow. (A and B) 5-Bromo-4-chloro-3-indolyl β-d-galactoside whole-mount staining of embryonic day 12.5 mice. Compared with the wild-type embryo (A), an embryo carrying the 6E5-βgeo-3′En transgene (SCLβgeo) shows predominant expression of lacZ in the fetal liver (B). Approximately 1–2% of mouse bone marrow cells are positive for the SCLβgeo transgene (C). (D–F) Expression of the SCLβgeo transgene in mouse bone marrow analyzed by staining with Hoechst 33342 and the β-galactosidase substrate FDG. SP cells account for ≈0.06% of bone marrow cells (D). Among SCLβgeo transgene-positive cells (FDG-positive gated), ≈1.5% are SP cells (E). (F) SCLβgeo and lineage-expression profile of the bone marrow SP cells (SP cells gated); among all the gated SP cells, ≈30% are positive for the SCLβgeo transgene and approximately half of these are lineage-negative or -low.
To examine whether the SCLβgeo reporter can enrich HSCs from bone marrow SP cells, we first stained bone marrow cells with Hoechst 33342 dye and FDG. The percentage of SP cells among the SCLβgeo-positive bone marrow cells (≈1.5%) is ≈25-fold higher than that in total bone marrow (≈0.06%), suggesting that SCLβgeo-positive bone marrow cells are enriched for HSCs (Fig. 1 D and E). Interestingly, we observed that ≈30% of the bone marrow SP cells expressed the SCLβgeo transgene (Fig. 1F), indicating that the SCLβgeo reporter can fractionate SP cells into two distinctive populations. Because the 3′En was able to target reporter gene expression to both adult and embryonic LTR HSCs (17, 24), HSC activity is likely enriched in the SCLβgeopos SP cells but not in the SCLβgeoneg SP cell population. Furthermore, we found that SCLβgeopos SP cells are heterogeneous for lineage marker expression (Fig. 1F). Because LTR HSCs are lineage-negative or -low (25), the SCLβgeopos SP Linneg/low cells, which account for 0.01% nucleated bone marrow cells, are likely more enriched for LTR HSCs.
To estimate the frequency of HSCs within the SCLβgeopos SP Linneg/low cell population, we carried out competitive repopulating assays using NOD-SCID mice as recipients. We injected 10 SCLβgeopos SP Linneg/low cells along with 2 × 105 competitor cells from NOD-SCID mice into each of 10 recipient mice. Because NOD-SCID mice express the Ly5.1 antigen and the transgenic mice (FVB strain) express the Ly5.2 antigen, donor white blood cell (WBC) contributions can be quantified by staining Ly5.1 and Ly5.2 antigens. Two months posttransplantation, we analyzed the donor WBC contribution of the seven surviving recipients. The percentages of donor (Ly5.2) WBCs in these recipients were 1.1%, 0%, 0.8%, 1.12%, 0%, 2.03%, and 25.83%, respectively. This result demonstrated that the SCLβgeopos SP Linneg/low cells indeed are highly enriched for LTR-HSC activity.
Identification of HSC-Specific Genes by CR-PCR and Oligonucleotide Microarray Analysis.
To provide control cell populations for SCLβgeopos SP Linneg/low cells, we fractionated lineage-negative or -low bone marrow cells into four cell populations by using the SCLβgeo marker and the SP phenotype: (i) SCLβgeopos SP Linneg/low, (ii) SCLβgeoneg SP Linneg/low, (iii) SCLβgeopos non-SP Linneg/low, and (iv) SCLβgeoneg non-SP Linneg/low. These populations represent ≈0.01, 0.01, 0.6, and 0.6% of total bone marrow cells, respectively. Linneg/low cells, which comprise ≈2% of total bone marrow cells, are quite heterogeneous and are mostly early myeloid or lymphoid progenitor cells. No LTR activity was found in SCLβgeoneg cell fractions (data not shown). The frequency of HSCs in SCLβgeopos non-SP Linneg/low cells, as measured by the CRU frequency, was ≈38 times lower than that in SCLβgeopos SP Linneg/low cells (data not shown). Therefore, genes specifically expressed in SCLβgeopos SP Linneg/low cells but not the other three populations are likely to be HSC-specific.
The frequency of SCLβgeopos SP Linneg/low cells is only ≈0.01% of nucleated bone marrow cells. Because of the low abundance of the these cells and the toxic nature of the Hoechst 33342- and FDG-staining procedures, only ≈1,000 SCLβgeopos SP Linneg/low cells could be isolated by FACS from the bone marrow of one transgenic mouse. Less than 1 ng of total RNA was obtained from 1,000 cells. Significant amplification is needed to identify genes that are differentially expressed in SCLβgeopos SP Linneg/low cells by oligonucleotide microarray analysis. To maintain the ratio of individual genes between samples during PCR amplification, we developed an amplification protocol, termed CR-PCR and depicted in Fig. 2. The key feature of CR-PCR is that sample and control cDNAs are amplified in the same PCR such that the ratio of individual genes between the sample and controls will not be skewed.
Fig 2.
Schematic diagram of CR-PCR and its application in gene-expression profiling using oligonucleotide microarrays. The key feature of this protocol is that sample and control cDNAs are amplified in the same PCR such that the ratio of individual genes between the sample and control RNAs will not be skewed during amplification. The poly(T) primers consist of oligo(dT), unique sequences (T-1, dark blue; T-2, dark orange), and universal forward sequences (black). These primers are designed to label an individual RNA sample with a specific sequence tag (T1 or T2) in the RT step. A universal primer (SMART oligo, dark green with GGG at the 3′ end) was added to the end of the newly synthesized cDNA by a template-switching mechanism (33). Thus the cDNA products contain common 5′ and 3′ flanking sequences (universal forward, black/gray; universal reverse, dark green/light green) that allow PCR amplification. Equal amounts of RT products from each sample were mixed and amplified with the universal forward and reverse PCR primers. Amplified samples were split into two (or more) equal fractions. A specific T7 promoter containing either the T1 or T2 sequence tag (T7-1 or T7-2) matched to the corresponding poly(T) primer [poly(T-1) or poly(T-2)] was used to selectively add a T7 promoter to the corresponding fraction of cDNAs in the mixture by linear extension reaction using DNA polymerase. cRNA samples were generated by in vitro transcription and subjected to oligonucleotide microarray analysis. Differentially expressed genes were identified based on the fluorescent intensity ratio of individual genes. Throughout, complementary sequences are shown as a lighter color of the matching strand (e.g., light blue versus dark blue).
Two sets of CR-PCRs were established to identify genes that are differentially expressed in SCLβgeopos SP Linneg/low cells, the enriched HSC population. cDNAs were synthesized by using 1 ng of total RNA from each cell population. The first PCR contained cDNAs from SCLβgeopos SP Linneg/low (HSC) and SCLβgeoneg SP Linneg/low cells. The second PCR contained cDNAs from SCLβgeopos SP Linneg/low, SCLβgeopos non-SP Linneg/low, and SCLβgeoneg non-SP Linneg/low cells. PCR amplifications were done in duplicates, and samples were amplified and subjected to oligonucleotide microarray hybridization.
To identify HSC-specific candidate genes, we focused on those abundantly expressed in SCLβgeopos SP Linneg/low cells but not in the other three cell populations; a selected set is shown in Table 1. Among those genes, c-fos was identified previously as an HSC-specific gene in a cDNA subtraction and microarray screen (19) and may play an important role in maintaining stem cells in a dormant state (26). Moreover, c-fos is also differentially expressed in the population of human CD34pos CD38neg cells that are enriched in primitive HSCs but not in CD34pos CD38pos cells that do not contain primitive HSCs (27, 28).
Table 1.
Genes differentially expressed in the enriched HSC population
| GenBank accession no.
|
Gene description
|
Amplification set 1 | Amplification set 2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| (1) SCLβgeo+ SP Lin− | (2) SCLβgeo− SP Lin− | Fold (1)/(2) | (3) SCLβgeo+ SP Lin− | (4) SCLβgeo+ non-SP Lin− | (5) SCLβgeo− non-SP Lin− | Fold (3)/(4) | Fold (3)/(5) | ||
| AI840975 | EST | 1,383.45 | 9.58 | 144 | 1,673.96 | 18.7 | 3.79 | 90 | 442 |
| X77952 | Endoglin | 996.39 | 78.36 | 13 | 3,501.58 | 48.71 | 90.36 | 72 | 39 |
| J05479 | Calcineurin | 746.15 | 36.58 | 20 | 1,016.78 | 18.84 | 2.48 | 54 | 410 |
| V00727 | c-fos | 4,645.57 | 143.68 | 32 | 433.82 | 16.97 | 8.01 | 26 | 54 |
| AW125272 | EST | 5,781.49 | 263.45 | 22 | 1,640.78 | 227.01 | 55.24 | 7 | 30 |
| AI853703 | EST | 234.64 | 9.67 | 24 | 2,003.22 | 156.98 | 214.93 | 13 | 9 |
| AV293396 | EST | 2,107.39 | 208.26 | 10 | 313.34 | 43.26 | 24.95 | 7 | 13 |
Columns (1)–(5) show the normalized fluorescence intensity for the indicated genes in the indicated sample. “Fold” lists the ratios of fluorescence intensity between the indicated samples and is a measure of enrichment of these mRNAs in the SCLβgeopos SP Linneg/low cells relative to the indicated control cell population.
Verification of Endoglin Expression on Highly Enriched HSCs by FACS Analysis.
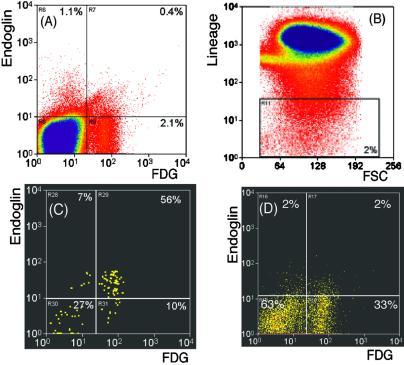
We also found that endoglin, an ancillary transforming growth factor β receptor, was highly differentially expressed on SCLβgeopos SP Linneg/low cells but not in the other cell populations (Table 1). To verify this, we used antibody staining and FACS analysis to examine surface endoglin expression on SCLβgeopos SP Linneg/low cells and the three control cell populations. To this end bone marrow cells from SCLβgeo transgenic mice were stained with Hoechst 33342, FDG, antiendoglin antibody, and antibodies against lineage markers. Approximately 0.4% of the total bone marrow cells are positive for endoglin and SCLβgeo (Fig. 3A). As before, SP cells comprised ≈0.06% of total bone marrow cells. The Linneg/low gate was set such that these cells account for ≈2% of the total bone marrow cells (Fig. 3B).
Fig 3.
FACS analysis shows that endoglin is differentially expressed on the enriched HSC SCLβgeopos SP Linneg/low population. Bone marrow cells from SCLβgeo transgenic mice were stained with antiendoglin antibody, Hoechst 33342, and lineage markers. (A) Endoglin and SCLβgeo expression in total bone marrow cells. (B) Gate for Linneg/low cells. FSC, forward scatter. (C) Endoglin and SCLβgeo expression profile of gated Linneg/low SP cells. (D) Endoglin and SCLβgeo expression profile of gated Linneg/low non-SP cells.
We then examined the expression of endoglin and SCLβgeo in gated SP and Linneg/low cells (Fig. 3C) or gated non-SP and Linneg/low cells (Fig. 3D). As shown in Fig. 3C, 66% (=56 + 10) of the SP Linneg/low cells were SCLβgeo-positive, and 85% (=56/66) of these SCLβgeopos SP Linneg/low cells also expressed endoglin. In contrast, 34% (=27 + 7) of the SP Linneg/low cells were SCLβgeo-negative, and only 20% (=7/34) of these SCLβgeoneg SP Linneg/low cells were positive for endoglin (Fig. 3C). If all the endoglin-positive (Endopos) cells express equal amounts of endoglin protein and mRNA, these FACS data indicate that endoglin expression on SCLβgeopos SP Linneg/low cells is 4.3-fold that in SCLβgeoneg SP Linneg/low cells. The mRNA ratio determined by array analysis is 13-fold (Table 1). Of the non-SP Linneg/low cells (Fig. 3D), 35% (=33 + 2) were SCLβgeo-positive, and only 6% (=2/35) of these SCLβgeopos non-SP Linneg/low cells also expressed endoglin. Similarly, 65% (=63 + 2) of the non-SP Linneg/low cells were SCLβgeo-negative, and 3% (=2/65) of SCLβgeoneg non-SP Linneg/low cells were positive for endoglin (Fig. 3D). Applying the same calculation, endoglin expression on SCLβgeopos SP Linneg/low cells should be 14-fold of that of SCLβgeopos non-SP Linneg/low cells and 28-fold of that of SCLβgeoneg non-SP Linneg/low cells, respectively. The mRNA ratios determined by array analysis were 72 and 39, respectively (Table 1).
Thus, the FACS analysis confirmed the ratio of endoglin expression in the enriched HSC population versus the other control population, suggesting that endoglin is highly differentially expressed on the surface of the enriched HSC population. This result further demonstrated that CR-PCR can maintain the ratio of differentially expressed genes when amplifying as little as 1 ng of total RNA. Because our competitive repopulation studies showed that SCLβgeopos SP Linneg/low cells are highly enriched for HSC activity, these results suggest that endoglin is likely to be a marker for HSCs within the SP cells.
Endopos SP Cells Contain Essentially All the LTR-HSC Activity Among SP Cells.
Although endoglin is expressed specifically on the SCLβgeopos SP Linneg/low cells, it marks a slightly smaller cell population of the total SP cells than does SCLβgeo. Approximately 30% and 20% of SP cells were positive for the SCLβgeo marker or endoglin expression, respectively (Fig. 4B). As indicated by the lineage profile of Endopos SP and SCLβgeopos SP cells (Fig. 4 A and B), this is largely due to the difference in lineage-marker expression between Endopos SP and SCLβgeopos SP cells. The majority of Endopos SP cells also expressed the SCLβgeo transgene (Endopos cells are shown as yellow dots in Fig. 4 A and B). Endopos SP cells are mostly lineage-negative or -low (Fig. 4B). In contrast, ≈33% of the SCLβgeopos SP cells are predominantly lineage-high and are endoglin-negative (Endoneg, red dots on right side of Fig. 4A), and the SCLβgeopos SP cells that are endoglin-positive (yellow dots in right side of Fig. 4A) are all Linneg/low.
Fig 4.
Endopos cells are enriched for LTR-HSC activity in bone marrow SP cells. (A and B) Correlation of endoglin and SCLβgeo expression in bone marrow SP cells. (A) Lineage and SCLβgeo (FDG) profile of bone marrow SP cells. (B) Lineage and endoglin profile of bone marrow SP cells. In both A and B Endopos SP cells are depicted as yellow dots and Endoneg SP cells are red dots. Arbitrary gates were set for lineage-marker expression: neg, low, and high. The blue quadrant in B was the sorting gate used to divide bone marrow SP cells into four cell populations: Endopos Linneg, Endopos Linlow, Endoneg Linneg, and Endoneg Linlow/hi cells. (C) Competitive repopulation analysis of Endopos SP cells (solid orange line and error bar) and Endoneg SP cells (dotted green line and error bar). Shown here is the donor contribution (% Ly5.1) by Endopos SP cells and Endoneg SP cells at 4, 16, and 26 weeks posttransplantation. One hundred Endopos SP or 375 Endoneg SP cells were injected into each recipient. (D) Competitive repopulation analysis of Endopos SP Linneg (orange bar) and Endopos SP Linlow (green bar) cells. Shown here is the CRU per 105 donor cells in the Endopos SP Linneg (orange bar) and Endopos SP Linlow (green bar) groups at 4, 14, and 28 weeks posttransplantation. Forty-two Endopos SP Linneg and 139 Endopos SP Linlow cells were injected into each recipient.
To demonstrate that endoglin is indeed a marker for LTR HSCs, we examined the reconstitution potential of Endopos SP and Endoneg SP cells using a competitive repopulation assay (29). We injected 100 Endopos SP cells or 375 Endoneg SP cells isolated from a Ly5.1 donor along with 2 × 105 Ly5.2 competitor cells into lethally irradiated congenic Ly5.2 mice. The number of Endopos SP and Endoneg SP cells injected was proportional to their abundance in the bone marrow SP-cell population. At least five recipient mice were used for each cell sample. We observed distinct reconstitution kinetics in the recipients that received Endopos SP or Endoneg SP cells, as indicated by the fraction of white blood cells expressing the donor Ly5.1 antigen (Fig. 4C). At 4 weeks posttransplantation, the donor contribution by 100 Endopos SP and 375 Endoneg SP cells was comparable (Fig. 4C). However, the contribution by Endoneg SP cells decreased significantly after 4 weeks, whereas the contribution by Endopos SP cells increased dramatically (Fig. 4C). The distinct reconstitution kinetics indicated that the Endoneg SP cells contain mostly short-term HSCs, which can only maintain transient reconstitution. Significant long-term multilineage reconstitution, as indicated by the percentage chimerism in T-lymphoid, B-lymphoid, and myeloid cells, was seen only in recipient mice that received Endopos SP cells (not shown). Our results established that Endopos SP cells contain essentially all the LTR activity within bone marrow SP cells (Fig. 4C). Because Endopos SP cells account for ≈20% of total SP cells (Fig. 4B), a 5-fold enrichment of LTR HSCs from the SP cells was achieved by using endoglin as a marker.
Some of the Endopos SP cells are still expressing lineage markers at a low level. We suspected that Endopos SP Linneg cells may be enriched further for LTR HSCs and that the Endopos SP Linlow cells may contain more short-term HSCs (25). We fractionated SP cells into Endoneg Linneg, Endoneg Linlow/hi, Endopos Linlow, and Endopos Linneg fractions (Fig. 4B, blue lines); these represent ≈3%, 50%, 34%, and 13% of the SP cells, respectively. No LTR-HSC activity was found in the two endoglin-negative fractions (data not shown). We injected 139 Endopos Linlow SP or 42 Endopos Linneg SP cells into lethally irradiated mice along with 2 × 105 Ly5.2 competitor cells. The number of cells injected was proportional to the cell distribution within each quadrant. At 4 weeks posttransplantation, Endopos Linlow SP and Endopos Linneg SP cells contained an equivalent amount of CRU activity per 105 cells (Fig. 4D). However, at 14 weeks posttransplantation, the competitive repopulating activity of the Endopos Linneg SP cells was >2-fold that of the Endopos Linlow SP cells. Notably, the CRU activity per 105 cells in the Endopos Linlow SP cells decreased significantly from 4 to 28 weeks posttransplantation, whereas that of the Endopos Linneg SP cells increased (Fig. 4D). The distinct reconstitution kinetics indicated that, as anticipated, the Endopos SP Linlow cells contained a significant number of short-term HSCs, but the Endopos SP Linneg cells did not (Fig. 4D). Based on this study, one can further enrich for LTR HSCs by selecting the lineage-negative fraction of Endopos SP cells.
To evaluate the self-renewal potential of Endopos SP cells, we carried out secondary transplantations using Ly5.1 SP cells isolated from primary Ly5.2 recipients effectively reconstituted with Ly5.1 Endopos SP cells at 14 weeks posttransplantation. Each lethally irradiated secondary recipient was injected with 400 bone marrow Ly5.1 SP cells from the primary recipient along with 2 × 105 Ly5.2 competitor bone marrow cells. Four of the five secondary recipients were reconstituted with Ly5.1 SP cells (1.8 ± 0.6% of donor contribution). These results confirmed that Endopos SP cells are enriched for LTR HSCs that are capable of self-renewal.
Discussion
Here we developed a strategy to isolate highly enriched HSCs from the bone marrow SP cells using a transgenic stem cell reporter driven by the SCL 3′En (17, 24). Furthermore, we developed an amplification protocol, CR-PCR, that made it possible to analyze the gene-expression profile by oligonucleotide microarray analysis using as little as 1 ng of total RNA. With these technical advances we identified endoglin as a protein differentially expressed on SCLβgeopos SP Linneg/low cells, a highly enriched HSC population. Most importantly, we demonstrated that endoglin is a functional marker for LTR HSCs and can be used to enrich ≈5-fold LTR HSCs from SP cells. These results validate our approach for enrichment of HSCs and identification of stem cell-specific genes or markers and suggest that our overall strategy may be applicable for the identification of markers for other types of tissue-specific stem cells.
Isolation of highly enriched LTR HSCs by fractionating bone marrow SP cells with a transgenic stem cell reporter represents an alternative HSC-purification approach that requires minimal knowledge about specific surface markers. This approach was validated by the identification of endoglin as a marker for LTR HSCs. Endoglin was not found in two previous large-scale expression analyses using HSCs enriched by conventional surface markers (9, 10). Moreover, because the SP phenotype is shared by many other adult stem cell types, our results also suggest that a similar strategy can be devised to enrich stem cells of different tissue origins with surface markers that are largely unknown.
The CR-PCR protocol, in which coamplification of cDNA samples was used to maintain the ratio of individual genes in different RNA samples, was also key to the identification of endoglin as an HSC-specific marker. This protocol allows the use of DNA microarrays to identify differentially expressed genes even when mRNA is available from only a minute number of cells, in our case ≈1,000 cells (≈1 ng of total RNA). As evidence, we were able to identify endoglin as a gene that is specifically expressed on the majority of SCLβgeopos SP Lin neg/low cells but not in three control bone marrow cell populations using 1 ng of total RNA from each population. Compared with the two-round in vitro-transcription protocol used in other studies (15, 16), the CR-PCR amplification protocol requires at least 10–50 times less RNA. Moreover, the relative abundance of endoglin expression in the HSC-enriched populations versus control cell groups were comparable at the mRNA level, as determined by array analysis, and at the level of cell-surface protein expression (Table 1 and Fig. 3). This result demonstrated that CR-PCR can maintain the ratio of individual genes during amplification. In two recent studies endoglin was identified as one of many genes expressed in enriched HSC populations. In one, endoglin was found to be highly expressed on many bone marrow cell populations, as shown by quantitative PCR (15). In the other, gene expression in LTR HSCs was not compared with that of any differentiated hematopoietic progenitor cells (16).
As with all methodologies, the CR-PCR protocol has its limitations. We noticed that many genes were undetectable on the chips, presumably because of inefficient PCR amplification of these transcripts. We also observed relatively low replicate correlation coefficients (≈0.65) that may be due to the quantum effects in amplifying low numbers of mRNA molecules isolated from very small numbers of cells. Variations in the early manipulations are amplified during the later PCR steps. Despite these limitations, we demonstrated that CR-PCR did provide a simple protocol that can be used to discover differentially expressed genes by oligonucleotide microarray analysis when the number of cells available is extremely limited.
Differential expression of endoglin in the enriched LTR-HSC population but not the control populations (Fig. 3) suggested that endoglin could be used as an effective surface marker to enrich LTR HSCs. Indeed, we demonstrated that endoglin can be used to further enrich LTR HSCs from bone marrow SP cells. This extends earlier work, suggesting that endoglin may have important functions in angiogenesis and hematopoiesis. Mutations in the endoglin gene that lead to haploinsufficiency are associated with the inherited human disorder hereditary hemorrhagic telangiectasia type 1, a disease characterized by bleeding caused by vascular malformations (30). Endoglin null embryos die at embryonic day 10.0–10.5 because of defects in blood vessel and heart development and show severe defects in yolk-sac hematopoiesis (31). A recent study of in vitro differentiation of Endo−/− embryonic stem cells suggested that endoglin also may be important in definitive hematopoiesis, particularly myelopoiesis and erythropoiesis (32). Together with our work showing specific expression of endoglin on LTR HSCs, this result indicates that endoglin and transforming growth factor β signaling may play important roles in regulating HSC self-renewal and/or differentiation. However, because endoglin null mice die around embryonic day 10.5, the time when adult repopulating cells appear, it is unclear whether endoglin plays an essential role in regulating HSC self-renewal and differentiation (31). To definitively establish a role of endoglin in regulating HSC function it may be necessary to examine the developmental potential of endoglin null embryonic stem cells in chimeric mice. Nonetheless, endoglin is a functional marker that defines and enables enrichment of LTR HSCs.
Note Added in Proof.
It was noted recently by another group that endoglin may be differentially expressed in enriched HSCs (34).
Supplementary Material
Acknowledgments
We thank Glenn Paradis and Michael Doire (Flow Cytometry Core Facility, Center for Cancer Research, Massachusetts Institute of Technology) for invaluable help with FACS sorting and C. Ladd (Whitehead Genome Center) and Dr. K. Chen (Transgenic Facility, Albert Einstein Medical School, New York) for excellent technical assistance. We also thank Drs. M. Xun, G. Wang, S. Ghaffari, C. Zhang, and B. Luo for helpful discussion and critical reading of the manuscript. Support to H.F.L. was from the Engineering Research Centers Program of the National Science Foundation under Award EEC 9843342 through the Biotechnology Process Engineering Center (Massachusetts Institute of Technology). Work in the A.R.G. laboratory is supported by the Wellcome Trust, Leukaemia Research Fund, and Medical Research Council. This work is supported in part by Affymetrix and Bristol–Myers Squibb. C.-Z.C. holds a Cancer Research Institute/Donaldson, Lufkin, and Jenrette postdoctoral fellowship.
Abbreviations
HSC, hematopoietic stem cell
SP, side population
LTR, long-term repopulating
NOD-SCID, nonobese diabetic/severe combined immunodeficient
FACS, fluorescence-activated cell sorter
FDG, di-β-galactopyranoside
CR, constant ratio
SCL, stem cell leukemia
CRU, competitive repopulation units
References
- 1.Abramson S., Miller, R. G. & Phillips, R. A. (1977) J. Exp. Med. 145 1567-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Till J. E. & McCulloch, E. A. (1961) Radiat. Res. 14 213-222. [PubMed] [Google Scholar]
- 3.Jordan C. T. & Lemischka, I. R. (1990) Genes Dev. 4 220-232. [DOI] [PubMed] [Google Scholar]
- 4.Osawa M., Hanada, K., Hamada, H. & Nakauchi, H. (1996) Science 273 242-245. [DOI] [PubMed] [Google Scholar]
- 5.Zhong R. K., Astle, C. M. & Harrison, D. E. (1996) J. Immunol. 157 138-145. [PubMed] [Google Scholar]
- 6.Spangrude G. J., Heimfeld, S. & Weissman, I. L. (1988) Science 241 58-62. [DOI] [PubMed] [Google Scholar]
- 7.Goodell M. A., Brose, K., Paradis, G., Conner, A. S. & Mulligan, R. C. (1996) J. Exp. Med. 183 1797-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uchida N., Fleming, W. H., Alpern, E. J. & Weissman, I. L. (1993) Curr. Opin. Immunol. 5 177-184. [DOI] [PubMed] [Google Scholar]
- 9.Phillips R. L., Ernst, R. E., Brunk, B., Ivanova, N., Mahan, M. A., Deanehan, J. K., Moore, K. A., Overton, G. C. & Lemischka, I. R. (2000) Science 288 1635-1640. [DOI] [PubMed] [Google Scholar]
- 10.Park I. K., He, Y., Lin, F., Laerum, O. D., Tian, Q., Bumgarner, R., Klug, C. A., Li, K., Kuhr, C., Doyle, M. J., et al. (2002) Blood 99 488-498. [DOI] [PubMed] [Google Scholar]
- 11.Duggan D. J., Bittner, M., Chen, Y., Meltzer, P. & Trent, J. M. (1999) Nat. Genet. 21 10-14. [DOI] [PubMed] [Google Scholar]
- 12.Luo L., Salunga, R. C., Guo, H., Bittner, A., Joy, K. C., Galindo, J. E., Xiao, H., Rogers, K. E., Wan, J. S., Jackson, M. R. & Erlander, M. G. (1999) Nat. Med. 5 117-122. [DOI] [PubMed] [Google Scholar]
- 13.Phillips J. & Eberwine, J. H. (1996) Methods 10 283-288. [DOI] [PubMed] [Google Scholar]
- 14.Wang E., Miller, L. D., Ohnmacht, G. A., Liu, E. T. & Marincola, F. M. (2000) Nat. Biotechnol. 18 457-459. [DOI] [PubMed] [Google Scholar]
- 15.Ivanova N. B., Dimos, J. T., Schaniel, C., Hackney, J. A., Moore, K. A. & Lemischka, I. R. (2002) Science 298 601-604. [DOI] [PubMed] [Google Scholar]
- 16.Ramalho-Santos M., Yoon, S., Matsuzaki, Y., Mulligan, R. C. & Melton, D. A. (2002) Science 298 597-600. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez M., Gottgens, B., Sinclair, A. M., Stanley, M., Begley, C. G., Hunter, S. & Green, A. R. (1999) Development (Cambridge, U.K.) 126 3891-3904. [DOI] [PubMed] [Google Scholar]
- 18.Micklem H. S., Ford, C. E., Evans, E. P., Ogden, D. A. & Papworth, D. S. (1972) J. Cell. Physiol. 79 293-298. [DOI] [PubMed] [Google Scholar]
- 19.Jackson K. A., Mi, T. & Goodell, M. A. (1999) Proc. Natl. Acad. Sci. USA 96 14482-14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hulspas R. & Quesenberry, P. J. (2000) Cytometry 40 245-250. [PubMed] [Google Scholar]
- 21.Zhou S., Schuetz, J. D., Bunting, K. D., Colapietro, A. M., Sampath, J., Morris, J. J., Lagutina, I., Grosveld, G. C., Osawa, M., Nakauchi, H. & Sorrentino, B. P. (2001) Nat. Med. 7 1028-1034. [DOI] [PubMed] [Google Scholar]
- 22.McKinney-Freeman S. L., Jackson, K. A., Camargo, F. D., Ferrari, G., Mavilio, F. & Goodell, M. A. (2002) Proc. Natl. Acad. Sci. USA 99 1341-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Begley C. G. & Green, A. R. (1999) Blood 93 2760-2770. [PubMed] [Google Scholar]
- 24.Sanchez M. J., Bockamp, E. O., Miller, J., Gambardella, L. & Green, A. R. (2001) Development (Cambridge, U.K.) 128 4815-4827. [DOI] [PubMed] [Google Scholar]
- 25.Morrison S. J. & Weissman, I. L. (1994) Immunity 1 661-673. [DOI] [PubMed] [Google Scholar]
- 26.Okada S., Fukuda, T., Inada, K. & Tokuhisa, T. (1999) Blood 93 816-825. [PubMed] [Google Scholar]
- 27.Oh I. H., Lau, A. & Eaves, C. J. (2000) Blood 96 4160-4168. [PubMed] [Google Scholar]
- 28.Okada S., Wang, Z. Q., Grigoriadis, A. E., Wagner, E. F. & von Ruden, T. (1994) Mol. Cell. Biol. 14 382-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrison D. E., Jordan, C. T., Zhong, R. K. & Astle, C. M. (1993) Exp. Hematol. (Charlottesville, Va) 21 206-219. [PubMed] [Google Scholar]
- 30.McAllister K. A., Grogg, K. M., Johnson, D. W., Gallione, C. J., Baldwin, M. A., Jackson, C. E., Helmbold, E. A., Markel, D. S., McKinnon, W. C., Murrell, J., et al. (1994) Nat. Genet. 8 345-351. [DOI] [PubMed] [Google Scholar]
- 31.Arthur H. M., Ure, J., Smith, A. J., Renforth, G., Wilson, D. I., Torsney, E., Charlton, R., Parums, D. V., Jowett, T., Marchuk, D. A., Burn, J. & Diamond, A. G. (2000) Dev. Biol. 217 42-53. [DOI] [PubMed] [Google Scholar]
- 32.Cho S. K., Bourdeau, A., Letarte, M. & Zuniga-Pflucker, J. C. (2001) Blood 98 3635-3642. [DOI] [PubMed] [Google Scholar]
- 33.Chenchik A., Moqadam, F. & Siebert, P., (1996) A New Method for Full-Length cDNA Cloning by PCR (Wiley–Liss, New York).
- 34.Akashi, K., He, X., Chen, J., Iwasaki, M., Niu, C., Steenhard, B., Zhang, J., Haug, J. & Li, L. (September 5, 2002) Blood, 10.1182/blood-2002-06-1780.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.