Abstract
We have quantitatively monitored the sporulation and germination efficiencies of ≈4,200 yeast deletion strains in parallel by using a molecular bar coding strategy. In a single study, we doubled the number of genes functionally implicated in sporulation to ≈400, identifying both positive and negative regulators. Our set of 261 sporulation-deficient genes illustrates the importance of autophagy, carbon utilization, and transcriptional machinery during sporulation. These general cellular factors are more likely to exhibit fitness defects when deleted and less likely to be transcriptionally regulated than sporulation-specific genes. Our postgermination screening assay identified recombination/chromosome segregation genes, aneuploid strains, and possible germination-specific factors. Finally, our results facilitate a genome-wide comparison of expression pattern and mutant phenotype for a developmental process and suggest that 16% of genes differentially expressed during sporulation confer altered efficiency of spore production or defective postgermination growth when disrupted.
Studies of the Saccharomyces cerevisiae life cycle have provided insight into the action of signaling pathways and transcriptional induction in higher eukaryotes. One stage of the yeast life cycle involves the production of haploid gametes from a parental diploid in a process known as sporulation. Sporulation encompasses the two meiotic divisions, followed by ascospore development around the four haploid nuclei. To date, more than 100 genes have been functionally implicated in sporulation as determined by mutant phenotype (1). These genes were identified largely through classical genetic screens (2) and more recently through the analysis of targeted gene deletions (3, 4), the results of which demonstrate that the isolation of sporulation genes has not been saturated. In contrast to the positive regulators required for spore production, relatively few genes are known to inhibit the process. These genes, such as the cell-type specific repressor RME1 (5), play a key role in the cell fate decision of yeast and ensure that sporulation occurs in the correct cell type and under appropriate nutrient conditions. Despite their importance, a comprehensive identification of genes that function in the negative regulation of sporulation has not been reported.
A second aspect of the yeast life cycle is spore germination. Germination is the process by which a dormant spore, in the presence of the appropriate nutrient conditions, resumes mitotic division. However, the genes and pathways that function in germination remain unknown because of difficulty in recovering mutants specifically defective in the process. One key issue is the determination of the extent to which novel germination-specific factors distinguish this process from normal cell division.
The completion of the yeast genome sequence has led to the development of new technologies for whole-genome analysis. One such technology is the DNA microarray used to measure mRNA transcript levels of all genes under a particular condition. Genome-wide expression analysis of yeast sporulation suggests that up to one-quarter of the yeast genome is transcriptionally regulated (6, 7). The analysis and conclusions drawn from these studies are largely based on the assumption of correlation between mRNA level and cellular function. However, the extent to which expressed genes are important to the process has not been extensively investigated on a genome-wide scale. Recently, precise deletions were constructed for all ORFs in the yeast genome (8). A molecular bar-coding strategy enables pooling of deletion mutants and parallel phenotypic analysis (9). This system has been successful in studies of UV sensitivity (10), nonhomologous DNA end-joining (11), and mitochondrial function (12).
In the present study we determined the sporulation efficiency of ≈4,200 deletion strains in parallel by using the molecular bar-coding approach. Our results are in excellent agreement with single gene studies and identify ≈200 additional genes that affect sporulation efficiency, both positively and negatively. A genome-wide comparison of sporulation transcription profiles to mutant phenotype suggests that a small percentage of differentially expressed genes alter sporulation efficiency when disrupted. We also developed a comprehensive screen for strains defective in postgermination growth. Our results with 4,200 deletion strains identified known meiotic mutants, mutated parental strains, and possible germination mutants.
Materials and Methods
Strains and Growth Conditions.
Homozygous diploid deletion strains in the BY4743 background (13), isogenic to the sequenced strain S288c, were used in all pool experiments. Systematic strain and pool construction was described previously (8). Fitness profiling of the homozygous diploid deletion pool and postgermination growth of spores were performed in standard rich media [yeast extract/peptone/dextrose (YPD)] at 30°C. In both experiments, four or five time points were taken over 20 generations of pool growth. The batch-transfer method (14) was used to ensure that the culture density never exceeded 2 × 107 cells per ml. At least 4 × 107 cells were diluted into fresh YPD at each time point to ensure that each deletion strain was adequately represented in the pool and to minimize sampling error. Aliquots of 4 × 107 cells collected at each time point were processed for array hybridization.
Sporulation Conditions and Spore Isolation.
Sporulation of the deletion pool was performed as described for single strains (8). The efficiency of sporulation was ≈15%. Pure spore suspensions were isolated by using a protocol that takes advantage of the hydrophobicity and zymolyase-resistance of spores relative to unsporulated vegetative cells (15). We collected >8 × 107 spores from the deletion pool at >99% purity (microscopic examination). Half of the spore sample was used to determine the sporulation efficiency of each strain in the pool. The second half was grown in YPD to determine the postgermination growth rate of all strains in the pool.
Sample Processing and Array Hybridization.
For samples collected at each time point, genomic DNA was extracted by using a glass-bead lysis protocol (16). This protocol was slightly altered for genomic DNA preparation from spores, given their relative resistance to standard vortexing. To overcome this problem, we used a multiple tube shaker (VWR Scientific) set to the maximum level to lyse spores. PCR amplification of the tag modules and hybridization to an oligonucleotide array (TAG3, Affymetrix) containing their complements is described (8).
Data Analysis.
For each scanned array, we extracted the intensity values for each probe on the array. For a single experiment, we normalized to the average of the perfect match signals across all time points to avoid bias associated with differences in overall array intensity. We then determined which deletion strains in the pool were detectable in the given experiment. For a strain to be “present,” at least two of its corresponding perfect match probes had to have intensity values greater than two-fold over the array background intensity in a single time point. Any tag that did not fit the “present” criteria was not used in subsequent analysis. For determining the sporulation efficiency of each strain in the deletion pool, we measured the abundance of molecular tags in the pool on the array from both a presporulation culture and a pure spore sample. For each informative tag of a given strain, we calculated the ratio of presporulation probe intensity to the spore probe intensity. We then averaged the ratios across all tags for a single strain to obtain a single measure of its sporulation efficiency, the PreSpo/Spore ratio. The correlation coefficient between two independent experiments was 0.80. To determine the fitness of the deletion strains in rich media, we hybridized the molecular tags from five separate time points. After normalization, the perfect match signals were log2 transformed. We then used a simple linear regression model to assess the change in abundance of each molecular tag over time (17). We averaged the growth rates obtained from all meaningful tags for a given strain to generate a single value. The tag intensities for most strains do not change over time resulting in a relative growth rate of 1, equal to that of the pool in general. Conversely, strains diminishing over time correspond to a relative growth rate less than 1. We used the same analysis to calculate postgermination growth rates for all strains present in the spore suspension. We then classified strains as postgermination defective by identifying those with a homozygous diploid growth rate to postgermination growth rate ratio greater than 1.05 in two independent experiments.
Results
A Whole-Genome Screen to Identify Sporulation Mutants.
A pool of 4,745 homozygous diploid deletion mutants, covering more than 95% of all nonessential ORFs in the yeast genome, was constructed for parallel phenotypic analysis. Each deletion strain contains two unique 20-bp sequences, termed molecular tags, flanked by common PCR priming sites. The relative abundance of a single deletion strain can be quantitatively monitored in a large pool of strains by the hybridization of its molecular tags to an oligonucleotide array containing the tag complements (9). We monitored the sporulation efficiency of each strain in the pool by comparing the hybridization intensities of its molecular tags before and after sporulation (Fig. 1).
Fig 1.
A parallel genetic approach to identify sporulation mutants. (A) Two-color overlay of scanned images from oligonucleotide arrays hybridized with fluorescently labeled molecular tags from a presporulation culture (red channel) and a pure spore suspension (green channel). Red (VAC8) and green (NOT3) probes indicate enrichment in the presporulation sample and the spore sample, respectively. Yellow probes correspond to deletion strains equally represented in the presporulation and spore cultures. (B) The ratio of presporulation signal to spore signal for 4,162 deletion strains is shown. The majority of genes have no effect on sporulation efficiency (PreSpo/Spore ratio of 1). The red and green bars correspond to sporulation-deficient and enhanced sporulation gene sets, respectively.
For each deletion strain, we calculated the ratio of presporulation signal to spore signal averaged across all informative tags. By using the PreSpo/Spore ratio as a quantitative measure of sporulation efficiency, each strain was categorized into one of three phenotypic classes. Strains enriched in the presporulation sample compared with the spore sample exhibited a PreSpo/Spore ratio greater than 1, indicative of an inability to make spores. Conversely, strains that sporulate better than average correspond to PreSpo/Spore ratios less than 1. Finally, deletion strains corresponding to genes whose disruption has no effect on the strain's capacity to make spores, display a PreSpo/Spore ratio at or near 1.
We reliably detected 4,162 deletion strains in two independent experiments. Two factors explain the inability to detect over 500 strains in this analysis. The first factor is mutations in the tag sequence or priming sites leading to poor hybridization properties or inability to amplify the tag sequences. The second factor, responsible for the majority of undetected strains (data not shown), was the growth of the deletion pool on presporulation media for several days before sporulation. During the growth on presporulation media, many strains with a slow growth phenotype were lost from the deletion pool and hence undetected in our analysis.
The majority of yeast genes play no role in sporulation, and therefore their deletion has no effect on sporulation efficiency. This observation was evident in our study as most deletion strains exhibit a PreSpo/Spore ratio around 1 (Fig. 1). We identified 102 strains with enhanced sporulation proficiency (PreSpo/Spore ratio of 0.8 or less in two independent experiments) and 261 sporulation-deficient strains (PreSpo/Spore ratio of 1.5 or greater in two independent experiments) in our analysis. These thresholds were chosen to include known positive controls that influence sporulation both positively (e.g., IME1) and negatively (e.g., RME1). Three lines of evidence suggest that these strains have true sporulation defects. First, the majority of known negative and positive regulators of sporulation fall into the proficient and deficient gene sets, respectively. We observed >80% (66 of 81) of nonessential genes required for sporulation (1) in our set of 261 sporulation-defective strains. Second, visual inspection of spore formation in mutants for 50 unclassified genes revealed 42 (84%) with sporulation efficiencies significantly under the normal frequency (www-sequence.stanford.edu/group/yeast_deletion_project/Sporulation.html). Third, for a subset of six unclassified genes, we deleted the ORFs (YJR120W, YML066C, YOR298W, YLL033W, YPL013C, YOL071W) in SK1, a genetic background that sporulates to high efficiency, and observed the same sporulation-deficient phenotype.
General Cellular Processes Are Critical for Efficient Sporulation.
To identify cellular processes important for sporulation, we searched for over represented functional classes among the 261 genes critical for sporulation (Table 1). As expected, a significant number of genes are known sporulation and meiosis factors (54 genes). However, the majority of characterized genes that we found to be required for sporulation are general cellular factors, involved in autophagy/vacuole targeting (29 genes), carbon utilization (37 genes), vesicle transport (9 genes), and general transcription (26 genes). Autophagy, a cellular pathway by which proteins and organelles are degraded in the vacuole under starvation conditions, has previously been implicated in sporulation (18). Respiration genes, including 10 components of the tricarboxylic acid cycle, account for over half of the carbon utilization genes identified as sporulation-deficient in the study. Respiration competence is required for efficient sporulation (19) and may reflect a requirement for additional energy from a nonfermentable carbon source during the process. Finally, 71 of the 261 sporulation-deficient genes (27.2%) are unclassified. Given the biased functional breakdown of the classified genes, it is likely that a portion of the 71 unclassified ones function in the categories listed in Table 1.
Table 1.
Overrepresented functional classes among 261 sporulation-deficient strains
| Functional class | No. of genes (fold enrichment over genome) | No. regulated transcriptionally | No. of slow growers |
|---|---|---|---|
| Total | 261 | 73 | 92 |
| Sporulation/meiosis | 54 (8.25) | 29 | 6 |
| Autophagy/vacuole function | 29 (3.55) | 3 | 9 |
| Carbon utilization/energy | 37 (3.60) | 11 | 11 |
| Vesicle transport | 9 (1.71) | 2 | 2 |
| Transcription | 26 (1.27) | 3 | 17 |
| Unclassified | 71 (0.72) | 16 | 21 |
Functional classes are from the MIPS database (1).
We calculated the number of genes expected in each functional class if a random sample of 261 genes was collected. The expected numbers were divided into our observed numbers to calculate the fold enrichment over genome value.
Gene shows a differential expression pattern during sporulation in both the SK1 and W303 backgrounds (7).
A strain is considered a slow grower if its growth rate is less than 0.95 of the pool average.
Identification of Sporulation-Specific Factors By Using Fitness and Expression Profiling.
Two distinct classes of genes essential to sporulation exist based on previous data and this study. The first class includes sporulation-specific genes whose function is limited to sporulation only. The second are general factors involved in the key processes shown in Table 1. We were interested in distinguishing between these two classes to predict the number of sporulation-specific genes among our unclassified set. We used two approaches to distinguish sporulation-specific genes from general factors. The first method was fitness profiling of the entire pool of homozygous diploid deletion strains in rich media (YPD) for quantitative growth defects as genes involved only in sporulation are expected to be dispensable for growth in YPD, whereas loss of a general factor should in many cases lead to a growth defect. The second method was to query public databases containing sporulation gene expression data for genes with significant expression changes. Here, our hypothesis was that sporulation-specific genes are more likely to be transcriptionally regulated during meiosis than general factors.
We grew the homozygous diploid deletion pool of 4,745 strains in YPD to determine a relative growth rate for each strain by using a linear regression model. When we examined the growth rates of the 261 sporulation-deficient strains, we identified 92 strains with growth rates <0.95 of the pool average (Fig. 2B). This slow growing fraction accounts for over one-third of the total sporulation-deficient strains identified (Table 1). There is a pronounced bias toward general cellular factors among the slow growers. Only six (11.1%) of the genes classified as sporulation/meiosis had a slow growth phenotype, and all six (UME6, NEM1, SIN3, SSN3, SSN8, SPO7) are involved in multiple cellular processes including sporulation. As expected, no gene involved only in sporulation is required for optimal growth in YPD. Conversely, deletions in 17 (65.4%) of the transcription factors confer a slow growth phenotype. Consistent with this result, gene disruptions in all functional classes other than sporulation are more likely to have quantitative growth defects in YPD (Table 1). Therefore, we estimate that 21 of the 71 uncharacterized genes identified in our study as sporulation-deficient are strong candidates for critical general cellular factors, while 50 are possible sporulation-specific genes.
Fig 2.
Sporulation-specific genes are transcriptionally regulated and not required for optimal fitness in YPD. (A) The 261 sporulation-deficient strains were separated into two classes based on YPD fitness profiling: slow growers (with growth rates less than 0.95 of the pool average) and normal growers. We then hierarchically clustered the genes from each set based on sporulation expression data (7) obtained from strains SK1 and W303. The expression time courses are reflected by the triangles above the cluster: the large end of the triangle represents the end of the corresponding time course. Rows represent individual genes and columns reflect single time points in each experiment. (B) The average YPD fitness profile of the slow growth and normal growth class as a function of original tag signal. (C) Average mRNA expression level of the slow growth and normal growth class during SK1 sporulation. (D) Same as C for the W303 background.
Our second approach to distinguish general factors from sporulation-specific genes was to examine the transcriptional profile of each gene during sporulation. We first separated the set of 261 sporulation-defective strains into two sets as determined by fitness profiling: slow and normal growers. We then hierarchically clustered the genes from each set by using sporulation expression data obtained from the SK1 and W303 strains (Fig. 2A) (7). In addition to being dispensable for optimal growth in rich media, the majority of known sporulation-specific genes are strongly induced during meiosis (Fig. 2A, top cluster). Conversely, few genes that confer a slow growth phenotype are differentially expressed (Fig. 2 A Lower, C, and D), suggesting that the general cellular factors are under less transcriptional control than the sporulation-specific genes during the process. These results are quantified in Table 1. Over half of the known sporulation genes (54%) are contained in the core meiotic transcriptome, which is defined as the set of genes differentially expressed during sporulation in both the SK1 and W303 backgrounds (7). Similarly, the carbon utilization genes are under strong transcriptional regulation during sporulation, whereas autophagy/vacuole genes are not extensively regulated (Table 1). A total of 16 of the 71 unclassified genes identified as sporulation-deficient are included in the core meiotic transcriptome. Of these, 14 are not required for optimal growth in rich media (Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). These are the best candidates for new, sporulation-specific genes.
Identification of Negative Regulators of Sporulation.
In contrast to the genes that promote sporulation, the factors that negatively regulate the process are less well understood. It has been demonstrated that the loss of negative meiotic regulators such as Cln3 (20) and Sok2 (21) results in increased sporulation efficiency. To identify previously undescribed negative regulators, we examined the 102 strains that appear to sporulate better than the average of 15%. The positive controls RME1, CLN3, and SOK2 are present in the list together with seven negative regulators of transcription (GAL80, NOT3, SET2, HTZ1, SIF2, SIR1, OPI1), six cell cycle control genes (CLN1, CLB2, BCK2, SAP185, PPH21, HSL1), and four genes involved in pseudohyphal differentiation (GPR1, DIG1, RSC1, ELM1).
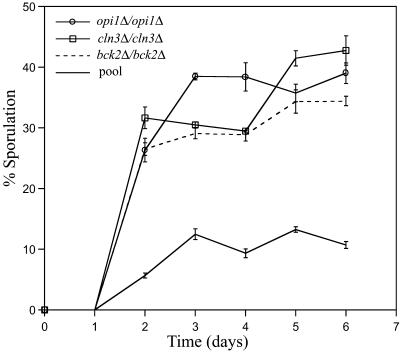
To verify our findings and determine the effect of single gene deletions on sporulation efficiency, we examined the kinetics and efficiency of spore production in cln3Δ/cln3Δ, bck2Δ/bck2Δ, and opi1Δ/opi1Δ deletion strains. All three strains sporulate three times better than the average for the entire deletion pool (Fig. 3). Because the cln3Δ/cln3Δ strain had the smallest PreSpo/Spore ratio in both experiments, a sporulation efficiency of 40% is the maximum for strain S288c carrying a single gene deletion.
Fig 3.
Identification of negative regulators of sporulation. Strains were added to sporulation media and the percentage of mature asci (2, 3, or 4 spore) was calculated at 1-day intervals. The curves represent the average of three independent experiments.
A Screen to Identify Strains Defective in Postgermination Growth.
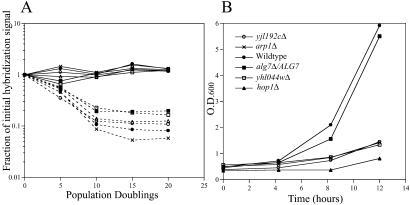
We identified mutants defective in postgermination growth by comparing growth rates obtained as homozygous diploids in YPD from those obtained postgermination (Fig. 4A). A suspension of spores derived from the homozygous diploid deletion pool was germinated in YPD and a postgermination growth rate was calculated for each gene. We identified 158 strains with a ratio of homozygous diploid to postgermination growth rate ≥1.05 (corresponding to a postgermination growth defect of ≈5%) in two independent experiments. This cutoff was chosen to include many known meiotic mutants, which sporulate but produce inviable spores, in the postgermination-defective class. The effectiveness of our screen was confirmed by outgrowth of spores derived from a subset of deletion strains identified by array analysis (Fig. 4B).
Fig 4.
A screen to identify strains defective in postgermination growth. (A) Homozygous diploid YPD growth (solid lines) and postgermination growth (dotted lines) of deletion strains [hop1Δ/hop1Δ (open circles), mam1Δ/mam1Δ (open squares), arp1Δ/arp1Δ (filled squares), wsc2Δ/wsc2Δ (open triangles), yhl044wΔ/yhl044wΔ (filled circles), and yjl192cΔ/yjl192cΔ (crosses)] with a postgermination growth defect. The curves are plotted as a function of the initial TAG hybridization signal over time. By 15 generations of postgermination growth, all six strains are absent from the pool. (B) A pure spore suspension (≈2 × 107 cells) derived from each diploid deletion strain was germinated in rich media (YPD). We monitored growth by assaying the culture turbidity at 3-hour intervals. Each curve represents the average of three independent experiments. The strain heterozygous for ALG7, an essential gene, is used as a control.
The 158 strains defective in postgermination growth were selected on basis of growth rates together with their ability to sporulate. The set of 261 sporulation-deficient strains were not included in the analysis because a subset are likely to produce a small percentage of spores that will drop out in a postgermination competitive growth assay. Despite this, about one-fifth of the germination-defective class (30 genes) is at the threshold of sporulation-deficient, as a PreSpo/Spore ratio >1.5 was obtained for that strain in one of the two experiments. Included in this set are genes involved in sporulation (MCK1, IDS2), mitochondrial function (IMP1, IMP2, TOM5), and autophagy/vacuole function (VPS30, VTC1). The presence of sporulation-deficient strains at PreSpo/Spore values below our original threshold suggests few false positives in our set of 261 genes critical for efficient sporulation.
As expected, about one-sixth (24 genes) of the genes identified as postgermination defective are involved in meiotic chromosome behavior including the double strand break enzyme Spo11 (22) and the kinetochore associated protein Mam1 (23). Our assay also uncovered genes involved carbon metabolism (GLC3, GPM2, FBP26), vesicle transport (SED4, TLG2, PEP8, BST1), and nutrient sensing/cell wall integrity (WSC2, BCK1, SLT2), suggesting a possible role for these processes in promoting either spore viability or germination. In addition, our set of 158 strains includes two strains, ECM18 and YAP3 homozygous diploids, that were previously identified as aneuploid for entire chromosomes (24). It is likely that spores derived from these strains are aneuploid and hence inviable, similar to what is observed with meiotic mutants. Finally, we identified 51 genes of unknown function that fit our postgermination defective criteria.
Analysis of Postgermination Defective Strains.
For the majority of the 158 strains identified as postgermination defective, the underlying cause of the defect remains unclear. The possibilities include meiosis mutants, aneuploid strains, strains with recessive mutations, and true germination mutants. To address this issue, we further analyzed six strains identified by our analysis: ecm18Δ/ecm18Δ, yap3Δ/yap3Δ, ybr027cΔ/ybr027Δ, ybl083cΔ/ybl083cΔ, pma2Δ/pma2Δ, and yjl192cΔ/yjl192cΔ. None of these genes are differentially expressed during sporulation (7), suggesting that none functions during meiosis. We determined whether each strain is aneuploid by hybridization of total genomic DNA derived from each strain onto a yeast expression array (Affymetrix) and searching for chromosome-wide signal bias. Using this method, we verified the aneuploidy of ecm18Δ/ecm18Δ (monoploid chromosome 7) and yap3Δ/yap3Δ (triploid chromosome 3) (24), whereas no aneuploidy was detected in the other four strains (data not shown).
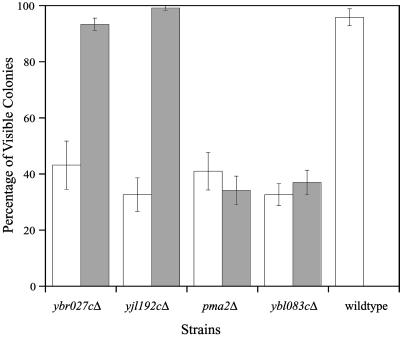
We dissected tetrads derived from each of the four nonaneuploid deletion strains and observed a small fraction of spores forming visible colonies (Fig. 5). A characteristic 2:2 segregation pattern was not observed for any of the strains, indicating that a single recessive mutation was not responsible for growth defect. To determine whether multiple recessive mutations were responsible for the growth defect, we backcrossed haploid deletions to an isogenic wild-type strain and dissected tetrads from the heterozygote. Resulting haploids that formed normal colonies and carried the correct deletion were crossed to make a new homozygous diploid deletion strain. Tetrads were then dissected from these new strains and analyzed. Spores derived from the backcrossed YBR027C and YJL192C strains behaved like the wild-type control (Fig. 5), suggesting that multiple recessive mutations in the original deletion strains were responsible for the spore growth defect. In contrast, the backcrossed pma2Δ and ybl083cΔ spores retained their growth defect, suggesting that these genes may play a specific role during germination.
Fig 5.
Analysis of postgermination defective strains. For the genes shown, tetrads were dissected from the original homozygous diploid deletion strain (white bars) and backcrossed strain (gray bars). Each bar represents the average of three independent tetrad dissections (10 tetrads each) per strain. Plates were scored for the presence of visible colonies after 2 days of growth.
Pma2 is a glucose-regulated proton pump postulated to function under starvation conditions consistent with a role during germination (25). The functionally unclassified YBL083C ORF physically overlaps the RHK1 gene encoded on the opposite strand. Interestingly, the rhk1Δ/rhk1Δ strain also exhibited a postgermination growth defect implying that the YBL083C phenotype is possibly due to a partial deletion of RHK1, a mannosyltransferase involved in protein N-glycosylation. Supporting this hypothesis is the observation of a similar germination defect in strains diminished in Alg7 activity, another N-glycosylation protein (26).
Discussion
An examination of the processes critical for efficient sporulation provides a glimpse into the entire genetic network required for a complex process. Our unbiased screen for sporulation-deficient strains identified a set of 261 genes required for optimal sporulation. The sporulation-specific genes, defined as those required only for sporulation, have received the most attention experimentally. However, our results suggest that the majority of genes that influence sporulation are those with a more general cellular function including those involved in the starvation response and carbon utilization. The general factors are more likely to exhibit a quantitative growth defect when deleted and less likely to be transcriptionally regulated during sporulation. We believe that the majority of the 71 unclassified genes identified in the sporulation-deficient set are general factors indirectly involved in sporulation. However, the role of genes with a slow growth phenotype should be investigated as a handful may a play a direct role in sporulation. Despite its slow growth phenotype, Ume6 acts as a key transcriptional activator of early meiotic genes and is essential for sporulation (27).
Diploid yeast can assume multiple cell fates including mitosis, sporulation, and filamentous growth depending on the nutrient conditions available at the time. Although the pathways that promote these developmental responses are well characterized, the inhibitory systems that make multiple responses incompatible are largely unknown. Our set of 102 strains with enhanced sporulation efficiency contains regulatory components that inhibit sporulation. The G1-cyclin Cln3 plays a key role in the initiation of mitosis and is known to block sporulation through inhibition of the early meiotic transcription factor Ime1 (20). Bck2 shares a role with Cln3 in the activation of the G1–S transition (28). Our results demonstrate that deletions of CLN3 and BCK2 increase sporulation efficiency 3-fold. However, it is unclear whether the sporulation role of Bck2 is manifested through Ime1 or indirectly through Cln3 activation. We also identified strains lacking components of a nutrient-sensing cAMP pathway that regulates filamentous growth as sporulation-proficient. A model where the pathways that activate mitosis and filamentous growth inhibit sporulation is suggested.
Whole-genome expression analysis has been used to provide insight into the function of thousands of previously uncharacterized genes. We observe a significant correlation between the 916 genes of the core meiotic transcriptome (7) and the 261 genes essential for efficient sporulation (χ2 test, P < 0.005). These results demonstrate that whole-genome expression profiling can enrich for genes functionally important for a developmental process. However, one cannot infer gene function directly from expression profiles, as we observe 551 (of 656) core meiotic transcriptome genes with no effect on sporulation efficiency or postgermination growth. Why are 84% of genes differentially expressed during sporulation not critical, based on mutant phenotype? One possibility is redundancy in the yeast genome at the individual gene or pathway level. A second possibility is that drastic cellular changes rework the transcription of much of the genome even though many of the genes are not required.
Of all phases of the yeast life cycle, germination has received the least attention experimentally, and hence is the least understood. A confounding aspect to the study of germination has been the inability to screen for genes critical in the process. Here, we describe a recently developed, comprehensive screen to identify strains that are defective in postgermination growth. Although a subset of our germination-defective strains derives from aneuploid spores or mutations in the parental diploid, we were successful in identifying genes, YBL083C/RHK1 and PMA2, which likely play a direct role in germination. The precise function of these genes during germination requires further investigation of the mutant phenotype in addition to the isolation of more germination mutants. A candidate gene approach based on our postgermination defective gene set together with additional methods, such as expression profiling, should provide a framework to understand the molecular basis of germination. Such efforts may open a new door in the functional classification of all 6,000 genes in the yeast genome.
Supplementary Material
Acknowledgments
We thank Anne Villeneuve, Tim Stearns, Brian Washburn, and members of the Davis laboratory for helpful comments. This work was supported by National Institutes of Health Grant P01HG00205 (to R.W.D.).
Abbreviations
YPD, yeast extract/peptone/dextrose
References
- 1.Mewes H. W., Albermann, K., Heumann, K., Liebl, S. & Pfeiffer, F. (1997) Nucleic Acids Res. 25 28-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esposito M. S. & Esposito, R. E. (1969) Genetics 61 79-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabitsch K. P., Toth, A., Galova, M., Schleiffer, A., Schaffner, G., Aigner, E., Rupp, C., Penkner, A. M., Moreno-Borchart, A. C., Primig, M., et al. (2001) Curr. Biol. 11 1001-1009. [DOI] [PubMed] [Google Scholar]
- 4.Briza P., Bogengruber, E., Thur, A., Rutzler, M., Munsterkotter, M., Dawes, I. W. & Breitenbach, M. (2002) Yeast 19 403-422. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell A. P. & Herskowitz, I. (1986) Nature 319 738-742. [DOI] [PubMed] [Google Scholar]
- 6.Chu S., DeRisi, J., Eisen, M., Mulholland, J., Botstein, D., Brown, P. O. & Herskowitz, I. (1998) Science 282 699-705. [DOI] [PubMed] [Google Scholar]
- 7.Primig M., Williams, R. M., Winzeler, E. A., Tevzadze, G. G., Conway, A. R., Hwang, S. Y., Davis, R. W. & Esposito, R. E. (2000) Nat. Genet. 26 415-423. [DOI] [PubMed] [Google Scholar]
- 8.Giaever G., Chu, A. M., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., Arkin, A. P., et al. (2002) Nature 418 387-391. [DOI] [PubMed] [Google Scholar]
- 9.Shoemaker D. D., Lashkari, D. A., Morris, D., Mittmann, M. & Davis, R. W. (1996) Nat. Genet. 14 450-456. [DOI] [PubMed] [Google Scholar]
- 10.Birrell G. W., Giaever, G., Chu, A. M., Davis, R. W. & Brown, J. M. (2001) Proc. Natl. Acad. Sci. USA 98 12608-12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ooi S. L., Shoemaker, D. D. & Boeke, J. D. (2001) Science 294 2552-2556. [DOI] [PubMed] [Google Scholar]
- 12.Steinmetz L. M., Scharfe, C., Deutschbauer, A. M., Mokranjac, D., Herman, Z. S., Jones, T., Chu, A. M., Giaever, G., Prokisch, H., Oefner, P. J. & Davis, R. W. (2002) Nat. Genet. 31 400-404. [DOI] [PubMed] [Google Scholar]
- 13.Brachmann C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P. & Boeke, J. D. (1998) Yeast 14 115-132. [DOI] [PubMed] [Google Scholar]
- 14.Smith V., Botstein, D. & Brown, P. O. (1995) Proc. Natl. Acad. Sci. USA 92 6479-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rockmill B., Lambie, E. J. & Roeder, G. S. (1991) Methods Enzymol. 194 146-149. [DOI] [PubMed] [Google Scholar]
- 16.Burke D., Dawson, D. & Stearns, T., (2000) Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 17.Winzeler E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285 901-906. [DOI] [PubMed] [Google Scholar]
- 18.Takeshige K., Baba, M., Tsuboi, S., Noda, T. & Ohsumi, Y. (1992) J. Cell Biol. 119 301-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Codon A. C., Gasent-Ramirez, J. M. & Benitez, T. (1995) Appl. Environ. Microbiol. 61 630-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colomina N., Gari, E., Gallego, C., Herrero, E. & Aldea, M. (1999) EMBO J. 18 320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan X. & Heitman, J. (2000) Mol. Cell. Biol. 20 8364-8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keeney S. (2001) Curr. Top. Dev. Biol. 52 1-53. [DOI] [PubMed] [Google Scholar]
- 23.Toth A., Rabitsch, K. P., Galova, M., Schleiffer, A., Buonomo, S. B. & Nasmyth, K. (2000) Cell 103 1155-1168. [DOI] [PubMed] [Google Scholar]
- 24.Hughes T. R., Roberts, C. J., Dai, H., Jones, A. R., Meyer, M. R., Slade, D., Burchard, J., Dow, S., Ward, T. R., Kidd, M. J., Friend, S. H. & Marton, M. J. (2000) Nat. Genet. 25 333-337. [DOI] [PubMed] [Google Scholar]
- 25.Supply P., Wach, A. & Goffeau, A. (1993) J. Biol. Chem. 268 19753-19759. [PubMed] [Google Scholar]
- 26.Kukuruzinska M. A. & Lennon, K. (1995) Biochim. Biophys. Acta 1247 51-59. [DOI] [PubMed] [Google Scholar]
- 27.Steber C. M. & Esposito, R. E. (1995) Proc. Natl. Acad. Sci. USA 92 12490-12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wijnen H. & Futcher, B. (1999) Genetics 153 1131-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.