Abstract
The Notch-signaling pathway controls cellular differentiation, including proliferation and cell death in all higher metazoans (including flies and men). Signal transduction through activated Notch involves the CSL group of transcriptional regulators. Notch signals need to be tightly regulated, and in Drosophila they are antagonized by the Hairless (H) protein. H silences the activity of Notch target genes by transforming the Drosophila CSL protein, Suppressor of Hairless [Su(H)], from a transcriptional activator into a repressor while recruiting one of the corepressors dCtBP or Groucho. The H protein has a calculated molecular mass of ≈110 kDa and contains several functional domains apart from the two small corepressor-binding domains. However, although there is no indication for alternative splicing, two Hairless protein isoforms, Hp120 and Hp150, are observed throughout development. Here, we show that the smaller isoform derives from an internal ribosome entry site (IRES) within the ORF. The IRES is active in a heterologous assay and contains an essential, conserved structural element. The two Hairless isoforms have residual activity in vivo which is, however, reduced compared to a combination of both, which implies that both protein isoforms are necessary for WT function. In larval tissues, translation of the two isoforms is cell-cycle regulated: whereas the Hp150 isoform is translated during interphase, Hp120 is enriched during mitosis. Thus, the presence of either H isoform throughout the cell cycle allows efficient inhibition of Notch-regulated cell proliferation.
Cellular differentiation including proliferation and cell death is under the control of the Notch-signaling pathway in all of the higher metazoans studied so far in greater detail, ranging from invertebrates like Drosophila to mammals like human (for review, see ref. 1). As a consequence of ligand binding, the Notch receptor is cleaved and the intracellular domain of Notch becomes part of a transcriptional activation complex together with a DNA-binding protein named CBF1 or RBP-Jκ in mammals, Suppressor of Hairless [Su(H)] in Drosophila and Lag1 in Caenorhabditis (hence, CSL-type proteins). Notch-signaling events in Drosophila are antagonized most efficiently by Hairless (H), which silences Notch target genes by binding to Su(H) (2) and thereby transforming Su(H) from an activator to a repressor while recruiting the corepressors dCtBP or Groucho (ref. 3 and unpublished data). In agreement with its role as a general Notch antagonist, H protein is ubiquitously expressed throughout development. Like its target Su(H), H protein is cytoplasmic and nuclear with a major focus of activity within the nucleus (3, 4). The antagonistic activity of H appears largely dose sensitive. Genetic analysis of loss or gain of H function are compatible with the idea that H antagonizes most Notch-dependent processes during fly imaginal development including the aspects of Notch-induced overproliferation (refs. 5–8 and our own unpublished observations).
Aside from internal cleavage, the intracellular Notch domain of the human Notch 2 (hN2) receptor might be produced independently of signaling events by means of internal translation initiation starting from a potential internal ribosome entry site (IRES; ref. 9). Originally identified and particularly well studied in picorna viruses, IRES sequences have been found in a number of cellular mRNAs. In most instances, they are located within the 5′-untranslated region but also rarely occur in the midst of the coding region (10). The functional significance of internal ribosome entry in cellular mRNAs is little understood; however, it might link to cell-cycle-regulated protein translation and cellular-stress situations, respectively. Normally, translation of capped mRNAs is inhibited during mitosis as eukaryotic translation initiation factors are inactivated (overview in ref. 11). However, as two examples of human ornithine decarboxylase (ODC) and PITSRLE genes suggest, mRNAs bearing an IRES might escape this inhibition because translation initiation is cap independent, allowing for translation during mitosis (12, 13). Thus, IRES sequences might be a general feature of mRNAs that encode proteins with functions during mitotic stages of the cell cycle, a prediction that correlates well with the observation that many genes involved in tumorigenesis bear IRES elements (14).
Here, we show that two H protein isoforms are produced during development, whereby the shorter is generated by internal translation initiation preferentially during mitosis. The two isoforms are both indispensable for normal fly development, suggesting a requirement for H protein throughout the cell cycle to antagonize Notch signaling properly.
Materials and Methods
Sources for Hairless Protein.
WT proteins were extracted from S2 cell culture, staged embryos, dissected imaginal discs or ovaries, or whole larvae (4, 6). Cytoplasmic and nuclear extracts were prepared from 0- to 3-h-old Drosophila embryos. All steps were performed in the cold; all buffers contained protease inhibitors [final concentration: 1 μg ml−1 tosyl-l-phenylalanine chloromethyl ketone/0.5 μg ml−1 leupeptin/1 μg ml−1 pepstatin/1 mM PMSF/1 mM EDTA]. Embryos (5 ml) were homogenized in 10 ml of HB (10 mM Hepes, pH 7.6/10 mM KCl/1 mM MgCl2/0.5 mM DTT) and filtered through a 100-μm nylon mesh. The dirt was pelleted for 5 min at 200 × g. The supernatant was cleared by centrifugation for 10 min at 3,000 × g, resulting in a crude nuclear pellet and cytoplasmic extract (the supernatant), which was frozen until use. The pellet was washed twice in HB, sedimented in between as above. Then, it was resuspended in PA (20 mM Hepes, pH 7.6/100 mM KCl/2 mM MgCl2/0.5 mM DTT with 0.3 M sucrose) and centrifuged through a cushion of PA with 1.7 M sucrose for 30 min at 50,000 × g. The nuclear pellet was extracted with 1 ml of 10 mM Hepes, pH 7.6/1 mM MgCl2/1 mM CaCl2/150 mM NaCl and frozen until use. Schneider S2 cells were collected by centrifugation and boiled for 5 min in sample buffer [250 mM Tris⋅HCl, pH 7.6/0.01% bromophenol blue/5% (vol/vol) SDS/5% (vol/vol) 2-mercaptoethanol/40% (vol/vol) glycerol]. In vitro protein, nuclear, and cytoplasmic extract were handled likewise. The other tissues were homogenized in lysis buffer [10 mM Hepes, pH 7.6/5 mM EDTA/5 mM EGTA/5% (vol/vol) SDS/1 mM PMSF], boiled for 5 min, and cleared by full-speed centrifugation in a microcentrifuge. Supernatant was mixed with sample buffer, boiled as above, and run on standard 7% or 10% SDS polyacrylamide gels. In vitro H protein was made from mutagenized or FL-H cDNAs by using the TnT-coupled transcription/translation assay (Promega).
Immunohistochemistry.
Ab stainings of imaginal tissues and Western blots were performed as described (4, 6, 8). Production of central Hairless domain A-GST fusion protein was as described (4). For N-terminal Hairless (NTH)-GST, H codons 19–140 were amplified by PCR and cloned into pGEX vectors. Protein expression and purification was done as for A-GST. Antisera directed against Hairless protein fragments A and NTH were raised in rabbits (pAB Productions, Munich). Anti-A Abs detect WT protein preferentially in the cytoplasm and only overexpressed protein in the nucleus (4, 6). Anti-β-galactosidase and anti-firefly luciferase Abs were purchased from Promega and Chemicon, respectively; anti-phospho-Histone H3 was obtained from Upstate Biotechnology (Lake Placid, NY). Secondary Abs coupled to alkaline phosphatase, FITC, or Cy3 were obtained from The Jackson Laboratory. Imaginal discs were mounted in Vectashield (Vector Laboratories) and analyzed by confocal microscopy by using a Bio-Rad MRC1024.
Plasmid Constructions and Fly Work.
For site-directed mutagenesis, either Excite PCR based or QuikChange kits (Stratagene) were used according to the supplier's protocol. Codons for methionines M1, M2, and M3 were changed to isoleucines (GTG) and UUUUU were changed to penta-adenine (codons 135/136) in the A-box. The frame-shift mutation (Cfs) was introduced by digestions with AflII at codon 113, polishing with T4-polymerase and religation. This change shifts the frame from b into c, resulting in multiple stop 45 codons behind M3. All constructs were verified by sequence analysis before shuttling as KpnI–XbaI fragments into the modified CaSpeR-RX8 heat-shock vector (6). To generate the dicistronic constructs, Met-19 was exchanged to either Leu (UUG; I) or stop (TGA; IS). The interval between Met-19 and Gly-160 was amplified by PCR and cloned in-frame into the SalI–BamHI sites of pBGLPLCys (15) between lacZ and luciferase genes. The lac-H-IRES-luc insert was excised by HindIII–BglII and shuttled into HindIII–BamHI of pBluescript (Stratagene) to give BT I, which was also used as promoterless control. Control construct pA LL, which contains the intercistronic sequences of pBGLPLCys (15), was shuttled alike. Finally, the insert was excised with KpnI–SacII and cloned into pAC5.1B (Invitrogen) under the control of the strong and constitutive actin 5C promoter. All clones were sequence-verified. Primer sequences are available on request. The dicistronic reporter construct, here named CI, was obtained from D. Niessing and H. Jäckle (Max-Planck-Institut für Biophysikalische Chemie, Göttingen, Germany; ref. 16). Fly culture and transformation were according to standard protocols. A minimum of three independent lines were tested in which all behaved the same; heat shock was given for 30 min at 39°C, as described (6).
Analysis of the IRES Activity.
S2 cells grown in Schneider's medium to log phase were transiently transfected with the respective constructs (5 μg of DNA) by using Superfect according to the suppliers protocol (Qiagen, Valencia, CA). After 2 days of further culture, the cells were lysed with lysis buffer taken from the luciferase assay system (Promega). This assay system was also applied on 10 μl of cell lysate per 100 μl of substrate for luciferase activity assay, according to the manufacturer's protocol. Quantification was with a luminometer (Lumat LB 9507, EG & G, Salem, MA). The mean of two measurements is given in each case. Seven independent experiments gave similar relative numbers; a representative example is shown. β-galactosidase activity measurements were on the respective cell lysates (20 μl) in 500 μl of lacZ buffer (60 mM Na2HPO4/10 mM KCl/1 mM MgCl2/12.5 mM 2-mercaptoethanol) plus 100 μl of o-nitrophenyl β-d-galactoside solution (4 mg ml−1; Roche Molecular Biochemicals). Constructs I, IS, and pA LL gave comparable values of ≈0.3 OD when measured at 405 nm against the buffer. For Western blots, cells from 1 ml of transformant culture were collected by centrifugation and lysed in loading dye by boiling for 5 min. Proteins were detected with anti-luc (1:1,000, Chemicon), preabsorbed on embryos, anti-β-galactosidase (1:10,000, Cappel), and respective alkaline phosphatase-coupled secondaries (1:1,000, The Jackson Laboratory).
Results
Two Hairless Protein Isoforms Are Found in Drosophila.
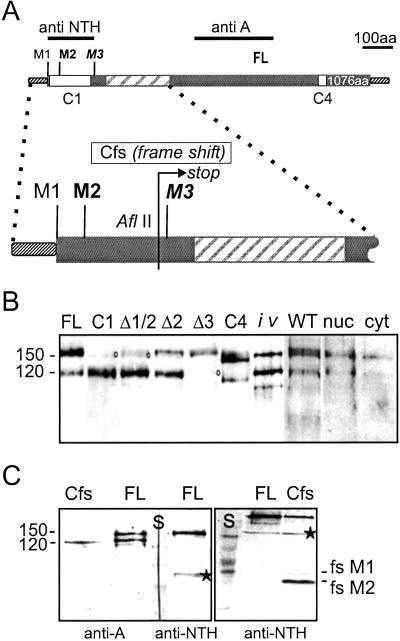
The Hairless (H) gene has a single large ORF encoding a protein of 1,077 aa with a calculated molecular mass of ≈110 kDa (Fig. 1A; refs. 5 and 17). However, the first start codon M1 does not conform well with the translation initiation consensus site for Drosophila genes (18) in contrast to the second start codon M2 at position 19, which is therefore the predicted start for a protein of 1,059 aa (5, 17). On Western blots two protein variants are detected, one of approximately the expected size with 120 kDa, Hp120, and a much larger species of ≈150 kDa, Hp150 (Fig. 1B; ref. 6). However, all H transcripts analyzed so far differ only in their trailer length, and apart from a 85-nt short intron within the leader, there is no evidence of alternative splicing (refs. 5 and 17; flybase), indicating that the two isoforms are presumably generated posttranscriptionally. This conclusion is confirmed by the fact that a full-length H cDNA clone transcribed in vitro with T3 polymerase and translated in a reticulocyte lysate gives rise to both H isoforms (Fig. 1B), which are both immunoprecipitated with anti-H Abs (not shown). We have shown before that the H protein is detected in the nucleus as well as the cytosol (4). However, there is not much difference between nuclear and cytoplasmic extracts with regard to the two H isoforms (Fig. 1B). Furthermore, both isoforms are detected after overexpression of the H protein (Fig. 1B), despite the fact that ectopic H protein is targeted to the nucleus (4, 6). Both isoforms appear at a fairly constant ratio throughout development of the fly, with the exception of ovaries and preblastoderm embryos (not shown), indicating that the maternal complement consists primarily of the longer protein isoform Hp150.
Fig 1.
The shorter Hairless Hp120 isoform is translated from the third start codon. (A) Schematic drawing of the H protein with potential start sites M1, M2, and M3. FL represents the full-length construct; the translated region is shown as the thicker bar, and the Su(H)-binding domain is crosshatched. Anti-A and anti-NTH show fragments used for production of respective antisera. C1 is an N-terminal truncation deleting the interval between M1 and M3; C4 is a 50-codon internal deletion in the C-terminal domain and was used as control. The blow-up of the N-terminal region shows the site of mutations that were introduced in respective constructs. The three potential start codons M1, M2 (at position 19), and M3 (at position 148) were altered individually or in combination into isoleucine (GTG). Mutant constructs (Δ) were cloned under heat-shock promoter control and transformed into flies. The same was done for Cfs, which contains a frame shift at position 113 (AflII), resulting in termination of translation 45 codons downstream of M3 (arrow). (B) Two H isoforms, Hp120 and Hp150, are detected in Drosophila proteins of different sources. WT, protein extracts from WT embryos; nuc, nuclear; cyt, cytoplasmic extracts from embryos; iv, in vitro-translated protein derived from an FL-H cDNA clone. Mutation of potential start sites eliminates formation of specific H isoforms. Protein extracts from embryos bearing the respective transgene and induced by a heat pulse were detected in Western blots with anti-A Abs. Mutation of M1 plus M2 (Δ1/2) interferes with Hp150 isoform formation just like the N-terminal truncation construct C1, whereas mutation of M3 (Δ3) eliminates formation of the short Hp120 isoform. Mutation of M2 (Δ2) still gives rise to the long isoform but at a reduced ratio compared to Hp120, suggesting that M1 is used less efficiently as start site. FL-H (full-length construct) and C4 serve as controls. Please note that the WT protein, denoted with a circle, is also detected at low levels. (C) A frame shift at codon 113 in construct Cfs results in a translation stop 45-aa downstream of M3. However, the Hp120 isoform is detected with anti-A antiserum, demonstrating that the short isoform is generated by internal translation initiation at M3 and not by site-specific cleavage or leaky scanning. The full-length construct (FL) serves as control. Anti-NTH Abs detect only the long H isoform in FL and, in addition, the N-terminal peptide derived from Cfs. Two peptides are seen because of translation start at M1 (fsM1) and M2 (fsM2), respectively. S, protein standard; *, unspecific crossreactivity of anti-NTH.
Hp120 Is Derived from Internal Translation Initiation.
We have no evidence that Hp150 arises by secondary protein modifications like phosphorylation or glycosylation (data not shown), suggesting that it corresponds to the full-length size. Presumably, H migrates slower than expected because it is very basic (overall pI of 10.4) and has a rather unusual amino acid composition (17). In this case, the smaller isoform Hp120 might be a shortened protein version resulting from cleavage or internal translation initiation. The latter hypothesis seemed curious because there are only a couple of examples of ribosome entry within coding regions (9, 12), and it appeared difficult to accommodate with the ribosome scanning model (19). The favorable context of the second start codon M2 renders leaky scanning rather unlikely. Moreover, the next start site M3 at codon 148 is separated by ≈400 nt and four additional AUGs in another reading frame, some with good start-site consensus.
However, we noted that the N-terminally truncated H-C1 construct, which was designed to start at M3 (6), gives rise to only one H protein of about the same size as Hp120 (Fig. 1 A and B). This result raised the possibility that the Hp120 isoform is produced by translation initiation at M3. To test this hypothesis, we altered each of the three methionines separately into isoleucines, generating the constructs ΔM1, ΔM2, ΔM1/2, and ΔM3, respectively. They were tested in an in vitro translation assay (data not shown) as well as in vivo in transgenic Drosophila lines that harbored the respective constructs under heat-shock control. Both sets gave identical results. As shown in Fig. 1B in the absence of M3, only the long Hp150 protein isoform is detected, whereas mutation of the first two methionines allows only the Hp120 isoform to be produced. Construct ΔM2 further reveals that M1 is used as a start site, less efficiently however, as predicted by the unfavorable sequence context (19). These results suggested internal initiation at M3 as source of the short Hp120 isoform but did not exclude cleavage directly at M3. Thus, construct Cfs was designed, which bears a frame shift mutation at codon 113, causing premature translation stop 45 codons downstream of M3 (Fig. 1A). From Cfs, the small Hp120 isoform is still produced, despite the fact that initiation occurs at M2 and to a lesser degree at M1 (Fig. 1C). These experiments allow us to exclude cleavage as a main source of the Hp120 isoform and renders initiation at an IRES very likely.
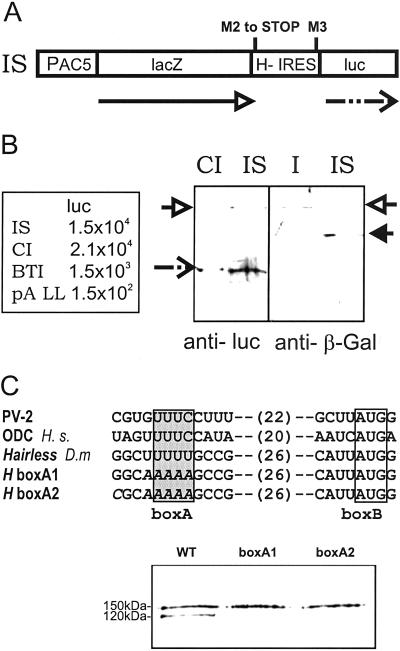
The M2–M3 Interval Serves as IRES.
A number of potential and bona fide IRESs have been identified in the past in cellular RNAs (reviewed in refs. 10 and 20). However, in most instances the IRES is located within the leader sequences and exceptionally within the coding region. To verify the hypothesis that the Hp120 isoform derives from internal translation initiation, we cloned the M2–M3 DNA interval between lacZ and luciferase (luc) genes, to generate dicistronic constructs (Fig. 2A). The resulting in-frame fusion in construct I was split into two cistrons in construct IS by mutating M2 into a stop codon. For negative controls, we used construct pA LL, which contains few intercistronic sequences (15), and BTI, which lacks its own promoter; for positive control, we used construct CI, which contains the well characterized IRES of the Drosophila Antp gene (16). The constructs were transfected into Drosophila S2 cells, and the enzymatic activities of luciferase and β-galactosidase were determined. High luciferase activity was determined for construct IS and found to be slightly lower than for the Antp-IRES control construct CI (Fig. 2B; ref. 16), which strongly suggests that H-IRES sequences allow internal ribosome entry. This conclusion is also supported by the negative controls, which show much lower luciferase activity: the starting vector pA LL provides little intrinsic luciferase activity, e.g., from ribosomal scanning, and BTI demonstrates that the M2–M3 interval does not contain a strong hidden promoter for monocistronic luc-transcripts. To visualize internal translation initiation products, the respective β-galactosidase and luciferase proteins were detected on Western blots, shown in Fig. 2B. These experiments demonstrate that the sequences located between the second and third methionine within the H ORF can serve as IRES in a heterologous assay.
Fig 2.
Hairless contains a bona fide IRES. (A) The interval between M2 and M3 was cloned between lacZ and luciferase (luc) genes under the control of a ubiquitous promoter. In construct IS, the two cistrons were split by converting M2 into a stop codon; this should result in β-galactosidase (arrow) and, only in the case of internal translation initiation at M3, luciferase would be expected (dashed arrow). (B) Enzymatic activity of luciferase (luc) was measured in S2 cells transfected with dicistronic and control constructs. Transfection was controlled by β-galactosidase activity. Experiments were repeated sevenfold; a representative example is shown. IS gives activity similar to construct CI, which contains the Antp-IRES (16) and several times above pA LL lacking the IRES (16) and BTI lacking a promoter. Luciferase also can be detected in Western blots from IS and CI (dotted arrow). IS also produces low amounts of a lac/luc fusion product, most likely resulting from repression of the stop codon (open arrow). However, the levels are rather low; probing the same extracts with anti-β-galactosidase Abs shows that the in-frame construct I produces similar levels of the lac/luc fusion protein (open arrow), as construct IS produces the lac-product (arrow) but no fusion protein. (C) Comparison among defined IRES sequences from polio virus 2 (PV-2), human ornithine decarboxylase (ODC H.s.), and Drosophila Hairless (D.m). Both defined boxes A and B, as well as the spacing, are roughly conserved. Two different mutations were introduced into box A of the H-IRES, here shown in italics (boxA1 and boxA2). In vitro translation products of full-length Hairless (WT) and the mutated constructs boxA1 and boxA2, detected with anti-A antiserum, are shown. Please note that the short Hp120 isoform is only produced from the WT construct and is essentially absent from the mutants, indicating the uridines in box A are essential for the activity of the H-IRES.
Structural Features of the H-IRES.
IRESs from various origins share little sequence homology except for those of picorna viruses (19). These contain a typical short pyrimidine-rich tract with the common sequence motif UUUC, located ≈25 bases upstream of the actual ribosome entry site. Pyronnet et al. (13) found a similar sequence in the IRES of the human ornithine decarboxylase (ODC) gene, called box A, and showed that it is required for efficient ribosome entry. Close inspection of the H-IRES sequence revealed a penta-uridine box ≈30 nt upstream of the M3 start codon embedded in a GC-rich sequence (Fig. 2C). This structure is reminiscent of the picorna virus and ODC IRESs (Fig. 2C) and indeed seems to be essential for H-IRES activity. Changing the uridines into adenines (H boxA1 and A2) inhibited Hp120 isoform formation nearly completely in an in vitro transcription/translation assay (Fig. 2C), indicating that box A is an essential sequence element for the activity of the H-IRES. Moreover, these results render the formal possibility of hidden promoter sequences within the M2–M3 interval rather unlikely and exclude the possible M3 usage by ribosomal scanning.
Both Isoforms Are Required for Normal H Activity.
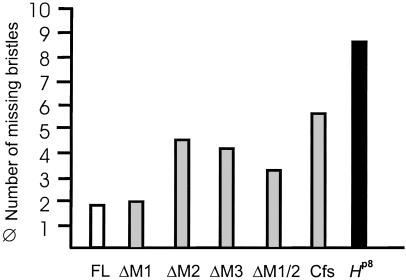
Mutations in H are haplo-insufficient and cause a dominant loss of mechano-sensory bristles and gaps in wing veins in the adult fly (21). This phenotype can be rescued by a single copy of an H full-length transgene (H-FL) under heat-shock promoter control even at ambient temperatures (5, 6, 17). To investigate the relevance of the H-IRES, we determined the biological activity of the mutated constructs, ΔM1, ΔM1/2, ΔM2, ΔM3, and Cfs in the fly. Without any exception, all of the mutated constructs were able to rescue to some degree bristle loss in the H heterozygotes (Fig. 3). However, only the ΔM1 construct, which is able to produce both H protein isoforms, gave results similar to the full-length construct. All other constructs, with the exception of the strongest expressing lines, showed markedly reduced activity (see Fig. 3). In addition, a considerable degree of variation among the various lines was observed. This result indicates that those constructs, which provide only one of the two isoforms, lack some aspects of normal H activity. As a consequence of binding to Su(H), overexpression of H interferes with Notch signaling, thereby disturbing bristle formation at several steps (5, 22). All mutant constructs caused typical overexpression phenotypes, however, with a considerably lower efficiency when compared to the full-length construct (data not shown; ref. 6). These phenotypes were expected, as each of the two H isoforms retains the Su(H)-binding domain (Fig. 1A). However, the reduced activity in both sets of experiments suggests that the two H protein isoforms are required together for normal H activity.
Fig 3.
Both H isoforms are required in vivo. Transformant flies carrying the respective mutant heat-shock constructs (Fig. 1) were crossed to Hairless loss of function allele HP8. Heterozygous H offspring were allowed to develop at ambient temperature and analyzed for dominant loss of large bristles. The HP8 mutant has ≈10 fewer macrochaetae than WT. An average of at least three independent strains of each construct is shown. Any one is able to rescue this heterozygous phenotype, however, with significantly lower efficiency and greater variability when compared to the FL-H and ΔM1 constructs, which allow the translation of both protein isoforms.
Cell-Cycle Dependence of IRES Usage.
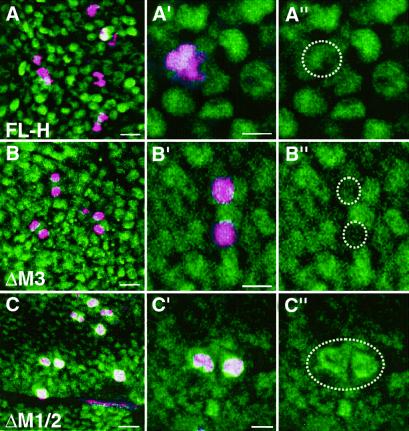
Recently it was proposed that IRES elements, which serve as cap-independent entry point for ribosomes, allow translation initiation also during mitosis when global rates of protein synthesis are reduced (overview in ref. 11). If this paradigm were a general one, it might also apply to the H-IRES. To demonstrate this assumption in situ in the fly, we induced either FL-H, ΔM3, or ΔM1/2 during larval development by a short, heat-shock pulse. H protein was revealed with anti-A antiserum, which preferentially detects H within the cytoplasm and only after overexpression within the nucleus (4). Thus, ectopic H protein can be specifically visualized as nuclear protein in this experiment. Within the same tissue, mitotic cells were marked with phospho-Histone H3 Abs. As can be seen in Fig. 4A, FL-H protein is detected at an intermediate level in all nuclei, including mitotic nuclei. In contrast, the long Hp150 isoform, which is produced from ΔM3, is largely absent from mitotic cells (Fig. 4B), whereas Hp120, which is produced by internal initiation from ΔM1/2, is strongly enriched in these cells (Fig. 4C). These results suggests that Hp120 is synthesized also during mitosis, whereas Hp150 is produced during interphase. Differential stability of the two H isoforms appears not very likely, because it would apply only to a small time window. As recently shown for two other IRES-containing cellular mRNAs (12, 13), we prefer the idea that the IRES allows for H translation during mitotic phase when normal, cap-dependent translation is inhibited. This conclusion is in agreement with the observation that maternal H consists primarily of Hp150, because maternal protein is produced in polytenic nurse cells which are mitotically quiescent (23).
Fig 4.
Hp120 but not Hp150 accumulates in mitotic cells. Transformant larvae carrying the respective constructs, FL-H (A), ΔM3 (B), and ΔM1/2 (C), were subjected to a brief heat shock, and imaginal discs were dissected ≈4 h later. Staining was performed with anti-A H Abs (green). This Ab specifically detects WT H protein in the cytoplasm; only ectopic H protein is detected in the nucleus (4). Thus, ectopically induced H protein can be specifically visualized. Mitotic cells were labeled with anti-phospho-Histone H3 Abs (pink). The blade region of wing imaginal discs is shown in an overview (Left, six sections crossing ≈10 μm; bar = 2 μm) and as close up (A′ and A" traverse ≈16 μm; B′ and B" and C′ and C" each ≈30 μm; bar = 1 μm). Double stainings are shown; the right column shows the respective single stainings of the close-ups. Note that ΔM3 gives rise only to Hp150 and ΔM1/2 gives rise only to Hp120 isoforms (compare to Fig. 2A). Hp150 is largely absent from mitotic cells (see B′ and B”); instead, these cells accumulate Hp120 (see C′ and C”).
Discussion
H Contains a Bona Fide IRES.
During all stages of the Drosophila life cycle except oogenesis, two H isoforms are produced, the long Hp150 isoform from the second M2, and less efficiently, from the first start codon M1 and the short Hp120 isoform from the third start codon M3. We provide strong evidence that translation from M3 is directed by internal ribosome entry and not, e.g., by cleavage of the longer protein isoform. In the latter case, no Hp120 isoform would be expected if M1 and M2 are mutated because translation should not occur at all; and secondly, the Cfs frame-shift construct should only provide the N-terminal peptides. The Cfs construct also excludes a leaky scanning mechanism, whereby the ribosome would pass by earlier start codons. Here, translation would terminate 45 codons behind M3 and 20 codons before the next in-frame start codon M4. Because backward scanning is less likely, the more closely located M4 should be preferred, which is not the case. Therefore, leaky scanning might lead to the production of Hp120 from the ΔM1/M2 construct, but not from Cfs.
Could the two isoforms possibly derive from different transcripts (20)? Our data provide good evidence to exclude both a hidden promoter for a shorter transcript as well as alternatively spliced mRNAs. As both H isoforms appear at fairly constant ratios (Fig. 1B), the corresponding mRNAs should be as frequent, but neither a shortened mRNA nor alternative splice products have been isolated (refs. 5 and 17; flybase). Moreover, the M2–M3 interval contains no obvious candidate sequence elements. There are just five in frame AG dinucleotides, one of which lies between boxA and M3, but none could serve as splice acceptor lacking the relevant polypyrimidine stretch. Not surprisingly, a dicistronic construct without actin promoter shows rather low luciferase activity (Fig. 2B). Also, in vitro transcription of the full-length H cDNA by a T3 promoter still gives rise to both H isoforms. Leaky scanning in this case can again be excluded by the fact that mutation of boxA prevents Hp120 production (Fig. 3C). Finally, ectopic induction of the WT transcripts from a heat-inducible promoter gives rise to both isoforms in the expected ratio (Fig. 1B; ref. 6).
Other dipteran flies also show two H protein isoforms, and the largely diverged species Drosophila hydei produces two H proteins only slightly bigger than the Drosophila melanogaster proteins (24). In D. hydei, the third methionine is well conserved, and both isoforms are also generated in an in vitro transcription/translation assay (24), suggesting internal translation initiation as well. Quite strikingly, the H-IRES shares similarity with IRESs from the human ODC gene and from picorna viruses regarding the pyrimidine-rich region harboring the A-box in close proximity of the actual ribosome entry site (13, 19). Our experiments indicate that the A-box is essential for H-IRES recognition. In addition, as predicted by the program mfold (version 10.2, GCG), there is a conspicuous Y-loop of 85 nt located 200 bases upstream of M3. Whether this structure, which is reminiscent of other cellular IRES sequences including that of the Drosophila Antp gene (25), is a relevant feature of the H-IRES remains to be determined.
Translation from the H-IRES results in a protein which is ≈20% shortened at the N terminus. The N terminus itself seems not to contain a specific functional domain, as either isoform retains some WT activity. However, only constructs that provide both isoforms have normal activity, whereas those that provide only one isoform are significantly less active. This observation strongly implicates that both H isoforms are required together. Two, nonexclusive scenarios can be envisaged to accommodate this observation. First, H might act as a homo- or heterodimer (or multimer) involving the two isoforms, and heteromers might be functionally distinct from homomers. Second, H activity might also be required during times when normal translation is inhibited, like in cellular stress situations and/or during the mitotic phase of the cell cycle.
IRES and Cell-Cycle-Dependent Translation.
It is generally accepted that IRES sequences allow for cap-independent translation through G2/M phase, when cap-dependent translation is inhibited. Several cases were described where an IRES directs cell-cycle-dependent translation of the respective mRNA (12, 13), suggesting that this might represent a general paradigm (19). Lacking the possibility of synchronizing Drosophila cell cultures, we used whole larval tissue to address this question in situ. In agreement with the above idea, the Hp120 isoform, which is initiated from the IRES, is specifically enriched in mitotic cells that largely exclude the Hp150 translation product. We note that this experiment was only possible by taking advantage of anti-A antisera, which allow the distinction between WT and overexpressed H protein. As cell-cycle-dependent differential stability of the two H isoforms seems not very likely, we favor the idea that Hp120 is translated during mitotic phases when Hp150 cannot be produced. In Drosophila, other functional IRES sequences have been identified before (26), but not with regard to cell-cycle-regulated translation. It will be interesting to see whether all of these mRNAs show a biphasic translation pattern also in Drosophila. After all, one might expect many more genes to contain IRES sequences if they need to be active throughout the cell cycle (11).
Requirement of H as Notch Antagonist Through the Entire Cell Cycle.
H acts as an antagonist of many Notch-dependent processes during Drosophila imaginal development including the regulation of cell proliferation. Overexpression of the activated Notch receptor results in profound overproliferation of a variety of tissues during imaginal development of Drosophila (ref. 7 and our own observations). This overgrowth is, in part, a result of the activation of Notch target genes, which themselves promote tissue growth, e.g., of vestigial or of the morphogen wingless (7, 27). In mammals, Notch has been implicated more directly in the regulation of cell proliferation as a number of neoplasms involve ectopic Notch activation (overview in refs. 1 and 28). Many genes involved in tumorigenesis are regulated at the translational level (overview in ref. 14). Although not directly demonstrated for Notch, recent work suggests that the activated form of the Notch receptor might be generated independently of signaling by internal ribosome entry (9). It is tempting to speculate that this process is under cell-cycle control, and that H is regulated alike to antagonize the Notch pathway properly.
Acknowledgments
We thank C. Alexief-Damianof for in vitro-mutagenized constructs, I. Beck for excellent technical help, and D. Niessing and H. Jäckle for their helpful input on IRES construction and analysis and for the CI construct and vectors. This work was supported by Deutsche Forschungsgemeinschaft Grant DFG DM 1328/4-2 (to D.M.).
Abbreviations
IRES, internal ribosome entry site
NTH, N-terminal Hairless
Cfs, frame-shift construct
References
- 1.Artavanis-Tsakonas S., Rand, M. D. & Lake, R. J. (1999) Science 284 770-776. [DOI] [PubMed] [Google Scholar]
- 2.Brou C., Logeat, F., Lecourtois, M., Vandekerckhove, J., Kourilsky, P., Schweisguth, F. & Israël, A. (1994) Genes Dev. 8 2491-2503. [DOI] [PubMed] [Google Scholar]
- 3.Morel V., Lecourtois, M., Massiani, O., Maier, D., Preiss, A. & Schweisguth, F. (2001) Curr. Biol. 11 789-792. [DOI] [PubMed] [Google Scholar]
- 4.Maier D., Nagel, A. C., Johannes, B. & Preiss, A. (1999) Mech. Dev. 89 195-199. [DOI] [PubMed] [Google Scholar]
- 5.Bang A. G. & Posakony, J. W. (1992) Genes Dev. 6 1752-1769. [DOI] [PubMed] [Google Scholar]
- 6.Maier D., Marquart, J., Thompson-Fontaine, A., Beck, I., Wurmbach, E. & Preiss, A. (1997) Mech. Dev. 67 97-106. [DOI] [PubMed] [Google Scholar]
- 7.Go M. J., Eastman, D. S. & Artavanis-Tsakonas, S. (1998) Development (Cambridge, U.K.) 125 2031-2040. [DOI] [PubMed] [Google Scholar]
- 8.Nagel A. C., Wech, I. & Preiss, A. (2001) Mech. Dev. 109 241-251. [DOI] [PubMed] [Google Scholar]
- 9.Lauring A. S. & Overbaugh, J. (2000) Mol. Cell 6 939-945. [DOI] [PubMed] [Google Scholar]
- 10.Carter M. S., Kuhn, K. M. & Sarnow, P. (2000) in Translational Control, eds. Hershey, J. W. B., Mathews, M. B. & Sonenberg, N. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 615–635.
- 11.Sachs A. (2000) Cell 101 243-245. [DOI] [PubMed] [Google Scholar]
- 12.Cornelis S., Bruynooghe, Y., Denecker, G., van Huffel, S., Tinton, S. & Beyaert, R. (2000) Mol. Cell 5 597-605. [DOI] [PubMed] [Google Scholar]
- 13.Pyronnet S., Pradaryol, L. & Sonenberg, N. (2000) Mol. Cell 5 607-616. [DOI] [PubMed] [Google Scholar]
- 14.Hershey J. W. B. & Miyamoto, S. (2000) in Translational Control, eds. Hershey, J. W. B., Mathews, M. B. & Sonenberg, N. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 637–654.
- 15.Kollmus H., Flohé, L. & McCarthy, J. E. G. (1996) Nucleic Acids Res. 24 1195-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niessing D., Dostatni, N., Jäckle, H. & Rivera-Pomar, R. (1999) EMBO J. 18 1966-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maier D., Stumm, G., Kuhn, K. & Preiss, A. (1992) Mech. Dev. 38 143-156. [DOI] [PubMed] [Google Scholar]
- 18.Cavener D. R. (1987) Nucleic Acids Res. 15 1353-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson R. H. (2000) in Translational Control, eds. Hershey, J. W. B., Mathews, M. B. & Sonenberg, N. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 127–183.
- 20.Kozak M. (2001) Mol. Cell. Biol. 21 1899-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bridges C. B. & Morgan, T. H., (1923) The Third Chromosome Group of Mutant Characters of Drosophila (Carnegie Inst. of Washington, Washington, DC), Publ. No. 327.
- 22.Nagel A. C., Maier, D. & Preiss, A. (2000) Mech. Dev. 94 3-12. [DOI] [PubMed] [Google Scholar]
- 23.Spradling A. (1993) in The Development of Drosophila, eds. Bate, M. & Martinez-Arias, A. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 1–70.
- 24.Marquart J., Alexief-Damianof, C., Preiss, A. & Maier, D. (1999) Dev. Genes Evol. 209 155-164. [DOI] [PubMed] [Google Scholar]
- 25.Le S.-Y. & Maizel, J. V., Jr. (1997) Nucleic Acids Res. 25 362-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye X., Fong, P., Iizuka, N., Choate, D. & Cavener, D. R. (1997) Mol. Cell. Biol. 17 1714-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Celis J. F., de Celis, J., Ligoxygakis, P., Preiss, A., Delidakis, C. & Bray, S. (1996) Development (Cambridge, U.K.) 122 2719-2728. [DOI] [PubMed] [Google Scholar]
- 28.Joutel A. & Tournier-Lasserve, E. (1998) Semin. Cell Dev. Biol. 9 619-624. [DOI] [PubMed] [Google Scholar]