Abstract
In East African Lake Victoria >200 endemic species of haplochromine fishes have been described on the basis of morphological and behavioral differences. Yet molecular analysis has failed to reveal any species-specific differences among these fishes in either mitochondrial or nuclear genes. Although the genes could be shown to vary, the variations represent trans-species polymorphisms not yet assorted along species lines. Nevertheless, fixed genetic differences must exist between the species at loci responsible for the adaptive characters distinguishing the various forms from one another. Here we describe variation and fixation at the long wavelength-sensitive (LWS) opsin locus, which is selection-driven, adaptive, and if not species- then at least population-specific. Because color is one of the characters distinguishing species of haplochromine fishes and color perception plays an important part in food acquisition and mate choice, we suggest that the observed variation and fixation at the LWS opsin locus may have been involved in the process that has led to the spectacular species divergence of haplochromine fishes in Lake Victoria.
The great lakes of the East African Rift Valley, Lakes Victoria, Malawi, and Tanganyika, harbor ≈200, 400, and 170 endemic species of haplochromine fishes of the family Cichlidae, respectively (1–3). These fishes have fascinated evolutionary biologists as spectacular examples of explosive radiations that have produced numerous new species adapted to different ecological niches. The youngest of these adaptive radiations is still in progress in the largest of the great lakes: Lake Victoria and its satellites. Lake Victoria dried out entirely at the end of the Pleistocene, and its basin did not begin to fill with water again until 12,400 years ago (4). Recent molecular evidence indicates that most of the endemic species now found in the lake originated from very few, perhaps only two, founding lineages that colonized the lake after its reconstitution (5). In agreement with their assumed young age, the endemic haplochromines of Lake Victoria have been shown to be genetically homogeneous in both mitochondrial (6) and nuclear (ref. 7 and Y.T., J.K., and N.O., unpublished data) genomes. The variation they display (5, 7, 8) is trans-specific in its distribution; all attempts to demonstrate species-specific genetic differences at the molecular level have failed thus far. This observation is in striking contrast to the conspicuous morphological and behavioral differentiation of the species (2, 3). Particularly striking are the species-specific differences in coloring patterns, especially in fishes inhabiting rocky shores in the lake (3). Because these differences seem to be genetically controlled, it can be expected that corresponding differences also exist in the genes responsible for them. Because these genes, however, have not been identified yet, the second best chance to identify species-specific genetic variations is to focus on some of the genes that allow the fishes to perceive the differences visually.
Visual systems have indeed been shown to be important in feeding (1) and mating (9) behavior of cichlid fishes. Visual pigments in photoreceptor cells of the retina consist of a light-absorbing component, the chromophore, and a protein moiety, the opsin (10, 11). In vertebrate visual pigments, there are two types of chromophore, 11-cis-retinal and 11-cis-3,4-dehydroretinal, and five types of opsin, one in the rods and four in the cones (12). The rod opsin (rhodopsin 1, RH1) makes it possible to form black-and-white images in dim light; the cone opsins mediate color vision in bright light. The four groups of color opsins differ in their light sensitivity: the short wavelength (UV)-sensitive 1 (SWS1), the short wavelength (blue)-sensitive 2 (SWS2), the rhodopsin-like 2 (RH2), and the middle (green)- and long (red)-wavelength sensitive (MWS/LWS) opsins have peak values of light absorption (λ max) at 360–430, 440–460, 470–510, and 510–560 nm, respectively. The light sensitivity of a visual pigment is determined not only by the chromophore itself but also by its interaction with the amino acid residues lining the pocket of the opsin in which the chromophore is embedded (13).
Representatives of all four groups of cone opsins have been found in haplochromine fishes (14, 15), and the SWS2 group has been subdivided into two subgroups, SWS2A and SWS2B (14). Molecular studies of cichlid visual pigments have concentrated thus far on species from Lake Malawi (14, 15), in which the clear water allows direct observation of fish behavior. Such observations are more difficult in Lake Victoria, in which the water is often turbid. This impediment notwithstanding, however, the haplochromine species flock of this lake attracts much attention because of its young age (4, 5) and the opportunity to study speciation in progress. Certain mutations in opsin genes might be expected to be adaptive and as such to have a higher probability of fixation in the populations and emerging species of cichlid fishes than neutral mutations. Opsin genes are therefore good candidates for finding fixed, presumably adaptive differences between populations and species of Lake Victoria haplochromines. The present study therefore was undertaken to find and characterize such fixed genetic differences among the Lake Victoria species.
Materials and Methods
Fish.
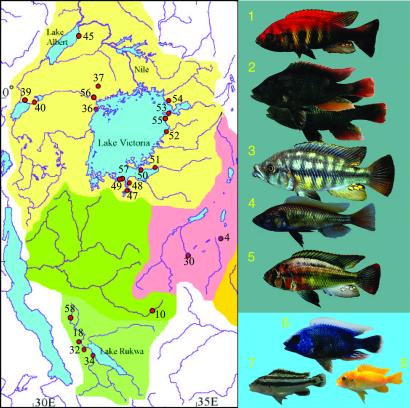
Specimens were collected by H.T. during expeditions in 1993, 1995, 1996, and 1998 at the locations shown in Fig. 1. Voucher specimens of the individual species have been deposited at the Museé Royal de l'Afrique Centrale (Turveren, Belgium), The Nationaal Natuurhistorisch Museum (Leiden, The Netherlands), and the Max-Planck-Institut für Biologie (Tübingen, Germany). Two nomenclatures of Lake Victoria haplochromines are in use currently. In one, most of the endemic species are lumped into a single genus, Haplochromis (2); in the other, the species are split into several genera (16). Here we adhere to the currently more widely used but not necessarily better splitters' nomenclature. Because the taxonomy of the riverine haplochromines remains unresolved, these specimens are referred to by the group number defined by mtDNA control region sequences (5) and the locality of their origin.
Fig 1.
(Left) River systems (differentiated by colors) and major lakes of East Africa. Arabic numerals indicate localities at which haplochromines examined in this study were collected. The names of the localities are: 4, Lake Babati; 10, Lake Nshere; 18, Kitilda/Rukwa; 30, Lake Singida; 32, Muze River; 34, Zimba River; 36, Lake Nabugabo, Lake Kayugi, and Lake Kayania; 37, Lake Wamala; 39, Lake Katwe/Lake Edward; 40, Katunguru Bridge/Kazinga Channel; 45, Butiaba/Lake Albert; 47, Buzumu Gulf, Kissenda Bay; 48, Mwanza Gulf, Butimba; 49, Chamagati Island; 50, Zue Island; 51, Speke Gulf; 52, Muhuru; 53, Rusinga, Gemba, Gingo; 54, Anyanga; 55, Mbassa; 56, Katonga; 57, Yetti, Nyegezi Rocks; 58, Katavi River. (Right) Representatives of the cichlid taxa used in this study. The background color indicates the different tinge and translucency of the water in Lake Victoria (Upper) and Lake Malawi (Lower). The cichlid fish are: 1, Haplochromis nyererei; 2, Astatotilapia nubilus from Lake Victoria (in front) and Lake Wamala (behind); 3, Paralabidochromis chilotes; 4, Haplochromis pyrrhocephalus; 5, Haplochromis katavi; 6, Aulonocara jacobfreibergi; 7, Melanochromis parallelus; 8, Pseudotropheus daktari.
PCR and Sequencing.
Genomic DNA was prepared and PCR and sequencing were performed as described (5, 7, 17).
Primers.
Sequences of primers are published as supporting information on the PNAS web site, www.pnas.org.
Sequence Analysis.
Sequences were aligned with the help of SEQPUP 0.6f software (ref. 18, http://iubio.bio.indiana.edu/soft/molbio) or by using the GENETYX-MAC 10.1 program. Pairwise distances and phylogenetic relationships of amino acid sequences were assessed with the help of the PAUP* 4.0b8 program (19) or the MEGA 2.1 program package (20) applying the neighbor-joining method (21) for P distances or Poisson-corrected distances or the maximum-parsimony method by using the heuristic search algorithm. Ancestral sequences were reconstructed by using the data set of Lake Victoria cichlids and the Oreochromis sequence as an outgroup and analyzing it with PAUP* 4.0b8 by heuristic searches using maximum likelihood as optimality criterion (transition/transversion ratio = 2, empirical base frequencies, equal substitution rate for all sites). Reconstructed ancestral states at internal nodes of the optimal tree were used for further evaluation. Frequencies of synonymous and nonsynonymous substitutions were assessed by using MEGA 2.1 (20).
Results
Characterization of SWS2B.
To identify cone opsin genes of Lake Victoria haplochromines, we designed oligonucleotide primers on the basis of Lake Malawi cichlid sequences in the database (15) and used PCR to amplify a 2-kb genomic DNA segment that encompassed the part of the gene extending from exon 1 to exon 5. The amplification product then was used as a template to amplify and sequence individual exons (and some of the introns) with the help of specific primers. We chose to study two opsin genes, SWS2B and LWS, the products of which absorb light near the two extremes of the visible spectrum. To study the SWS2B-encoding gene, we amplified the 2-kb segment from representatives of nine haplochromine species endemic to Lake Victoria and its satellite, Lake Nabugabo, and then sequenced, after secondary PCR, but without cloning, exons 2–5 from each of the nine templates. The nine sequences were identical at the nucleotide level except for one sequence, which differed from all the others by a single nucleotide substitution (data not shown). The sequences were very similar to the SWS2B sequence of the Lake Malawi species, differing from it by five and six substitutions at the nucleotide sequence level and two replacements at the amino acid level. Thus, the SWS2B-encoding gene behaved similar to other genes (5–7) in that it displayed little sequence variability.
Characterization of the LWS Locus.
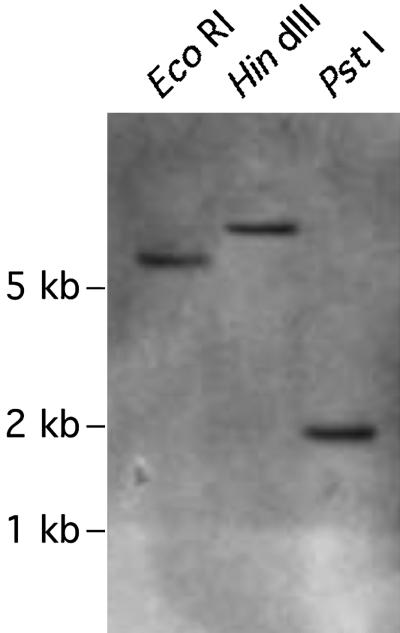
To check the copy number of LWS, we carried out Southern blot hybridization of EcoRI-, HindIII-, and PstI-digested genomic DNA, isolated from Neochromis nigricans tissues, using LWS intron 1 as a probe. Only a single hybridizing band was obtained on the blot in each of the digests (Fig. 2). Although this observation alone does not prove that LWS is a single locus, combined with the fact that we never found more than two different LWS sequences in one individual (see below), it makes the multilocus hypothesis unlikely.
Fig 2.
Southern blot of genomic DNA isolated from N. nigricans with labeled LWS intron 1 probe. The DNA was digested with the indicated restriction enzymes before fractionation by electrophoresis and blotting. Positions of size-marker bands are indicated to the left. Southern blot hybridization was performed as described (30).
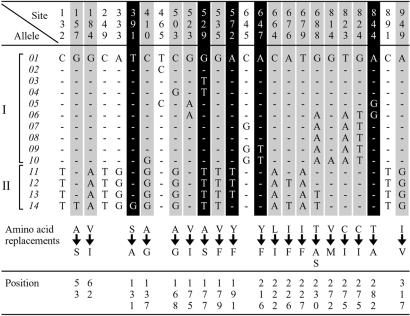
Preliminary tests revealed the LWS-encoding locus to be highly variable. These tests prompted us to extend our analysis on the variability of the LWS gene. Altogether, we obtained 58 sequences of the segment encompassing exons 2–5 from the genomic DNA of 31 individuals sampled from 11 localities along the entire eastern coast of Lake Victoria and one locality on the western coast. We also obtained 16 LWS sequences of nine fishes from Lake Nabugabo and, for comparison, 42 sequences of fishes from Lake Malawi. These samples represented 10 endemic and 1 nonendemic, widely distributed haplochromine species (Astatoreochromis alluaudi) from Lake Victoria, 3 endemic species from Lake Nabugabo, and 8 endemics from Lake Malawi (Fig. 1). The sequences revealed the existence of 14 LWS alleles in the Lake Victoria/Lake Nabugabo sample and 12 alleles in the Lake Malawi sample. For convenience, the alleles in Lake Victoria/Lake Nabugabo were numbered from 01–14 (Table 1). Fig. 3 shows variable nucleotide sites differentiating LWS alleles of Lake Victoria/Lake Nabugabo cichlids. The 25 variable sites include 6 synonymous and 19 nonsynonymous sites. To obtain further information about the distribution, frequencies, and origin of the major Lake Victoria alleles, we sequenced the most variable of the five exons (exon 4, 166 bp) from 184 Lake Victoria/Lake Nabugabo haplochromines representing 17 species as well as from 80 haplochromines collected in other East African lakes and rivers (Table 2). This sample revealed the existence of one additional allele in riverine haplochromines and three additional alleles in species from other lakes (Table 2). Thus the LWS locus in Lake Victoria/Lake Nabugabo displays a striking variability, representing a range of genetic distance from 0.0015 to 0.2294 (see Discussion).
Table 1.
Exon 2–5 LWS gene variability in cichlid fishes of Lakes Victoria/Nabugabo
| Species
|
n
|
Heterozygotes
|
Occurrence (frequency) of alleles in species | Undetermined
|
Locality
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 02 | 03 | 04 | 05 | 06 | 07 | 08 | 09 | 10 | 11 | 12 | 13 | 14 | |||||
| Paralabidochromis beadlei | 3 | 0 | 6 | LN | ||||||||||||||
| P. chilotes | 3 | 1 | 1 | 5 | LV | |||||||||||||
| N. nigrican | 3 | 0 | 4 | 2 | LV | |||||||||||||
| H. nyererei | 3 | 1 | 1 | 5 | LV | |||||||||||||
| Paralabidochromis plagiodon | 3 | 2 | 2 | 1 | 1 | 2 | LV | |||||||||||
| H. pyrrhocephalus | 4 | 0 | 4 | 2 | LV | |||||||||||||
| Gaurochromis simpsoni | 3 | 1 | 2 | 2 | 2 | LN | ||||||||||||
| Astatotilapia velifer | 3 | 0 | 2 | 4 | LN | |||||||||||||
| Haplochromis velvet black | 3 | 3 | 2 | 2 | 2 | LV | ||||||||||||
| Ptyochromis xenognathus | 3 | 0 | 6 | LV | ||||||||||||||
| Ptyochromis sauvagei | 3 | 0 | 6 | LV | ||||||||||||||
| Haplochromis rockkribensis | 3 | 1 | 1 | 5 | LV | |||||||||||||
| Yssichromis laparogramma | 3 | 0 | 2 | 2 | 2 | LV | ||||||||||||
Heterozygotes were revealed by the presence of two superimposed nucleotide signals in the sequence profiles obtained by direct sequencing of PCR fragments. “Undetermined” indicates the presence of more than two sites of two superimposed nucleotide signals. Of 264 individuals, 41 tested for exon 4 variability, and 12 of 40 individuals tested for variability in exons 2–5 (16% and 30%, respectively) were heterozygotes. The allelic composition of the heterozygotes could be deduced only when single nucleotide sites varied or when the heterozygosity pattern consisted unambiguously of two alleles present in the same population.
LN, Lake Nabugabo; LV, Lake Victoria.
Fig 3.
Variable nucleotide sites differentiating LWS alleles of Lake Victoria/Lake Nabugabo cichlids. A dash (—) indicates identity with the top sequence (allele 01). Nonsynonymous substitution sites in which amino acid replacement might change the absorption spectra of opsin proteins are highlighted in black; the remaining nonsynonymous sites are highlighted in gray. At 3 of the 19 nonsynonymous sites (391, 529, and 844) the amino acid replacements caused by the nucleotide substitutions have been shown in other systems to result in shifts of absorption sensitivity (λmax; refs. 26 and 27). At one of the three sites (391) and two other sites (572 and 647) substitutions cause amino acid replacements at positions 131, 191, and 216, respectively, which correspond to positions 118, 178, and 203 in the bovine rhodopsin molecule, respectively. From the crystal structure of bovine rhodopsin (28), the residues at positions 131, 191, and 216 in the cichlid LWS opsin can be expected to be close to each other and may be able to form hydrogen bonds among them. The replacements, in cichlid LWS, could be expected to disrupt these hydrogen bonds and thus lead to a shift in absorption sensitivity. Substitutions at sites 529 and 844 cause amino acid replacements at positions 177 and 282, respectively, which are among the five positions that, according to the “five-sites rule” of Yokoyama and Radlwimmer (27), are primarily responsible for the spectral sensitivities of the MWS and LWS pigments in humans and fishes. The two ancient allelic lineages (I and II) are indicated by brackets. Amino acid replacements are given in the International Union of Physiological Sciences single-letter code; the arrows indicate a change from the more common to the less common residue. “Positions” refers to the numbering of amino acid residues in the cichlid LWS opsin.
Table 2.
LWS exon 4 variability in cichlid fishes of Lakes Victoria/Nabugabo and other East African lakes and rivers
| Species
|
Occurrence (frequency) of alleles in taxa | Undetermined
|
Locality
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Heterozygotes | 01, 02, 03, 04, 05 | 09, 10 | 06, 08 | 07 | 11 | 12 | 13 | 14 | 17 | 18 | 19 | |||
| Lakes Victoria/Nabugabo | |||||||||||||||
| P. beadlei | 10 | 2 | 18 (0.90) | 2 (0.10) | 36 | ||||||||||
| P. chilotes | 22 | 3 | 17 (0.39) | 16 (0.36) | 11 (0.25) | 54, 52, 50, 55 | |||||||||
| Lipochromis melanopterus | 5 | 0 | 10 (1.0) | 51 | |||||||||||
| N. nigricans | 10 | 0 | 20 (1.0) | 50, 51 | |||||||||||
| Astatotilapia nubila | 13 | 4 | 9 (0.35) | 3 (0.12) | 4 (0.15) | 4 (0.15) | 6 (0.23) | 36, 36 | |||||||
| H. nyererei | 9 | 3 | 4 (0.22) | 2 (0.11) | 11 (0.61) | 1 (0.06) | 57 | ||||||||
| Haplochromis “piscivore” | 4 | 0 | 4 (0.50) | 4 (0.50) | 51, 39 | ||||||||||
| P. plagiodon | 9 | 7 | 2 (0.11) | 7 (0.39) | 6 (0.33) | 3 (0.17) | 47; 47 | ||||||||
| H. pyrrhocephalus | 7 | 0 | 14 (1.0) | 47 | |||||||||||
| 8 | 0 | 16 (1.0) | 51 | ||||||||||||
| 3 | 1 | 4 (0.66) | 2 (0.33) | 53 | |||||||||||
| G. simpsoni | 10 | 2 | 9 (0.45) | 4 (0.20) | 5 (0.25) | 1 (0.05) | 1 (0.05) | 36 | |||||||
| A. velifer | 10 | 2 | 9 (0.45) | 7 (0.35) | 1 (0.05) | 3 (0.15) | 36 | ||||||||
| H. velvet black | 10 | 0 | 20 (1.0) | 57, 50, 49 | |||||||||||
| Prognathochromis venator | 6 | 1 | 9 (0.75) | 3 (0.25) | 36 | ||||||||||
| P. xenognathus | 10 | 0 | 20 (1.0) | 57, 57, 48 | |||||||||||
| P. sauvagei | 10 | 3 | 5 (0.25) | 15 (0.75) | 57, 57, 53, 53 | ||||||||||
| H. rockkribensis | 19 | 1 | 33 (0.86) | 5 (0.13) | 51, 54, 52, 47, 55 | ||||||||||
| Y. laparogramma | 9 | 1 | 11 (0.61) | 5 (0.28) | 2 (0.11) | 48, 53, 48 | |||||||||
| Other lakes and rivers | |||||||||||||||
| 10 | 1 | 20 (1.0) | 45 | ||||||||||||
| 10 | 1 | 2 (0.10) | 17 (0.85) | 1 (0.05) | 18 | ||||||||||
| 9 | 0 | 18 (1.0) | 58 | ||||||||||||
| 5 | 0 | 10 (1.0) | 4 | ||||||||||||
| 9 | 3 | 1 (0.06) | 12 (0.67) | 5 (0.28) | 34 | ||||||||||
| 5 | 0 | 10 (1.0) | 30 | ||||||||||||
| 4 | 0 | 8 (1.0) | 10 | ||||||||||||
| 4 | 2 | 4 (0.50) | 2 (0.25) | 1 (0.25) | 32 | ||||||||||
| 5 | 2 | 2 (0.20) | 4 (0.40) | 4 (0.40) | 32 | ||||||||||
| 8 | 0 | 16 (1.0) | 56 | ||||||||||||
| 7 | 1 | 5 (0.36) | 6 (0.43) | 3 (0.21) | 40 | ||||||||||
| 4 | 1 | 6 (0.75) | 2 (0.25) | 37 | |||||||||||
Alleles fixed in species or population in Lakes Victoria/Nabugabo are underlined.
These alleles are distinguished by differences in exons 2, 3, and 5 but not 4.
Numbers in parentheses refer to the map in Fig. 1.
Fixation of the LWS Alleles.
The patterns of distribution of the LWS alleles among the species and populations of the Lake Victoria haplochromine flock can be divided into two categories. Some alleles, when present, appear to be fixed in some species or populations (Table 2, underlined), whereas the same or other alleles occur in the populations of the different species at polymorphic frequencies. For example, allele 14 appears to be fixed in the H. pyrrhocephalus population of Buzumu Bay but is present at a polymorphic frequency of 0.11 in Y. laparogramma (Table 2). Table 3 shows a compilation of the fixed LWS alleles (exon 4) among individuals in some species or populations, each of which is characterized by having particular nucleotides in the six variable sites comprising one synonymous site (645) and five nonsynonymous sites (see Discussion).
Table 3.
The fixed LWS gene variability (exon 4) in cichlid fishes of Lake Victoria
| Species
|
Locality
|
n
|
Frequency of alleles in species or populations
|
Alleles
|
Sites | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 6 | 6 | 6 | 6 | 6 | 6 | |||||
| 7 | 4 | 4 | 6 | 7 | 7 | 8 | |||||
| 2 | 5 | 7 | 4 | 6 | 9 | 8 | |||||
| N. nigricans | 10 | 1.0 | 01, 02, 03, 04, 05 | A | C | A | C | A | T | G | |
| H. pyrrhocephalus | Buzumu bay | 7 | 1.0 | 14 | A | C | A | A | T | A | T |
| Speke golf | 8 | 1.0 | 09, 10 | A | G | T | C | A | T | A | |
| H. velvet black | 10 | 1.0 | 01, 02, 03, 04, 05 | A | C | A | C | A | T | G | |
| P. xenognathus | 10 | 1.0 | 06, 08 | A | C | A | C | A | T | A | |
| L. melanopterus | 5 | 1.0 | 01, 02, 03, 04, 05 | A | C | A | C | A | T | G | |
Localities at which alleles were the same are not shown.
These alleles are distinguished by differences in exon 2, 3, and 5 but not 4.
Phylogenetic Analysis of LWS Genes.
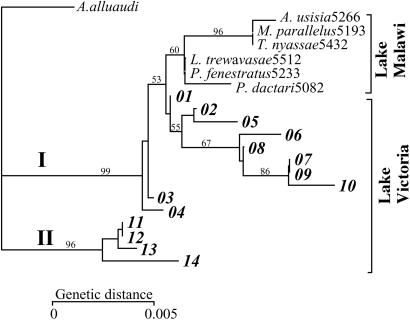
The phylogenetic analysis of 14 Victorian alleles of LWS genes indicates that they can be divided into two groups, designated I and II (Fig. 4). Alleles from Lake Malawi were shown to be included in group I, indicating that the distance between LWS alleles of the same species in Lake Victoria was comparable to or exceeded that observed in inter-lake comparisons (e.g., alleles 10 and 14 in Fig. 4). It also suggests that the divergence of these two groups predated the divergence of the Lake Victoria and Lake Malawi haplochromines (see Discussion).
Fig 4.
Phylogenetic relationship of LWS genes. The dendrogram was constructed by using the neighbor-joining method (21) on Kimura two-parameter distances (29) of exon 2–5 nucleotide sequences. Bootstrap values obtained in 500 replications are given on the branches. The two ancient allelic lineages (I and II) are indicated on the branches. The geographical origin of the samples is indicated. The alleles found in Lake Victoria are specified by numbers. The Lake Malawi sequences were from Aulonocara usisia, Labeotropheus trewavasae, M. parallelus, Protomelas fenestratus, P. daktari, and Taeniolethrinops nyassae.
Discussion
High Variability of the LWS Locus in Lake Victoria Haplochromine.
Compared with the SWS2B locus and other loci, the variability of the LWS locus in Lake Victoria haplochromines is extraordinary. Although variability has been found in this group of fish at both nuclear (7) and mitochondrial (control region; ref. 5) DNA segments, it is different (with one exception) from that observed at the LWS locus in terms of both the number of alleles and the genetic distances between the alleles. At the most polymorphic of the nuclear loci studied previously, only two alleles were found (7) in a sample comparable in size and composition to the one used in the present study. More significantly, however, alleles at the previously studied loci differed from each other by one nucleotide substitution only (7). By contrast, a pairwise comparison of the 14 LWS alleles identified in the Lake Victoria sample revealed a range of differences that extended from 1 nucleotide substitution (1 amino acid replacement) to 20 substitutions (15 replacements), which represents a range of genetic distances from 0.00115 to 0.2294. There is only one other set of loci that displays such striking variability in Lake Victoria haplochromines: the class I and class II loci of the major histocompatibility complex (Mhc; ref. 8). Although 12 LWS alleles were found in the Lake Malawi samples, there were only 9 variable sites in these alleles (data not shown), and the variation among the alleles was ≤0.0069. Thus, the high variability of the LWS locus is particularly conspicuous in the Lake Victoria haplochromine species flock.
Driving Force Behind the Variation of the LWS Gene and Fixation of Alleles.
These findings raise a question: What is the driving force behind the variation of the LWS gene? The following observations suggest that the answer is selection. First, extensive variability occurs at one locus (LWS) but not at another, closely related locus (SWS2B) and at other genes (5–7). Second, at the LWS locus, high variability occurs in the Lake Victoria but much less in the Lake Malawi species flocks (see below). Third, within the LWS locus of the Lake Victoria haplochromines, the high variability is restricted to the exons. We sequenced the longest intron of the LWS gene (intron 1, ≈800 bp long) from 38 individuals representing four Lake Victoria/Nabugabo species and the nonendemic A. alluaudi and found the sequences to be either identical (77%) or separated by a genetic distance of only 0.0014–0.0430. Fourth, within the exons, there is a preponderance of nonsynonymous (Dn) over synonymous (Ds) substitutions (Fig. 3). Because, however, the number of synonymous sites in the sequenced segments is low, most of the observed values were not statistically significant. Fifth, variation of the nonsynonymous sites is distributed unevenly. There are 25 variable sites in the sequenced coding part of the gene, 6 of them synonymous and the remaining 19 nonsynonymous (Fig. 3). At 5 of the 19 nonsynonymous sites (391, 529, 572, 647, and 844; Fig. 3, highlighted in black), the amino acid replacements caused by the nucleotide substitutions might lead to a shift in absorption sensitivity (see Fig. 3 legend). Substitutions at the remaining 14 nonsynonymous sites cause amino acid replacements at different positions in the transmembrane helices of the opsin molecule (Fig. 3). Whether any of these replacements affect the absorption sensitivity of the LWS pigment is not known. If they do not, they have other functions, or the polymorphism at these sites could be influenced, similar to that at the synonymous sites, by their vicinity to the sites under selection. Sixth, the sites that vary undergo substitutions repeatedly, a conclusion that follows from the results of phylogenetic analysis. We aligned the opsin protein sequences deposited in the databases with the sequences obtained by us and used the alignment to draw a phylogenetic tree of the entire opsin protein family (not shown). We then focused on the variable sites and used maximum-likelihood methods to reconstruct the most likely path of nucleotide substitutions during the evolution of the opsin genes. The analysis indicates that the same substitutions occurred more than once at some of these sites. Whether the substitutions occurred by mutations, recombination, or some other mechanism is not relevant to this conclusion. If the variants were the results of increased rates of these processes rather than selection, one would not expect the recurrence of the same change. Seventh, there is evidence for evolutionary convergence at some of the variable amino acid positions. The sequence alignment of the various opsin proteins reveals that all the amino acid residues accounting for the variation of the LWS opsin in Lake Victoria haplochromines also can be found in other opsins. The position of these proteins on the phylogenetic tree rules out the possibility that the sharing of residues is a consequence of a common descent. Codon analysis further supports convergence as an explanation for the sharing. Although taken individually some of these observations are open to alternative interpretations, taken together they strongly support the conclusion that selection is acting on the variable sites.
The LWS alleles in exon 4 that were distinguished by seven variable sites (572, 645, 647, 664, 676, 679, and 688) were fixed in four species and two populations in Lake Victoria/Lake Nabugabo (Tables 2 and 3), although as mentioned earlier molecular analysis showed trans-species persistence of neutral polymorphisms among Lake Victoria haplochromines; no species-specific substitution has been documented to date. The LWS thus is the first locus on record indicating genetic differentiation of populations and species constituting the Lake Victoria haplochromine flock. The LWS alleles seem to be fixed in these species or populations by selection acting on the variable sites. (The synonymous substitution at site 645 presumably was fixed by close linkage with sites under selection, i.e., by “hitchhiking.”)
Possible Relationship Between the Variation of LWS and the Adaptive Radiation of the Haplochromine Flock.
We note that the only known function of the cone opsins is color discrimination. The adaptiveness of the variation in the LWS gene therefore should be related to activities in which color sensation and discrimination, particularly in the longer wavelength segment of the visible spectrum, plays a part. Two such activities spring to mind, both related to the photic environment in which the fish live: food acquisition and mate choice. That perception of color is an important factor in these activities of haplochromine cichlids is well documented. Cichlids are known to search for food visually (1), and color discrimination enables them to recognize food sources against the background of a particular photic environment. Specialization to different food resources in a given ecological niche leads in turn to morphological adaptation to that niche and so contributes to speciation. As for mate selection, male cichlids are known for their display of bright breeding coloration, and the ability of females to choose conspecific males on the basis of visual cues has been documented extensively by observations in natural environments and behavioral experiments (9, 22, 23). It therefore is likely that the genetic variation and fixation at the LWS locus is connected to these two interrelated activities and through them to speciation. In this context, the fact that high variability occurs at the LWS but not SWS2B locus might be significant. The waters of Lake Victoria, in contrast to those of Lake Malawi (or Lake Tanganyika), are generally quite turbid and hence largely impenetrable to any great depth to light of shorter wavelengths (24). Color discrimination under these conditions therefore can be expected to shift to longer wavelengths of the visible spectrum. The observed variation and fixation in the LWS gene might be a reflection of just such a shift. If this were the case, one might expect to find a corresponding shift in the coloration of the fishes. Indeed, a difference in coloration between Lake Victoria and Lake Malawi haplochromines has been noted (ref. 25; Fig. 1b). Although the patterns of coloration of the fishes in the two lakes are similar; generally the color patches that are yellow in Lake Malawi are red in Lake Victoria.
Evolution of LWS Alleles.
If the endemic Lake Victoria haplochromine species flock is indeed as young as recent studies suggest (4, 5), it is unlikely that all the observed variation, involving differences of up to 20 substitutions between some of the alleles, arose in Lake Victoria within a span of 12,400 years. To search for the origin of the variants, we therefore sequenced LWS exon 4 of 80 haplochromine fishes inhabiting rivers and other lakes. The sample covered 12 localities in different regions of East Africa (Fig. 1). Among the 83 sequences obtained, 3 of the 11 alleles from both allelic lineages I and II found in Lake Victoria also were present in the riverine species (Table 2). Some of the Lake Victoria alleles therefore must have been present already in the riverine-founding populations from which the adaptive radiation in this lake took off. This conclusion is strengthened further by phylogenetic analysis, which suggests that the divergence of the two groups of LWS alleles predated the divergence of the Lake Victoria and Lake Malawi haplochromine flocks (Fig. 4). This conclusion also makes sense in terms of the adaptiveness of the variants. Because the waters of most of the East African rivers are no less turbid but much more variable and unstable in terms of photic conditions than those of Lake Victoria, the variations must have possessed an adaptive value in the East African rivers already, allowing greater color discrimination at longer wavelengths. In the founding populations that migrated from the rivers to the forming Lake Malawi, the variation might have been lost largely through fixations of a limited number of alleles, because the need for the discrimination in this part of the spectrum ceased to exist in the lake's clear waters. In the populations that founded the Lake Victoria flock, by contrast, the turbid waters might have favored the retention of the variation over the entire lake, but the greater stability of the various niches (in contrast to the instability in the riverine environments) favored a trend toward fixation of various alleles in different populations and species. Measurements of light absorption by the individual LWS variants are now in progress. They should either confirm or refute some of these inferences.
Supplementary Material
Acknowledgments
We thank Drs. Y. Shichida and H. Imai (Graduate School of Science, Kyoto University, Kyoto) for helpful discussion and information about opsin, Ms. N. Morikawa for technical assistance, and Ms. J. Kraushaar for editorial assistance. This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
References
- 1.Fryer G. & Iles, T. D., (1972) The Cichlid Fishes of the Great Lakes of Africa (Oliver & Boyd, Edinburgh).
- 2.Greenwood P. H., (1981) The Haplochromine Fishes of the East African Lakes (Cornell Univ. Press, Ithaca, NY).
- 3.Seehausen O., (1996) Lake Victoria Rock Cichlids: Taxonomy, Ecology, and Distribution (Verduijn Cichlids, Zevenhuizen, The Netherlands).
- 4.Johnson T. C., Scholz, C. A., Talbot, M. R., Kelts, K., Ricketts, R. D., Ngobi, G., Beuning, K., Ssemmanda, I. I. & McGill, J. W. (1996) Science 273 1091-1093. [DOI] [PubMed] [Google Scholar]
- 5.Nagl S., Tichy, H., Mayer, W. E., Takezaki, N., Takahata, N. & Klein, J. (2000) Proc. R. Soc. London Ser. B 267 1049-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer A., Kocher, T. D., Basasibwaki, P. & Wilson, A. C. (1990) Nature 347 550-553. [DOI] [PubMed] [Google Scholar]
- 7.Nagl S., Tichy, H., Mayer, W. E., Takahata, N. & Klein, J. (1998) Proc. Natl. Acad. Sci. USA 95 14238-14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein D., Ono, H., O'hUigin, C., Vincek, V., Goldschmidt, T. & Klein, J. (1993) Nature 364 330-334. [DOI] [PubMed] [Google Scholar]
- 9.Seehausen O. & van Alphen, J. J. M. (1998) Behav. Ecol. Sociobiol. 42 1-8. [Google Scholar]
- 10.Wald G. (1968) Nature 219 800-807. [DOI] [PubMed] [Google Scholar]
- 11.Shichida Y. (1999) in The Retinal Basis of Vision, ed. Toyoda, J. (Elsevier Science, Amsterdam).
- 12.Yokoyama S. (2000) Prog. Retin. Eye Res. 19 385-419. [DOI] [PubMed] [Google Scholar]
- 13.Yokoyama S. (1995) Mol. Biol. Evol. 12 53-61. [DOI] [PubMed] [Google Scholar]
- 14.Carleton K. L., Harosi, F. I. & Kocher, T. D. (2000) Vision Res. 40 879-890. [DOI] [PubMed] [Google Scholar]
- 15.Carleton K. L. & Kocher, T. D. (2001) Mol. Biol. Evol. 18 1540-1550. [DOI] [PubMed] [Google Scholar]
- 16.Greenwood P. H. (1979) Bull. Br. Mus. Nat. Hist. Zool. 35 265-322. [Google Scholar]
- 17.Sugawara T., Terai, Y. & Okada, N. (2002) Mol. Biol. Evol. 19 1807-1811. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert D. G., (1996) SEQPUP, A Biosequence Editor and Analysis Application (Indiana Univ., Bloomington), Version 0.6f.
- 19.Swofford D. L., (2001) paup, Phylogenetic Analysis Using Parsimony (Sinauer, Sunderland, MA), Version 4.0b8.
- 20.Kumar S., Tamura, K., Jakobsen, I. B. & Nei, M., (2001) MEGA 2, Molecular Evolutionary Genetic Analysis software (Arizona State Univ., Tempe). [DOI] [PubMed]
- 21.Saitou N. & Nei, M. (1987) Mol. Biol. Evol. 4 406-425. [DOI] [PubMed] [Google Scholar]
- 22.Dominey W. J. (1984) in Evolution of Fish Species Flocks, eds. Echelle, A. A. & Kornfield, I. (Univ. of Maine at Orono Press, Orono), pp. 231–249.
- 23.Knight M. E. & Turner, G. F. (1999) Anim. Behav. 58 761-768. [DOI] [PubMed] [Google Scholar]
- 24.van der Meer H. J. & Bowmaker, J. K. (1995) Brain Behav. Evol. 45 232-240. [DOI] [PubMed] [Google Scholar]
- 25.Seehausen O., van Alphen, J. J. M. & Witte, F. (1999) Belg. J. Zool. 129 43-60. [Google Scholar]
- 26.Janz J. M. & Farrens, D. L. (2001) Biochemistry 40 7219-7227. [DOI] [PubMed] [Google Scholar]
- 27.Yokoyama S. & Radlwimmer, F. B. (1998) Mol. Biol. Evol. 5 560-567. [DOI] [PubMed] [Google Scholar]
- 28.Palczewski K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., Le, T. I., Teller, D. C., Okada, T., Stenkamp, R. E., Yamamoto, M. & Miyano, M. (2000) Science 289 739-745. [DOI] [PubMed] [Google Scholar]
- 29.Kimura M. (1980) J. Mol. Evol. 16 111-120. [DOI] [PubMed] [Google Scholar]
- 30.Hamada M., Kido, Y., Himberg, M., Reist, J. D., Cao, Y., Hasegawa, M. & Okada, N. (1997) Genetics 146 355-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.