Abstract
Micro-RNAs (miR genes) are a large family of highly conserved noncoding genes thought to be involved in temporal and tissue-specific gene regulation. MiRs are transcribed as short hairpin precursors (≈70 nt) and are processed into active 21- to 22-nt RNAs by Dicer, a ribonuclease that recognizes target mRNAs via base-pairing interactions. Here we show that miR15 and miR16 are located at chromosome 13q14, a region deleted in more than half of B cell chronic lymphocytic leukemias (B-CLL). Detailed deletion and expression analysis shows that miR15 and miR16 are located within a 30-kb region of loss in CLL, and that both genes are deleted or down-regulated in the majority (≈68%) of CLL cases.
Chronic lymphocytic leukemia (CLL) is the most common form of adult leukemia in the Western world (1). Hemizygous and/or homozygous loss at 13q14 occur in more than half of cases and constitute the most frequent chromosomal abnormality in CLL (reviewed in ref. 2). Deletions at 13q14 also occur in ≈50% of mantle cell lymphoma, in 16–40% of multiple myeloma, and in 60% of prostate cancers (3), suggesting that one or more tumor suppressor genes at 13q14 are involved in the pathogenesis of these human tumors. The karyotyping of CLL samples identified relatively few chromosomal abnormalities, suggesting that the specificity and frequency of observed deletions at 13q14 have pathologic significance. Several groups have used positional cloning to identify the gene or genes targeted by the deletions. A region of more than 1 Mb has been fully sequenced and characterized in detail (4, 5). So far, a total of eight genes have been identified and screened for alterations at the DNA and/or RNA level in sporadic and familial cases of CLL: Leu-1 (BCMS or EST70/Leu-1), Leu 2 (ALT1 or 1B4/Leu-2), Leu 5 (CAR), CLLD6, KPNA3, CLLD7, LOC51131 (putative zinc finger protein NY-REN-34 antigen) and CLLD8. However, detailed genetic analysis, including extensive loss of heterozygosity (LOH), mutation, and expression studies, have failed to demonstrate the consistent involvement of any of the genes located in the deleted region (4–11).
Noncoding RNAs (ncRNA) range in size from 21 to 25 nt for the large family of micro-RNAs (miRNAs) that modulate development in several organisms including mammals, up to >10,000 nt for RNAs involved in gene silencing in higher eukaryotes (12). miRNAs represent a new class of gene products whose functions are generally unknown, and are typically excised from 60- to 70-nt fold-back RNA precursor structures. The miRNA precursor processing reaction requires Dicer RNase III and Argonaute family members (13). Here we show that two miR genes are located at 13q14 within a 30-kb region of loss in CLL, and that both genes are deleted or down-regulated in the majority of CLL cases.
Materials and Methods
Patient Samples and Cell Lines.
Patient samples were obtained after informed consent from patients diagnosed with CLL at the CLL Research Consortium institutions. Briefly, peripheral blood was obtained from CLL patients, and mononuclear cells were isolated through Ficoll/Hypaque gradient centrifugation (Amersham Pharmacia Biotech) and then processed for RNA and DNA extraction according to standard protocols (14). As normal controls for LOH studies, we used DNA from bucal mucosa from the corresponding patients included on small (1–2 mm2) pieces of paper.
We also used 30 human cell lines: AS283, BL2, Bla, BJAB, CA46, Namalva, P3HRI, PAPB 682, PABm, and Raji (Burkitt's lymphoma); Del1, SKDHL, and ST486 (T cell lymphoma); JM (immunoblastic B cell lymphoma); MC116 (undifferentiated lymphoma); Molt3 and Supt 11 (T-ALL); U266 (multiple myeloma); A549 and H1299 (lung carcinoma); TE2 and TE10 (esophageal carcinoma); HeLa (cervical carcinoma); RC48 (kidney carcinoma); and 2220, 2221, 11609, 11611, LNCAP, and TSUR (prostate carcinoma). The cell lines were obtained from the American Type Culture Collection and maintained according to their instructions.
CD5+ B Cell Separation.
Tonsils were obtained from patients in the pediatric age group (3–9 years) undergoing routine tonsillectomies. Purified B cells were obtained by rosetting the mononuclear cells with neuraminidase treated sheep erythrocytes. The B cells were further fractionated by discontinuous Percoll gradients (Pharmacia Biotech, Uppsala) as detailed (15). The B cells collected from the 50% Percoll fraction were incubated with anti-CD5 mAb, followed by goat anti-mouse Ig conjugated with magnetic microbeads. CD5+ B cells were obtained by positive selection by collecting the cells retained on the magnetic column MS with the MiniMACS system (Miltenyi Biotec, Auburn, CA).
Somatic Cell Hybrids.
Somatic cell hybrids were generated following conventional methods and selected in hypoxanthine-aminopterin-thymidine (HAT) medium as described (16). DNA derived from single-cell clones and subclones was isolated with the DNeasy tissue kit (Qiagen) and screened by PCR for the presence or absence of chromosome 13 and chromosome 2 markers (see Table 3, which is published as supporting information on the PNAS web site, www.pnas.org, for primer sequence). Fifteen clones were isolated from fusion of a CLL case (CLL-B) carrying a t(2;13)(q32; q14) translocation, and one clone was isolated from fusion of another CLL case (CLL-A) carrying a t(2;13)(q12; q13) translocation. Twelve CLL-B-derived clones carried a full complement of both chromosomes 13 and 2, whereas three carried the del(13q) and a full complement of chromosome 2. The single clone from CLL-A carried a chromosome 13 with a small deletion at 13q14, and no part of chromosome 2.
Northern Blotting.
Total RNA isolation was performed using the Tri-Reagent protocol (Molecular Research Center, Cincinnati). RNA samples (30 μg each) were run on 15% acrylamide denaturing (urea) Criterion precast gels (Bio-Rad), and then transferred onto Hybond-n + membrane (Amersham Pharmacia Biotech). The hybridization with [α-32P]ATP was performed at 42°C in 7% SDS/0.2M Na2PO4 (pH 7.0) overnight. Membranes were washed at 42°C, twice with 2× SSPE [standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA)]/0.1% SDS and twice with 0.5× SSPE/0.1% SDS. The probes were as published (17). Blots were stripped by boiling in 0.1% aqueous SDS/0.1× SSC for 10 min and were reprobed several times. As loading control we used 5S rRNA stained with ethidium bromide.
RT-PCR.
The RT-PCR was performed to analyze the levels of gene expression in normal CD5 cells and 23 B-CLL samples. One microliter of cDNA was used for each PCR with Advantage2 PCR kit (CLONTECH), and 10 pmol of each gene-specific primer for 35 cycles of 94°C for 20 s, 65°C for 30 s, and 68°C for 1 min (for a complete list of primers used in this study, see Table 3, which is published as supporting information on the PNAS web site). To ensure that the RNA was of sufficient purity for RT-PCR, a PCR assay with primers specific for GAPDH cDNA (CLONTECH) was used. RT-PCR products were separated by agarose gel electrophoresis following standard procedures (14).
Western Blotting.
SDS/PAGE gels of cell lysates from nine B-CLL patients were probed with GST-SLUG Middle antibody (a gift from Thomas Look, Harvard University, Cambridge, MA) and SNX2 (N17) antibody (Santa Cruz Biotechnology). Detection was performed using the ECL Western Blotting Detection Kit (Amersham Pharmacia) according to the manufacturer's instructions.
Database Analysis.
blast searches against nr, dbEST, and search for short nearly exact matches were done at the NCBI server (www.ncbi.nlm.nih.gov/). fasta searches for homology of short sequences were done at the Biology WorkBench site (http://workbench.sdsc.edu).
Results
A 30-kb Region of Deletion Was Identified by Exploiting Somatic Cell Hybrids of B-CLL Patients.
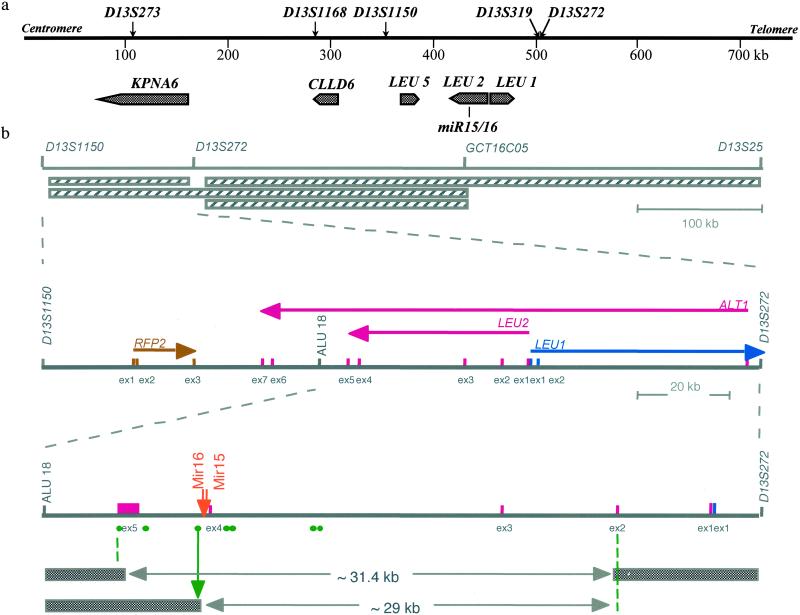
One of the difficulties first encountered during analysis of the CLL locus at 13q14 was the lack of a clear definition of the minimal region of loss, because various and relatively large (between 130 and 550 kb) such regions have been described in CLL (Fig. 1; refs. 18–22). In previous studies, using LOH and Southern blot analysis, we identified the centromeric boundary of homozygous loss at the Alu18 locus (Fig. 1; ref. 18), located between D13S1150 and D13S272, less than 65 kb centromeric to exon 5 of the LEU2 gene. However, at that time we did not find any small or overlapping homozygous deletions that would allow us to better localize the target tumor suppressor. Therefore, we generated somatic cell hybrids between mouse LM-TK− and CLLs cells carrying 13q14 translocations and/or deletions (see Materials and Methods). PCR screening of resulting hybrid clones allowed us to segregate the two copies of chromosome 13 present in the tumors. In this manner, we identified a 31.4-kb deletion in one case, and precisely localized the chromosomal breakpoint in the other (Fig. 1). Our results indicate that the 13q14 tumor suppressor gene(s) lies within a 29-kb region between exons 2 and 5 of the LEU2 gene. As shown in Fig. 1, this region is consistent with all reported regions of loss, including a 10-kb region reported several years ago by Liu et al. (6). Exons 1 and 2 of LEU2 lie within that region (and within the one defined here). However, LEU2 has been the subject of extensive study, and was excluded as a likely candidate tumor suppressor gene for B-CLL by us and others (4, 5, 9, 10).
Fig 1.
(a) Genes within the 13q14 tumor suppressor locus in CLL and localization of miR15/16 cluster. The positions of genetic markers and the positions of genes on the map are shown. (b Top) Map of the locus with previously reported 13q14 deletions marked by horizontally striped boxes. (Middle) Map of the locus between D13S1150 and D13S272 markers. The orientation of each gene is marked by an arrow under the gene's name. Colored vertical bars mark the position of corresponding exons for each gene. (Bottom) Map of the locus between Alu18 and D13S272 markers. Bars and boxes mark the position of exons for LEU2/ALT1 and LEU1 following colors in panels above. The orange arrow marks the position of miR15 and miR16 genes. Green circles mark the positions of PCR primers used to screen somatic cell hybrid clones derived from a fusion of two independent leukemia cases (CLL-A and CLL-B). Green circles mark the positions of oligonucleotides pairs used to screen hybrids by PCR. All oligonucleotides pairs shown, as well as exon-specific primers for each exon shown, and miR15/16-specific primers were used to screen the hybrids. The green arrow represents the position of the breakpoint in CLL-B carrying a t(2;13)(q32;q14) translocation. Filled boxes represent portions of chromosome 13 present in the hybrids. The 31.4-kb deletion was present in a clone derived from CLL-A, a patient with CLL carrying a t(2;13)(q12;q13) translocation, bilateral retinoblastoma, and ulcerative colitis.
Two Small Noncoding RNAs, miR15 and miR16, Are Localized in the Minimally Deleted Region and Are Highly Expressed in CD5+ Cells.
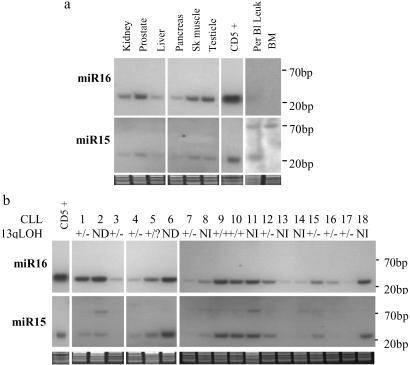
The delineation of the minimally region of loss at 13q14 led us to continue the search for genes within the region. Using publicly available sequence information and databases, we screened for new regulatory genes in this area. We found that a cluster of two recently cloned miR genes, miR15 and miR16 (17, 23–25), is located exactly in the deleted region (Fig. 1), and could therefore be involved in CLL pathogenesis. To evaluate the level of expression of miR15 and miR16 in normal tissues, we performed Northern blot analysis on a panel of normal tissues, including CD5+ B cells isolated from tonsils of normal individuals (Fig. 2). We used these cells as controls because B-CLL is characterized by a progressive accumulation of CD5+ B lymphocytes (26). We found ubiquitous expression of both genes with the highest level in normal CD5+ lymphocytes, suggesting that these genes play an important role in normal CD5+ B cell homeostasis. Also, in normal tissues, miR16 is consistently expressed at higher levels than miR15.
Fig 2.
(a) Analysis of miR15/16 cluster expression in human tissues by Northern blotting reveals that the two genes are ubiquitously expressed, with the highest levels in normal CD5+ cells. (b) The majority of CLL samples express lower levels of miR16 and miR15 as normal CD5+ cells. The LOH status for the presented samples is shown as: +/+, heterozygosity; +/−, LOH; −/−, homozygous deletion; NI, not informative; ND, not done. As normalization controls, we used staining with ethidium bromide of Northern gels.
miR15 and miR16 Are Frequently Deleted or Down-Regulated in CLL Samples with Deletions on 13q14.
To determine whether miR15 and miR16 are indeed involved in CLL pathogenesis, we analyzed 60 CLL samples and 30 human cancer cell lines by Northern blotting (Fig. 2). We found that 68% of CLL patients (41/60), as well as five of six analyzed prostate cancer cell lines, showed a significant reduction in expression when compared with their normal tissue counterparts. These findings demonstrate that miR15 and miR16 are down-regulated in the majority of B-CLL cases. In addition, we found that 23 of 60 samples (38%) present a clearly identifiable band of ≈70 nt representing the precursor miR15 RNA that was not found in any normal tissue analyzed except for bone marrow (Fig. 2), suggesting that miR15 could be inefficiently processed in CLL.
To determine whether the observed down-regulation of expression correlates with allelic loss in CLL, we performed LOH studies with microsatellite markers D13S272 and D13S273 on 46 patients from whom normal DNA was available. We found that 68% of informative samples display LOH in at least one marker (24 of 35 cases). In all but four samples (75%), the levels of miR15/16 were reduced (for 12 samples we were not able to obtain reproducible results for the normal samples because of the poor quality of the starting material). Additionally, expression levels were reduced in 6 of 11 cases (55%), without apparent LOH. In these cases, deletions may have been too small to be detected with the markers analyzed (as was the case in one of the patients used to generate somatic cell hybrids).
Northern analysis suggested that both miR15 and miR16 were expressed in cases with known large homozygous deletions at 13q14, and with <5% normal cells, pointing to the presence of other highly similar miRNA genes in the genome. Indeed, a cluster very similar to miR15 and miR16 (but with different precursors) was reported on chromosome 3q25–26.1 (23). To prove that the variation in miR15/16 expression is strictly related to deletions on chromosome 13q, we designed two specific probes for each precursor of miR16. We found that, whereas the premessenger from chromosome 13 can be detected at low levels, no specific hybridization with chromosome 3 probe can be found in the same samples. In addition, we performed an LOH study with two microsatellite markers spanning a region of 2 Mb located immediately centromeric to this cluster. Four of 17 informative samples showed LOH in at least one marker, and no correlation with the levels of expression of miR15/16 was found. These data clearly demonstrate that down-regulation of miR15 and miR16 expression in CLL correlates with allelic loss at 13q14, and point to a role for miR15 and miR16 in CLL pathogenesis.
The Mutation Study Did Not Reveal Point Mutations in CLL and Gastrointestinal Cancers.
To further evaluate involvement of these genes in CLL, we screened a set of 120 B-CLLs and 80 colorectal and gastric cancers for mutations by direct sequencing of PCR amplification products. We amplified a 720-bp genomic region containing the entire cluster. In three cases, we found the same alteration in the precursor of miR16, a T to C substitution at position 2. This change is not predicted to alter the hairpin structure. We also found several extragenic polymorphisms (data not shown). This paucity of mutations is not surprising given the small size (70 bp) of miRNA genes. To identify alternative mechanisms for inactivation of the remaining allele in CLL cases showing LOH, we used in silico cloning to identify a putative promoter region located about 215 bp downstream of the miR16 gene. Down-regulation by promoter hypermethylation was reported for several cancer-related genes, including p16INK4a, p73, hMLH1, or VHL (27). We therefore used methylation-specific PCR to analyze the methylation status of one CpG-rich region located 5′ from the putative promoter. There was no detectable difference in the methylation patterns in any of the analyzed CpG sites independent of the miR15 or miR16 levels of expression in 10 samples (8 with decreased expression and 2 with high expression). However, methylation of different regions or of small regions of CpG sites escaping detection by the methylation-specific PCR cannot be excluded.
The Arginyl-tRNA Synthetase Gene (RARS) Is a Putative Target of miR16.
To further evaluate possible mechanisms of miR15 and miR16 in CLL, we analyzed the level of expression of genes involved in miRNA processing by RT-PCR. miRNAs are typically excised from 60- to 70-nt fold-back RNA precursor structures (17, 23–25). The miRNA precursor processing reaction requires interaction with a protein complex including Dicer RNase III and Argonaute family members (28). We analyzed the expression of Dicer1, hiwi1, eIF2C1, and eIF2C2 in a set of 23 B-CLLs and in CD5+ cells. We found a relatively constant level of expression of Dicer1, hiwi1, and eIF2C1, and no expression of eIF2C2, indicating that variations in the quantity of precursor processing machinery do not influence variations in miR15/16 levels.
Using an analogy with the proven mechanism of action of lin-4 and let-7, it was proposed that miRNAs act as antisense repressors of messenger RNA translation by nonperfect base pairing with the 3′ UTRs of one or more target genes (29, 30). In fact, it was recently shown that several miRNAs are complementary to 3′ UTR sequence motifs that mediate negative posttranscriptional regulation (31). However, this is not the case for miR15 and miR16. Therefore, we searched the NCBI database for short and nearly exact matches with miR15 and miR16, and we found antisense complementary regions in several cDNAs (Tables 1 and 2). We performed Western blotting analysis to determine the levels of SLUG and SNX2 proteins in CLL samples and in CD5+ cells. SLUG is known to inhibit apoptosis (32), to which CLL cells are largely resistant, and SNX2 is involved in the epidermal growth factor receptor signaling pathway. We found that SLUG is expressed neither in normal CD5+ cells nor in B-CLLs cells, except in one patient with a very low expression of miR15/miR16. Sorting nexin, SNX2, is a protein interaction partner of FBP17, the human formin-binding protein 17, which is an MLL-fusion partner in acute myelogenous leukemia (33). We found variations in protein expression in ≈33% of samples, but without strong correlation with the levels of expression of the miR15/miR16 gene cluster (data not shown).
Table 1.
Human genes with nonperfect base pairing with miR15 gene
| Gene name | Gene symbol | Location | miR15 homology |
|---|---|---|---|
| Capping protein (actin filament) muscle Z-line, alpha 2 | CAPZA2; CAPZ, CAPPA2 | 7q31.2–q31.3 | 18-nt overlap, 89% |
| Chemokine (C-X-C motif), receptor 4 (fusin) | CXCR4; CD184 | 2q21 | 17-nt overlap, 94% |
| Coatomer protein complex, subunit alpha | COPA | 1q23–q25 | 19 nt overlap, 89% |
| Cytoplasmic FMRP interacting protein 2 | PIR121; CYFIP2 | 5q34 | 21-nt overlap, 90% |
| Growth-associated protein 43 | GAP43 | 3q13.1–q13.2 | 21-nt overlap, 81% |
| Integrin, beta 5 | ITGB5 | 3q21.2 | 21-nt overlap, 81% |
| Profilin 2 | PFN2 | 3q25.1–q25.2 | 20-nt overlap, 90% |
| Syntaxin binding protein 1 | STXBP1, hUNC18 | 9q34.1 | 19-nt overlap, 89% |
Table 2.
Human genes with nonperfect base pairing with miR16 gene
| Gene name | Gene symbol | Location | miR16 homology |
|---|---|---|---|
| Arginyl-tRNA synthetase | RARS | 5pter–q11 | 20-nt overlap, 85% |
| ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit | ATP5E | 20q13.3 | 19-nt overlap, 89% |
| Bactericidal/permeability-increasing protein | BPI | 20q11.23–q12 | 19-nt overlap, 89% |
| DC2 protein | DC2 | 4q25 | 17-nt overlap, 95% |
| Enhancer of polycomb 1 | EPC1 | 10p11 | 18-nt overlap, 89% |
| Insulin-like growth factor 1 (somatomedin C) | IGF1 | 12q22–q23 | 20-nt overlap, 95% |
| Keratin 18 | KRT18 | 12q13 | 18-nt overlap, 89% |
| Member of the cysteine-rich secretory protein (CRISP) family | TPX1, GAPDL5, CRISP-2 | 6p21–qter | 21-nt overlap, 86% |
| Phosphoribosylglycinamide formyltransferase | GART | 21q22.11 | 18-nt overlap, 89% |
| Protein phosphatase 2, regulatory subunit A, beta isoform | PPP2R1B | 11q23 | 21-nt overlap, 81% |
| Slug homolog, zinc finger protein | SLUG; SNAI2 | 8q11 | 21-nt overlap, 81% |
| Sorting nexin 2 | SNX2 | 5q23 | 21-nt overlap, 81% |
| Spectrin, beta, nonerythrocytic 1 | SPTBN1 | 2p21 | 18-nt overlap, 89% |
| WW domain binding protein 11 | WBP11 | 12p13.1 | 21-nt overlap, 81% |
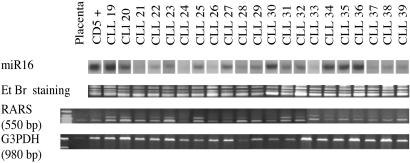
Because no function has been ascribed to any miRNA outside of Caenorhabditis elegans, we hypothesized that miRNA may not only act as translational regulators but also as modulators of mRNA stability (34). Therefore, we analyzed the expression of several genes with nonperfect base pairing with miR15 or miR16 messengers (Tables 1 and 2) in a set of 22 B-CLLs and in CD5+ B cells. We found that the levels of expression of the arginyl-tRNA synthetase gene (RARS), which encodes the protein that catalyzes the aminoacylation of tRNA by its cognate amino acid, correlates with the levels of expression of miR15 and miR16. In five of six samples with high levels of miRNAs, low expression of RARS was found. Eight of the remaining 16 samples (50%) with low/moderate expression of miR15/16 display high levels of RARS (Fig. 3). The significance of this finding remains to be determined, and functional assays with the different putative targets will be required to further test the mechanism(s) of action for miR15 and miR16 genes.
Fig 3.
Correlation between miR16 levels of expression (by Northern blotting) and RARS expression (by RT-PCR) in B-CLL samples. As normalization controls, we used staining with ethidium bromide (Et Br) for Northern gels and GAPDH for RT-PCRs.
Discussion
The presence of both clonal homozygous and heterozygous deletions, and the very high frequency of 13q14 loss suggest that these deletions are of pathogenic significance. The identification of pathogenic genes involved in the 13q14 deletions in CLL patients remains a difficult task despite the extensive efforts of different groups (4–11). There are several possible explanations for this fact. First, it is possible that the 13q14 tumor suppressor gene is normally expressed during a small window of B cell development, thus explaining its absence from current versions of the public database (4). Second, reduced dosage of the product of one or more of the candidate genes may be sufficient to contribute to B-CLL development (5). Finally, the responsible gene may be within the region of loss but may have escaped detection because of atypical features, such as the presence of relatively few exons separated by large introns or the lack of a polyadenylation signal (4). Indeed, the 13q14 region of homozygous loss is particularly interesting because it contains at least four noncoding genes (of the five known at present), including the two miRNAs reported in this paper and Leu-1 and Leu-2. Noncoding RNAs have been found to have roles in a great variety of processes, including transcription and chromosome structure, RNA processing and modifications, mRNA stability and translation, and protein stability and transport (12, 30, 34). Therefore, it is possible that the CLL gene(s) on 13q14 acts in a different way compared with the classical tumor suppressor genes.
Our findings that miR15 and miR16 lie within a small region of chromosome 13q14 that is deleted in more than 65% of CLL and that allelic loss in this region correlates with down-regulation of both miR15 and miR16 expression suggest that these genes represent the targets of inactivation by allelic loss in CLL. Although we could not determine the levels of miR15 or miR16 expression in CLL-A or CLL-B, the deletion pattern observed in these two cases is also consistent with inactivation through allelic loss and/or deregulation. CLL-A, which carries a 31-kb hemizygous deletion, was derived from a patient with both CLL and retinoblastoma. The deletion observed in the chromosome 13 isolated in the hybrid clone would result in a hemizygous deletion that includes both miR genes and the putative promoter, located immediately centromeric to the genes (Fig. 1). CLL-B cells derive from a patient with a t(2, 13)(q32;q14) translocation. The translocation breakpoint is located <3 kb centromeric to the locus, suggesting that perhaps the translocation separates the genes from as yet unrecognized regulatory sequences and results in abnormal miR15 and miR16 regulation.
The mechanism of action of miR genes remains to be elucidated. It is possible that miRNA levels are crucial in maintaining regulatory control over target genes during normal CD5+ B cell differentiation. It was shown that a large subset of Drosophila miRNAs with homologs in the human genome is perfectly complementary to several classes of sequence motifs previously demonstrated to mediate negative posttranscription regulation (31). Neither miR15 nor miR16 are complementary to such 3′ UTR sequence motifs. Moreover, the identification of putative targets for these two genes from 13q14 is complicated by the presence of a very similar cluster on chromosome 3. However, the lower levels of expression for the former cluster argue that the expression of the 13q cluster is of importance for the cell homeostasy. Thus, it is possible that inactivation of one allele at 13q, with the resulting decrease in miRNA levels, has physiological consequences that play a role in CLL initiation or progression. Recently, it was shown that the near-perfect complementarity between plant miRNAs and their targets suggests that many plant miRNAs act similarly to small interfering RNAs and direct RNA cleavage (35). In fact, we have found that miR16 has a homology of 85% on the 20-nt overlap with the RARS gene, and that the variation in expression levels of RARS can be explained by mRNA cleavage directed by miR16.
The 13q14 loss in CLL is frequently found as the sole genetic abnormality in a high percentage of cases (2). Such aberrations are generally of pathogenic significance in hematopoietic tumorigenesis (36). Clinical studies describe favorable outcomes for CLL patients with 13q deletions: patients with a normal karyotype or deletion of 13q14 as the sole genetic abnormality have a better prognosis than those with a complex karyotype or deletion of 11q23 or 17p13 (1, 37, 38). The presence of a very similar cluster of miR15/16-related genes on chromosome 3, which is active at lower levels but not deleted in CLL, can explain these data. We have found a low frequency of LOH at 3q26.1 (<25% when combining two markers), and, indeed, few CLL cases with involvement of chromosome 3 in any kind of cytogenetic abnormality have been described (39). Therefore, the absence of both alleles of miR15/16 on chromosome 13q14.3 is not associated with a complete loss of function, but only with a reduction of normal miR15/16 function. These findings can explain why the accumulation of secondary abnormalities (such as 12q trysomy, 11q deletion, or 17p deletion) associates with a poor prognosis.
Further studies aimed at the identification of miR15 and miR16 target genes will shed light on their mechanism of action and provide further clues about their role in B-CLL pathogenesis. This report presents evidence for the involvement of miRNA genes in human tumors, and provides further evidence of the significance of this growing family of regulatory genes.
Supplementary Material
Acknowledgments
We thank B. Perussia and J. Rothstein for valuable suggestions. This work was supported by National Cancer Institute Program Project Grants P01CA76259, P01CA81534, and P30CA56036, and by a Kimmel Scholar award to F.B. C.D.D. is a Special Fellow of the Leukemia and Lymphoma Society. M.K., K.R., L.R., T.K., M.N., F.B., and C.M.C. are members of the CLL Research Consortium.
Abbreviations
CLL, chronic lymphocytic leukemia
LOH, loss of heterozygosity
miRNA, micro-RNA
References
- 1.Dohner H., Stilgenbauer, S., Benner, A., Leupolt, E., Krober, A., Bullinger, L., Dohner, K., Bentz, M. & Lichter, P. (2000) N. Engl. J. Med. 343 1910-1916. [DOI] [PubMed] [Google Scholar]
- 2.Bullrich F. & Croce, C. M. (2001) in Chronic Lymphoid Leukemias, ed. Chenson, B. D. (Dekker, New York), pp. 9–32.
- 3.Dong J. T., Boyd, J. C. & Frierson, H. F., Jr. (2001) Prostate 49 166-171. [DOI] [PubMed] [Google Scholar]
- 4.Bullrich F., Fujii, H., Calin, G., Mabuchi, H., Negrini, M., Pekarsky, Y., Rassenti, L., Alder, H., Reed, J. C., Keating, M. J., et al. (2001) Cancer Res. 61 6640-6648. [PubMed] [Google Scholar]
- 5.Migliazza A., Bosch, F., Komatsu, H., Cayanis, E., Martinotti, S., Toniato, E., Guccione, E., Qu, X., Chien, M., Murty, V. V., et al. (2001) Blood 97 2098-2104. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y., Corcoran, M., Rasool, O., Ivanova, G., Ibbotson, R., Grander, D., Iyengar, A., Baranova, A., Kashuba, V., Merup, M., et al. (1997) Oncogene 15 2463-2473. [DOI] [PubMed] [Google Scholar]
- 7.Mabuchi H., Fujii, H., Calin, G., Alder, H., Negrini, M., Rassenti, L., Kipps, T. J., Bullrich, F. & Croce, C. M. (2001) Cancer Res. 61 2870-2877. [PubMed] [Google Scholar]
- 8.Rondeau G., Moreau, I., Bezieau, S., Petit, J. L., Heilig, R., Fernandez, S., Pennarun, E., Myers, J. S., Batzer, M. A., Moisan, J. P. & Devilder, M. C. (2001) Mutat. Res. 458 55-70. [DOI] [PubMed] [Google Scholar]
- 9.Wolf S., Mertens, D., Schaffner, C., Korz, C., Dohner, H., Stilgenbauer, S. & Lichter, P. (2001) Hum. Mol. Genet. 10 1275-1285. [DOI] [PubMed] [Google Scholar]
- 10.Mertens D., Wolf, S., Schroeter, P., Schaffner, C., Dohner, H., Stilgenbauer, S. & Lichter, P. (2002) Blood 99 4116-4121. [DOI] [PubMed] [Google Scholar]
- 11.Rowntree C., Duke, V., Panayiotidis, P., Kotsi, P., Palmisano, G. L., Hoffbrand, A. V. & Foroni, L. (2002) Leukemia 16 1267-1275. [DOI] [PubMed] [Google Scholar]
- 12.Storz G. (2002) Science 296 1260-1263. [DOI] [PubMed] [Google Scholar]
- 13.Hutvagner G., McLachlan, J., Pasquinelli, A. E., Balint, E., Tuschl, T. & Zamore, P. D. (2001) Science 293 834-838. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J., Frisch, E. F. & Maniatis, T., (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 15.Dono M., Zupo, S., Leanza, N., Melioli, G., Fogli, M., Melagrana, A., Chiorazzi, N. & Ferrarini, M. (2000) J. Immunol. 164 5596-5604. [DOI] [PubMed] [Google Scholar]
- 16.Negrini M., Sabbioni, S., Haldar, S., Possati, L., Castagnoli, A., Corallini, A., Barbanti-Brodano, G. & Croce, C. M. (1994) Cancer Res. 54 1818-1824. [PubMed] [Google Scholar]
- 17.Lagos-Quintana M., Rauhut, R., Lendeckel, W. & Tuschl, T. (2001) Science 294 853-858. [DOI] [PubMed] [Google Scholar]
- 18.Bullrich F., Veronese, M. L., Kitada, S., Jurlander, J., Caligiuri, M. A., Reed, J. C. & Croce, C. M. (1996) Blood 88 3109-3115. [PubMed] [Google Scholar]
- 19.Stilgenbauer S., Nickolenko, J., Wilhelm, J., Wolf, S., Weitz, S., Dohner, K., Boehm, T., Dohner, H. & Lichter, P. (1998) Oncogene 16 1891-1897. [DOI] [PubMed] [Google Scholar]
- 20.Corcoran M. M., Rasool, O., Liu, Y., Iyengar, A., Grander, D., Ibbotson, R. E., Merup, M., Wu, X., Brodyansky, V., Gardiner, A. C., et al. (1998) Blood 91 1382-1390. [PubMed] [Google Scholar]
- 21.Bouyge-Moreau I., Rondeau, G., Avet-Loiseau, H., Andre, M. T., Bezieau, S., Cherel, M., Saleun, S., Cadoret, E., Shaikh, T., De Angelis, M. M., et al. (1997) Genomics 46 183-190. [DOI] [PubMed] [Google Scholar]
- 22.Kalachikov S., Migliazza, A., Cayanis, E., Fracchiolla, N. S., Bonaldo, M. F., Lawton, L., Jelenc, P., Ye, X., Qu, X., Chien, M., et al. (1997) Genomics 42 369-377. [DOI] [PubMed] [Google Scholar]
- 23.Lagos-Quintana M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W. & Tuschl, T. (2002) Curr. Biol. 12 735-739. [DOI] [PubMed] [Google Scholar]
- 24.Lau N. C., Lim, L. P., Weinstein, E. G. & Bartel, D. P. (2001) Science 294 858-862. [DOI] [PubMed] [Google Scholar]
- 25.Lee R. C. & Ambros, V. (2001) Science 294 862-864. [DOI] [PubMed] [Google Scholar]
- 26.Caligaris-Cappio F. & Hamblin, T. J. (1999) J. Clin. Oncol. 17 399-408. [DOI] [PubMed] [Google Scholar]
- 27.Esteller M. (2002) Oncogene 21 5427-5440. [DOI] [PubMed] [Google Scholar]
- 28.Grishok A., Pasquinelli, A. E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D. L., Fire, A., Ruvkun, G. & Mello, C. C. (2001) Cell 106 23-34. [DOI] [PubMed] [Google Scholar]
- 29.Wightman B., Ha, I. & Ruvkun, G. (1993) Cell 75 855-862. [DOI] [PubMed] [Google Scholar]
- 30.Ambros V. (2001) Cell 107 823-826. [DOI] [PubMed] [Google Scholar]
- 31.Lai E. C. (2002) Nat. Genet. 30 363-364. [DOI] [PubMed] [Google Scholar]
- 32.Inukai T., Inoue, A., Kurosawa, H., Goi, K., Shinjyo, T., Ozawa, K., Mao, M., Inaba, T. & Look, A. T. (1999) Mol. Cell 4 343-352. [DOI] [PubMed] [Google Scholar]
- 33.Fuchs U., Rehkamp, G., Haas, O. A., Slany, R., Konig, M., Bojesen, S., Bohle, R. M., Damm-Welk, C., Ludwig, W. D., Harbott, J. & Borkhardt, A. (2001) Proc. Natl. Acad. Sci. USA 98 8756-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarz D. S. & Zamore, P. D. (2002) Genes Dev. 16 1025-1031. [DOI] [PubMed] [Google Scholar]
- 35.Rhoades M., Reinhart, B., Lim, L., Burge, C., Bartel, B. & Bartel, D. (2002) Cell 110 513-520. [DOI] [PubMed] [Google Scholar]
- 36.Gauwerky C. E. & Croce, C. M. (1993) Semin. Cancer Biol. 4 333-340. [PubMed] [Google Scholar]
- 37.Oscier D. G., Gardiner, A. C., Mould, S. J., Glide, S., Davis, Z. A., Ibbotson, R. E., Corcoran, M. M., Chapman, R. M., Thomas, P. W., Copplestone, J. A., et al. (2002) Blood 100 1177-1184. [PubMed] [Google Scholar]
- 38.Juliusson G., Oscier, D. G., Fitchett, M., Ross, F. M., Stockdill, G., Mackie, M. J., Parker, A. C., Castoldi, G. L., Guneo, A., Knuutila, S., et al. (1990) N. Engl. J. Med. 323 720-724. [DOI] [PubMed] [Google Scholar]
- 39.Specchia G., Albano, F., Anelli, L., Storlazzi, C. T., Monaco, M., Capalbo, S., Rocchi, M. & Liso, V. (2002) Cancer Genet. Cytogenet. 133 160-163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.