Abstract
Activin A has potent mesoderm-inducing activity in amphibian embryos and induces various mesodermal tissues in vitro from the isolated presumptive ectoderm. By using a sandwich culture method established to examine activin A activity, we previously demonstrated that activin-treated ectoderm can function as both a head and trunk-tail organizer, depending on the concentration of activin A. By using activin A and undifferentiated presumptive ectoderm, it is theoretically possible to reproduce embryonic induction. Here, we test this hypothesis by studying the induction of cartilage tissue by using the sandwich-culture method. In the sandwiched explants, the mesenchymal cell condensation expressed type II collagen and cartilage homeoprotein-1 mRNA, and subsequently, cartilage was induced as they are in vivo. goosecoid (gsc) mRNA was prominently expressed in the cartilage in the explants. Xenopus distal-less 4 (X-dll4) mRNA was expressed throughout the explants. In Xenopus embryos, gsc expression is restricted to the cartilage of the lower jaw, and X-dll4 is widely expressed in the ventral head region, including craniofacial cartilage. These finding suggest that the craniofacial cartilage, especially lower jaw cartilage, was induced in the activin–treated sandwiched explants. In addition, a normal developmental pattern was recapitulated at the histological and genetic level. This work also suggests that the craniofacial cartilage-induction pathway is downstream of activin A. This study presents a model system suitable for the in vitro analysis of craniofacial cartilage induction in vertebrates.
Keywords: chondrogenesis, differentiation, development, animal cap
Activin A is a crucial morphogen in the vertebrate body plan (1–5), as demonstrated by its potent mesoderm-inducing activity on Xenopus undifferentiated presumptive ectoderm (6, 7). Activin A induces nearly every mesodermal tissue from amphibian undifferentiated presumptive ectoderm in a dose-dependent manner, whereas untreated ectoderm forms an irregularly shaped epidermis (8–10). At low concentrations of activin A, ventral mesoderm, such as blood-like cells, coelomic epithelium, and mesenchyme are induced in the explants. At intermediate concentrations, muscle and neural tissue are induced. At high concentrations, notochord, the most dorsal mesoderm, is induced. These activin A-induced tissues are indistinguishable at the histological and molecular levels from those found in normal embryos. The most characteristic property of activin A is the induction of organizer activity. Previously, we established the sandwich culture method to examine activin A activity in this role, demonstrating that activin-treated presumptive ectoderm can function as a head or trunk-tail organizer, depending on the activin A concentration and preculture period after the activin treatment (8, 11, 12). Activin A triggers several gene-expression cascades in the presumptive ectoderm with a time course that mimics the sequence in normal embryonal development (2, 12). Therefore, it is theoretically possible to reproduce embryonic induction by using activin A and undifferentiated presumptive ectoderm and design a fundamental embryonic form in vitro.
To confirm this hypothesis, we investigated the induction of cartilage, which hitherto had not been identified in the activin-treated presumptive ectoderm. During chondrogenesis of craniofacial development, the mesoderm-derived or neural crest-derived mesenchyme cells initially migrate to the pharyngeal arches and condense (13–15). The mesenchyme cell condensation proceeds to the commitment of chondrocytes, producing type II collagen (Col2) and the large chondroitin-sulfate proteoglycans. Finally, chondrocytes differentiate into the mature cartilage tissues, including infrarostral cartilage, Meckel's cartilage, palatoquadrate, and branchial arch cartilage. This study demonstrates that craniofacial cartilage can be induced by using activin A and the presumptive ectoderm, and, therefore, represents an in vitro model to examine the craniofacial cartilage induction mechanisms in vertebrates.
Materials and Methods
Embryos.
Eggs of general Xenopus laevis were prepared as described (8, 10, 16). In brief, both males and females were injected with 600 units of gonadotropin (Gestron; Denka Seiyaku, Kawasaki, Japan), and fertilized eggs were obtained. The eggs were raised until the late blastula stage (stage 9; ref. 17), then the jelly coat was removed with Steinberg's solution (SS) containing 4.5% cysteine hydrochloride (pH 7.8). The vitelline membranes were manually removed from stage-9 embryos with forceps.
Sandwich Culture.
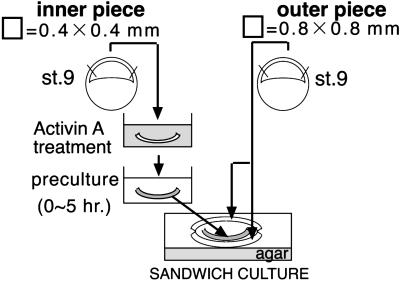
Presumptive ectodermal sheets (0.4 mm × 0.4 mm) cut from stage-9 embryos were treated with 100 ng/ml activin A in SS for 1 h. Presumptive ectoderm precultured for 0, 1, 2, 3, or 4 h in fresh SS was sandwiched between two untreated-presumptive ectoderm sheets (0.8 mm × 0.8 mm) cut from stage-9 eggs (Fig. 1). The sandwiched explants were cultured in SS containing 0.1% fatty acid-free BSA (Sigma) in 96-well plates (Sumitomo Bakelite, Tokyo) for 4 days at 20°C. On day 4, the culture medium was changed to fresh SS, and the explants were cultured in 24-well plates (Sumitomo Bakelite) for an additional 10 days.
Fig 1.
Schematic diagram of the procedure and sandwich-culture method.
Histological and Immunohistochemical Examination.
The general embryos and explants were fixed with cold acetone overnight at 4°C and then processed into paraffin for serial sectioning (6 μM). The sections were deparaffinized and then stained with Alcian blue and PAS and counterstained with hematoxylin for light microscopy. For immunohistochemistry, the serial sections were immunostained with mouse anti-Col2 antibody (Research Diagnostics, Flanders, NJ), visualized with peroxidase-conjugated Simple Stain MAX PO goat anti-mouse IgG (Nichirei, Tokyo) and 3-amino-9-ethylcarbazol (AEC, Nichirei). Nuclei were counterstained with hematoxylin.
RT-PCR.
Total RNA was extracted from explants and embryos by the guanidine-thiocyanate-phenol-chloroform extraction method (18). Total RNA from entire explant samples and 1 μg of total RNA derived from the whole embryo, the head region including the branchial arches, and the trunk-tail region, were treated with RNase-free DNase I (GIBCO/BRL). RT-PCR (19) was then performed according to the instruction of GIBCO RT-PCR applications. Each cDNA was amplified by PCR by using the following primer pairs and cycling conditions: Xenopus distal-less 4 (X-dll4) (20), 5′-tgcattccaaccacatgcc-3′, 5′-tttctgaaatcgccgctgg-3′, for 28 cycles; Col2 (21), 5′-aggcttggctggtcctcaaggt-3′, 5′-ggtccttgcataactcccatttgt-3′, for 25 cycles; cartilage homeoprotein 1 (Cart-1), 5′-aggagaacaacgcgaactacg-3′, 5′-tagcagcgaactggctcttagc-3′, for 30 cycles; and an internal control, ornithine decarboxylase (ODC) (22), 5′-cagctagctgtggtgtgg-3′, 5′-caacatggaaactcacacc-3′ for 25 cycles. Five microliters of each PCR product was resolved by 3% agarose gel electrophoresis and visualized with syber-gold (Wako Pure Chemical, Osaka).
Sequences.
PCR products of X-dll4, Col2, and Cart-1 were inserted directly into pGEM-T vector (Promega), according to the manufacturer's instructions. Recombinant plasmids were isolated by using GFXTM Micro plasmid prep kit (Amersham Pharmacia), and inserts were sequenced by using the dye terminator-cycle sequence method with a DTCS kit (Beckman Coulter) and CEQ 2000 DNA analysis system (Beckman Coulter). The cloned PCR products were confirmed by sequence analysis as X-dll4, Col2, and Cart-1 cDNA.
RNA Probe Preparation.
Recombinant plasmids of X-dll4, Col2, Cart-1, and goosecoid (gsc; generously provided by E. De Robertis, University of California, Los Angeles) were used as templates for the synthesis of RNA probes. The digoxigenin (DIG) or fluorescein-labeled RNA sense and antisense probes were prepared from template cDNA, according to the RNA-labeling kit instructions (Roche Molecular Biochemicals).
In Situ Hybridization.
The general embryos and explants were fixed with 4% (wt/vol) paraformaldehyde in phosphate buffer for 24 h at 4°C and then processed into paraffin for serial sectioning (10 μm). Except for the Col2, the deparaffinized sections were stained with Alcian blue for 2 min and cleared with 15% (vol/vol) acetic acid in methanol for 10 min. The sections were hybridized with DIG or fluorescein-labeled antisense or sense RNA probes at 60°C for 16 h. Then, the slides were washed once with 2× SSC/50% (vol/vol) formamide, two times with 2× SSC, and three times with 0.1× SSC, at 60°C for 15 min per wash. The DIG-labeled Col2 signal was detected according to DIG detection kit instructions by using NBT/BCIP (Roche Molecular Biochemicals). To enhance the signals in the embryos (except for the Col2), the DIG-labeled signals were detected with peroxidase-labeled anti-DIG antibody, biotinyl-tyramide (NEN), alkaline phosphatase-labeled streptavidin, and NBT/BCIP (Roche Molecular Biochemicals) with 5% (vol/vol) polyvinyl alcohol (Wako Pure Chemical). To analyze the signals in a section of the explant, DIG was detected by using tyramide signal amplification (TSA)-cyanine 3 system (NEN), and fluorescein was detected by using the TSA-fluorescein system (NEN). The sections were analyzed with fluorescence microscope and the images were captured with a charge-coupled device camera (Hamamatsu Photonics, Hamamatsu City, Japan) and AQUAMARINE software (Hamamatsu Photonics).
Results
Determination of Culture Conditions for Cartilage Induction in the Explants.
The sandwich-cultured explants were fixed with cold acetone on culture days 4 and 7 and then processed and sectioned for staining with Alcian blue/PAS. The induced tissues were analyzed as described (Table 1; ref. 8). In the explants cultured for 4 days, chondrocytes were not identified under any culture conditions. In the explants cultured for 7 days, chondrocyte-like cells were most frequently observed in the samples precultured for 1 h. The frequency of chondrocyte induction decreased with increasing preculture times, whereas the frequency of notochord induction increased with the time of preculture. Consequently, the presumptive ectoderm, which were treated with 100 ng/ml Activin A for 1 h, precultured for 1 h, and then sandwiched with untreated presumptive ectoderm, were cultured in SS until day 14 for further histological and genetic examination.
Table 1.
Induced tissue analysis
| Culture day | 4-day culture | 7-day culture | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Duration of preculture, hr | 0 | 1 | 2 | 3 | 4 | 0 | 1 | 2 | 3 | 4 |
| Numbers of cases | 11 | 10 | 11 | 12 | 11 | 24 | 23 | 22 | 21 | 21 |
| Epidermis | 10 (91) | 10 (100) | 6 (55) | 11 (92) | 10 (91) | 24 (100) | 23 (100) | 17 (77) | 14 (67) | 18 (86) |
| Cement gland | 1 (9) | 8 (80) | 11 (100) | 12 (100) | 11 (100) | 4 (17) | 13 (57) | 21 (96) | 19 (91) | 19 (91) |
| Neural tissue | 11 (100) | 9 (90) | 11 (100) | 12 (100) | 11 (100) | 24 (100) | 23 (100) | 22 (100) | 20 (95) | 19 (91) |
| Eye | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (9) | 0 (0) | 3 (13) | 3 (14) | 5 (24) | 1 (4.7) |
| Notochord | 2 (18) | 2 (20) | 5 (46) | 7 (58) | 9 (82) | 4 (17) | 4 (17) | 11 (50) | 16 (76) | 17 (81) |
| Muscle | 10 (91) | 9 (90) | 0 (0) | 7 (58) | 8 (73) | 24 (100) | 22 (96) | 21 (96) | 19 (91) | 10 (48) |
| Pronephros | 2 (18) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (25) | 1 (4) | 0 (0) | 0 (0) | 0 (0) |
| Mesenchyme | 11 (100) | 9 (90) | 3 (27) | 3 (25) | 4 (36) | 23 (96) | 23 (100) | 16 (73) | 13 (62) | 14 (67) |
| Endodermal cells | 2 (18) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 5 (21) | 4 (17) | 0 (0) | 0 (0) | 0 (0) |
| Cartilage | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (8) | 11 (48) | 5 (23) | 3 (14) | 0 (0) |
Percentages appear in parentheses.
Histology of the Explants.
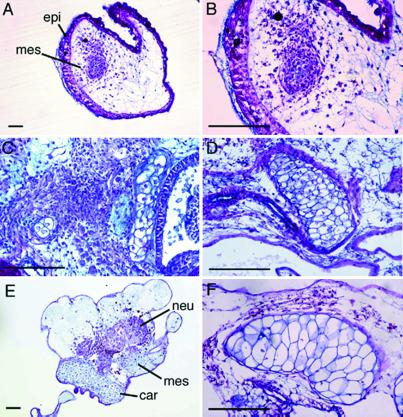
The Alcian blue-positive undifferentiated mesenchymal cell condensation was found in the explants on day 4 of culture (Fig. 2 A and B). Chondrocyte-like cells and cartilaginous tissue were apparent in the mesenchymal cell condensation on day 7 of culture (Fig. 2C). Some of the chondrocytes appeared matured, and the amount of cartilaginous tissues increased on day 10 of culture (Fig. 2D). Perichondrium was noted surrounding the cartilaginous tissues, and the cartilage appeared hypertrophic on day 14 (Fig. 2 E and F).
Fig 2.
Alcian blue and PAS staining of the explants. The acetone-fixed paraffin sections of the explants cultured for 14 days were stained with Alcian blue/PAS solution. (A) Four day-cultured explant. (B) Higher magnification of A. (C) Seven day-cultured explant. (D) Ten day-cultured. (E) Fourteen day-cultured explant. (F) Higher magnification of E. epi, epidermis; mes, mesenchyme; car, cartilage; neu, neural tissue. (Bars = 50 μm.)
Localization of Col2 Protein and mRNA in the Explants.
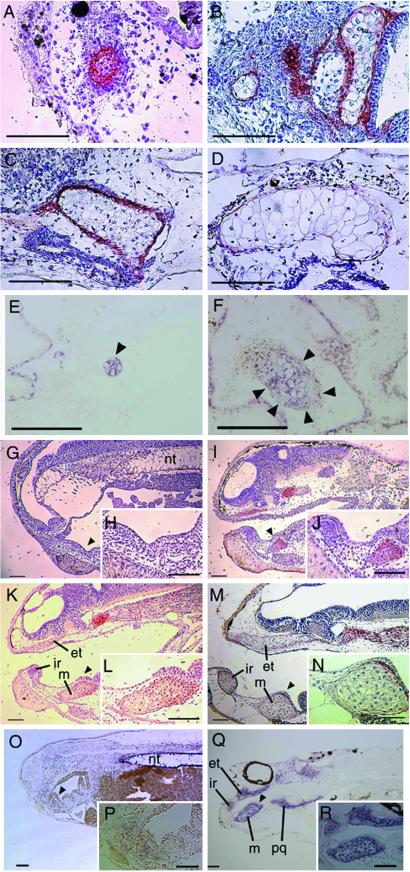
The serial sections of the histologically analyzed explant (Fig. 2) were immunostained with anti-Col2 antibody (Fig. 3). The Col2 immunostaining was found in the cells at the center of the mesenchyme condensation on day 4 of culture (Fig. 3A); it was intense in the immature chondrocytes on day 7 of culture (Fig. 3B) and in the mesenchymal perichondrium cells, but it was faint in the well differentiated chondrocytes on day 10 of culture (Fig. 3C). By day 14 of culture, the mature chondrocytes exhibited no immunopositive reactivity, and the perichondrium cells and immature chondrocytes were only slightly immunopositive (Fig. 3D). In situ hybridization analysis showed that Col2 mRNA was localized in immature chondrocytes on day 4 of culture (Fig. 3E) and in the chondrocytes on day 7 of culture (Fig. 3F). No reaction was detected when using the sense Col2 probe (data not shown).
Fig 3.
Type II Collagen protein and mRNA expression in the explants and general embryos. The serial sections of the histologically analyzed explants and the sections of general embryos were immunostained with anti-Col2 antibody which was visualized with AEC and counterstained with hematoxylin. The sections of explants and general embryos were hybridized with DIG-labeled Col2 RNA probe, and the localization was visualized with NBT/BCIP. (A) Immunolocalization of Col2 in the serial section of Fig. 2B, the 4 day-cultured explant. (B) Immunolocalization of Col2 in the serial section of Fig. 2C, 7 day-cultured explant. (C) Immunolocalization of Col2 in the serial section of Fig. 2D, 10 day-cultured explant. (D) Immunolocalization of Col2 in the serial section of Fig. 2F, 14 day-cultured explant. (E) Col2 mRNA expression in the section of 4 day-cultured explant. (F) Col2 mRNA expression in the section of 7 day-cultured explant. (G) Immunolocalization of Col2 in the section of Stage 35 embryo. (H) Higher magnification of the arrowhead indicating area in G. (I) Immunolocalization of Col2 in the section of Stage 40 embryo. (J) Higher magnification of the arrowhead indicating area in I. (K) Immunolocalization of Col2 in the section of Stage 42 embryo. (L) Higher magnification of the arrowhead indicating area in K. (M) Immunolocalization of Col2 in the section of Stage 44 embryo. (N) Higher magnification of the arrowhead indicating area in M. (O) Col2 mRNA expression in the section of Stage 35 embryo. (P) Higher magnification of indicating area in O. (Q) Col2 mRNA expression in the section of Stage 44 embryo. (R) Higher magnification of indicating area in Q. et, ethmoid trabecular cartilage; ir, infrarostral cartilage; m, Meckel's cartilage; nt, notochord; pq, palatoquadrate cartilage. (Bars = 50 μm.)
Col2 immunostaining was localized in the mesenchymal cell condensation, immature chondrocytes, and perichondrium of the craniofacial skeleton primordium and anterior notochord in the stage 35 (Fig. 3 G and H), 40 (Fig. 3 I and J), and 42 (Fig. 3 K and L) embryos but not in the mature chondrocytes in the stage 44 tadpole (Fig. 3 M and N). In situ hybridization analysis showed that Col2 mRNA was detected in the mesenchyme cell condensation in the mandibular and branchial arches in the stage 38 embryos (Fig. 3 O and P) and in the craniofacial cartilage and anterior notochord in the stage 42 tadpole (Fig. 3 Q and R).
Gene Expression in the Explants.
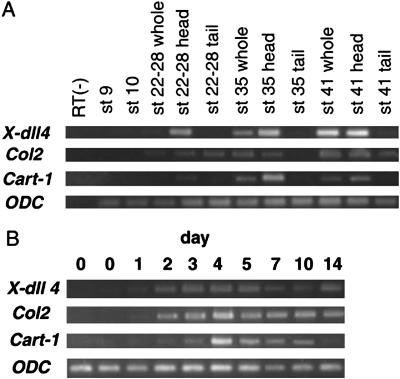
We examined the mRNA expression of X-dll4, Col2, Cart-1, and ODC in the explants (Fig. 4A) and embryos (Fig. 4B) with RT-PCR. The expression of X-dll4 mRNA was detected from day 1 to 14 of culture in all of the explants. In embryos, X-dll4 mRNA was expressed at the tailbud stage, stage 35, and tadpole stage 41, especially in the anterior region, as reported (17). Col2 mRNA expression was detected in the explants after 2 days of culture, increased to a maximum on day 4 of culture, and then decreased slightly by culture day 10. In embryos, Col2 mRNA expression was confirmed mainly in the anterior region at the tailbud stage (stage 22–28), stage 35, and tadpole stage 41. Cart-1 mRNA expression was detected in the explant samples after 2 days of culture, increased to a maximum on day 4, and then decreased. In embryos, expression of Cart-1 mRNA was detected in the anterior region at low levels at the tailbud stage, increased by stage 35, and subsequently decreased by stage 41. Expression levels of ODC mRNA in the explants did not change over the period of culturing.
Fig 4.
Expression of X-dll4, Col2, and Cart-1 mRNA in the explants and embryos detected by RT-PCR. RNA was extracted from embryos and explants. ODC was included as an internal RNA loading control. (A) Expression in the explants cultured for 14 days. (B) Expression in the embryos at stage 9, stage 10, and tailbud stages 22–28, 35, and 41.
Localization of Cart-1, X-dll4, and gsc mRNA in the Explants and Embryos.
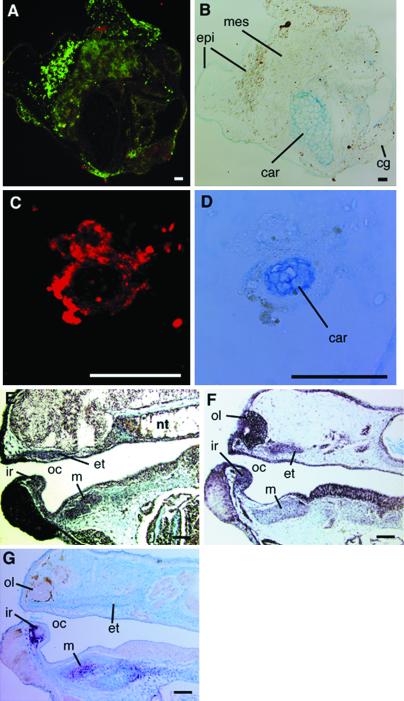
The Alcian blue-stained sections of 7-d cultured explants were double-stained with DIG-labeled Cart-1 probe and fluorescein-labeled X-dll4 probe or stained with a DIG-labeled gsc probe. Cart-1 mRNA expression (red) was detected slightly in cartilage and its surrounding mesenchyme, and X-dll4 mRNA expression (green) was detected throughout the explant (Fig. 5 A and B). The expression of gsc mRNA (Fig. 5 C and D, red) was detected prominently in the Alcian blue-positive immature chondrocytes and its surrounding mesenchyme in the serial section stained with Cart-1 and X-dll4 probes.
Fig 5.
Expression of Cart-1, X-dll4, and gsc mRNA in the cartilage of the explants and general embryos by in situ hybridization. The Alcian blue-stained section of a 7 day-cultured explant was double-stained with DIG-labeled Cart-1 probe and fluorescein-labeled X-dll4 RNA probes, visualized with Cyanine 3 and fluorescein, respectively. The serial section of above section was stained with DIG-labeled gsc RNA probe and visualized with Cyanine 3. The Alcian blue-stained sections of stage 41 general embryos were stained with DIG-labeled RNA probes and visualized with NBT/BCIP (purple). (A) Cart-1 mRNA (red) and X-dll4 mRNA (green) expression in a section of 7 day-cultured explant. (B) The Alcian blue staining of A. (C) gsc mRNA (red) in the serial section of A. (D) The Alcian blue-stained staining of C. (E) Cart-1 mRNA expression in embryo. (F) X-dll4 mRNA expression in embryos. (G) gsc mRNA expression in embryo. cg, cement gland; et, ethmoid trabecular cartilage; ir, infrarostral cartilage; m, Meckel's cartilage; nt, notochord; oc, oral cavity; ol, olfactory organ; pq, palatoquadrate cartilage. (Bars = 10 μm.)
In the stage 41 tadpole, Cart-1 mRNA expression was detected in olfactory organ, infrarostral cartilage, paltoquadrate and Meckel's cartilage, branchial arches cartilage, and notochord (Fig. 5E). X-dll4 mRNA expression was detected in olfactory organ, cement gland, infrarostral cartilage, ethmoid-trabecular cartilage, the epithelium of pharynx, and slightly in Meckel's cartilage (Fig. 5F). The expression of gsc mRNA was detected in the immature chondrocytes and condensed mesenchyme of infrarostral cartilage and Meckel's cartilage (Fig. 5G).
Discussion
Our sandwich-culture assay was previously established to examine the inducing activity of activin A, and we demonstrated that head and trunk-tail structures could be induced in the explants (8–12, 16, 23, 24). The time of preculture after the activin A treatment definitively affects the structures induced. The ectoderm precultured for a short time induced trunk-tail structures, whereas the ectoderm precultured for a long time induced anterior head structures. These studies suggest that the intermediate preculture term may initiate a gene-expression cascade, inducing the craniofacial region including cartilage tissues. Histological analysis in this study showed that cartilaginous tissue induction was most frequent in the ectoderm precultured for 1 h. Firstly, the mesenchymal cells condensed in the explants, and a few cells at the center of the condensation were heavily stained with Alcian blue. Subsequently, Col2-expressing chondrocyte-like cells and cartilaginous tissues were identified in the explants. This result indicates that the cartilage may be induced from the undifferentiated presumptive ectoderm.
Col2 expression characterizes chondrocyte differentiation. However, embryonic notochord also has been reported to express Col2 (25, 26). In this study, 17.4% of ectoderm samples precultured for 1 h exhibited notochord induction and 47.8% exhibited cartilage induction. RT-PCR analysis revealed that Col2 mRNA was expressed in all explants from day 2 to 14 of culture, and the expression was detected during stage 21–41 in the embryos. The expression of Col2 mRNA, therefore, reflected cartilaginous tissue development rather than notochord induction. The Col2 mRNA expression increased in the explants to a maximum level from day 4 to 7 of culture and then decreased from day 10. The result above is consistent with a decrease in Col2 mRNA expression when chondrocytes differentiate to the hypertrophic level (27, 28). Immunohistochemical examination revealed barely detectable levels of Col2 protein in the mature chondrocytes, and its localization was restricted to the perichondrium in the 14 day-cultured explants, as was the case in embryos. These findings demonstrate that the cartilaginous tissue in the explants differentiated into maturity.
Cart-1, a homeodomain-containing gene, is expressed in prechondrogenic condensation, and differentiating chondrocytes accompany with coexpression of type II collagen (29, 30). During mouse and rat embryonic development, Cart-1 is prominently expressed in craniofacial mesenchyme, weakly in mesenchymal cells of limb buds, and it is also found in lung bud, tendons, and mesonephros, which have the potential to undergo chondrogenesis (31, 32). Our RT-PCR and in situ hybridization results showed that Cart-1 mRNA expression localized mainly to the head region in Xenopus embryos: craniofacial cartilage and mesenchyme, cement gland, and anterior notochord. The Cart-1 mRNA expression increased to a maximum when mesenchymal cell condensation appeared in the explants. When the mesenchymal cells committed to chondrocytes, the Cart-1 mRNA expression levels decreased rapidly. In the explants, the Cart-1 mRNA was expressed in the cartilage and surrounding mesenchyme. These findings demonstrate that the gene cascade for the cartilage differentiation was induced in the explants.
The homeobox gene X-dll4 is related to the Drosophila Distal-less gene. Our in situ hybridization analysis revealed that X-dll4 was expressed throughout the explants. In Xenopus embryos, X-dll4 mRNA was widely expressed in the olfactory organ, cement gland, pharynx epithelium, and cartilage in the ventral head region but not in the trunk-tail region. Papalopulu and Kintner (20) reported that X-dll4 is expressed in anterior ectodermal derivatives: the ventral forebrain, the cranial neural crest cells of the branchial arches, and the cement gland. Our result was consistent with this report. The mouse Dlx-2 gene, which is related to X-dll4 (33), is expressed in the mandibular and maxillary primordial (34). Dlx-2 mutant mouse exhibited severe defects in the craniofacial bones derived from the mandibular and hyoid arches (35). These studies suggest that X-dll4 is a key molecule in the development of the craniofacial region. In summary, anterior ectodermal derivatives were induced in the explants used in this study.
gsc, a homeobox gene, is the first Spemann organizer-specific gene discovered, and it is activated by activin A during mesoderm induction (36, 37). gsc continues to be expressed in mesenchyme in the ventral head region in the late tailbud Xenopus embryos (38). Our in situ hybridization analysis revealed that gsc mRNA was expressed in lower jaw, infrarostral cartilage, and Meckel's cartilage in the Xenopus tadpole. The induced cartilage expressed gsc mRNA in the explants. These findings suggest that the ventral head mesenchyme was induced in the explants, and the mesenchyme differentiated into cartilage, which is equivalent to the lower jaw.
In this report, the cartilage derived from anterior ventral mesenchyme, which may be equivalent to lower jaw, was induced in vitro by using the undifferentiated presumptive ectoderm and activin A. The assay can reproduce not only cell differentiation but also pattern formation during craniofacial development. Activin A is expressed widely during mammalian development and highly conserved during vertebrate evolution. Matzuk et al. (39) reported that mice with mutation in activin-β A lacked whiskers and lower incisors and have defects in their secondary palates, including cleft palate, demonstrating that activin-β A must have a role during craniofacial development. They also reported that activin receptor II-deficient mice showed skeletal and facial abnormalities reminiscent of the Pierre-Robin syndrome in humans, which is characterized by brachygnathia/micrognathia (40). Activin accumulates gsc mRNA in early mouse embryos (41), and mice with inactivated gsc show craniofacial and rib cage defects (42). The findings presented here indicate that activin A regulates an induction cascade for craniofacial skeletal differentiation during vertebrate development. This model system represents a useful tool for the in vitro analysis of craniofacial cartilage development in vertebrates.
Acknowledgments
We thank Dr. Y. Eto (Central Research Laboratory, Ajinomoto Co., Kanagawa, Japan) for the gift of activin A. We thank Dr. Eddy M. De Robertis (University of California, Los Angeles) for the kind gift of the gsc probe. This work was supported in part by a grant-in-aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan, the Solution Oriented Research for Science and Technology Project of the Japan Science and Technology Corporation, and by a grant from the Smoking Research Foundation (to T.O.).
Abbreviations
SS, Steinberg's solution
DIG, digoxigenin
References
- 1.Asashima M., Kinoshita, K., Ariizumi, T. & Malacinski, G. M. (1999) Int. Rev. Cytol. 191 1-52. [DOI] [PubMed] [Google Scholar]
- 2.Asashima M., Ariizumi, T. & Malacinski, G. M. (2000) Comp. Biochem. Physiol. B Biochem. Mol. Biol. 126 169-178. [DOI] [PubMed] [Google Scholar]
- 3.Asashima M., Ariizumi, T., Takahashi, S. & Malacinski, G. M. (2000) Methods Mol. Biol. 136 15-26. [DOI] [PubMed] [Google Scholar]
- 4.Furue M., Okamoto, T., Hayashi, H., Sato, J. D., Asashima, M. & Saito, S. (1999) In Vitro Cell. Dev. Biol. Anim. 35 131-135. [DOI] [PubMed] [Google Scholar]
- 5.Furue M., Zhang, Y., Okamoto, T., Hata, R. I. & Asashima, M. (2001) Biochem. Biophys. Res. Commun. 282 745-749. [DOI] [PubMed] [Google Scholar]
- 6.Asashima M., Nakano, H., Shimada, K., Kinoshita, K., Ishii, K., Shibai, H. & Ueno, N. (1990) Roux's Arch. Dev. Biol. 198 330-335. [DOI] [PubMed] [Google Scholar]
- 7.Asashima M., Nakano, H., Uchiyama, H., Sugino, H., Nakamura, T., Eto, Y., Ejima, D., Nishimatsu, S., Ueno, N. & Kinoshita, K. (1991) Proc. Natl. Acad. Sci. USA 88 6511-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ariizumi T. & Asashima, M. (1994) Dev. Growth Differ. 36 499-507. [DOI] [PubMed] [Google Scholar]
- 9.Ariizumi T., Sawamura, K., Uchiyama, H. & Asashima, M. (1991) Int. J. Dev. Biol. 35 407-414. [PubMed] [Google Scholar]
- 10.Ariizumi Y., Moriya, N., Uchiyama, H. & Asashima, M. (1991) Roux's Arch. Dev. Biol. 200 230-233. [DOI] [PubMed] [Google Scholar]
- 11.Ariizumi T. & Asashima, M. (1995) Roux's Arch. Dev. Biol. 204 427-435. [DOI] [PubMed] [Google Scholar]
- 12.Ariizumi T. & Asashima, M. (2001) Int. J. Dev. Biol. 45 273-279. [PubMed] [Google Scholar]
- 13.Langille R. M. (1994) in Differentitaion and Morphogenesis of Bone, ed. Hall, B. K. (CRC, Boca Raton, FL), Vol. 9, pp. 1–64. [Google Scholar]
- 14.Hall B. K. & Miyake, T. (1995) Int. J. Dev. Biol. 39 881-893. [PubMed] [Google Scholar]
- 15.Shigetani Y., Nobusada, Y. & Kuratani, S. (2000) Dev. Biol. 228 73-85. [DOI] [PubMed] [Google Scholar]
- 16.Ariizumi T., Takano, K., Asashima, M. & Malacinski, G. M. (2000) Methods Mol. Biol. 135 89-112. [DOI] [PubMed] [Google Scholar]
- 17.Nieuwkoop P. D. & Faber, J., (1956) Normal Table of Xenopus laevis (Daudin) (North–Holland, Amsterdam).
- 18.Chomczynski P. & Sacchi, N. (1987) Anal. Biochem. 162 156-159. [DOI] [PubMed] [Google Scholar]
- 19.Dieffenbach C. W. & Dveksler, G. S., (1995) A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 20.Papalopulu N. & Kintner, C. (1993) Development (Cambridge, U.K.) 117 961-975. [DOI] [PubMed] [Google Scholar]
- 21.Su M. W., Suzuki, H. R., Bieker, J. J., Solursh, M. & Ramirez, F. (1991) J. Cell Biol. 115 565-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bassez T., Paris, J., Omilli, F., Dorel, C. & Osborne, H. B. (1990) Development (Cambridge, U.K.) 110 955-962. [DOI] [PubMed] [Google Scholar]
- 23.Ariizumi T. & Asashima, M. (1995) Zool. Sci. 12 509-521. [DOI] [PubMed] [Google Scholar]
- 24.Ariizumi T., Komazaki, S., Asashima, M. & Malacinski, G. M. (1996) Int. J. Dev. Biol. 40 715-718. [PubMed] [Google Scholar]
- 25.Seufert D. W., Hanken, J. & Klymkowsky, M. W. (1994) Anat. Embryol. 189 81-89. [DOI] [PubMed] [Google Scholar]
- 26.Bieker J. J. & Yazdani-Buicky, M. (1992) J. Histochem. Cytochem. 40 1117-1120. [DOI] [PubMed] [Google Scholar]
- 27.Pacifici M., Golden, E. B., Oshima, O., Shapiro, I. M., Leboy, P. S. & Adams, S. L. (1990) Ann. N.Y. Acad. Sci. 599 45-57. [DOI] [PubMed] [Google Scholar]
- 28.Nah H. D., Swoboda, B., Birk, D. E. & Kirsch, T. (2001) Dev. Dyn. 220 307-322. [DOI] [PubMed] [Google Scholar]
- 29.Zhao G. Q., Zhou, X., Eberspaecher, H., Solursh, M. & de Crombrugghe, B. (1993) Proc. Natl. Acad. Sci. USA 90 8633-8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loty S., Foll, C., Forest, N. & Sautier, J. M. (2000) Arch. Oral Biol. 45 843-856. [DOI] [PubMed] [Google Scholar]
- 31.Zhao G. Q., Eberspaecher, H., Seldin, M. F. & de Crombrugghe, B. (1994) Mech. Dev. 48 245-254. [DOI] [PubMed] [Google Scholar]
- 32.Qu S., Tucker, S. C., Zhao, Q., deCrombrugghe, B. & Wisdom, R. (1999) Development (Cambridge, U.K.) 126 359-369. [DOI] [PubMed] [Google Scholar]
- 33.Stock D. W., Ellies, D. L., Zhao, Z., Ekker, M., Ruddle, F. H. & Weiss, K. M. (1996) Proc. Natl. Acad. Sci. USA 93 10858-10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferguson C. A., Tucker, A. S. & Sharpe, P. T. (2000) Development (Cambridge, U.K.) 127 403-412. [DOI] [PubMed] [Google Scholar]
- 35.Qiu M., Bulfone, A., Martinez, S., Meneses, J. J., Shimamura, K., Pedersen, R. A. & Rubenstein, J. L. (1995) Genes Dev. 9 2523-2538. [DOI] [PubMed] [Google Scholar]
- 36.Blumberg B., Wright, C. V., De Robertis, E. M. & Cho, K. W. (1991) Science 253 194-196. [DOI] [PubMed] [Google Scholar]
- 37.Cho K. W., Blumberg, B., Steinbeisser, H. & De Robertis, E. M. (1991) Cell 67 1111-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newman C. S., Grow, M. W., Cleaver, O., Chia, F. & Krieg, P. (1997) Dev. Biol. 181 223-233. [DOI] [PubMed] [Google Scholar]
- 39.Matzuk M. M., Kumar, T. R., Vassalli, A., Bickenbach, J. R., Roop, D. R., Jaenisch, R. & Bradley, A. (1995) Nature 374 354-356. [DOI] [PubMed] [Google Scholar]
- 40.Matzuk M. M., Kumar, T. R. & Bradley, A. (1995) Nature 374 356-360. [DOI] [PubMed] [Google Scholar]
- 41.Blum M., Gaunt, S. J., Cho, K. W., Steinbeisser, H., Blumberg, B., Bittner, D. & De Robertis, E. M. (1992) Cell 69 1097-1106. [DOI] [PubMed] [Google Scholar]
- 42.Rivera-Perez J. A., Mallo, M., Gendron-Maguire, M., Gridley, T. & Behringer, R. R. (1995) Development (Cambridge, U.K.) 121 3005-3012. [DOI] [PubMed] [Google Scholar]