Abstract
The Mecp2 gene has been shown to be mutated in most cases of human Rett syndrome, and mouse models deleted for the ortholog have been generated. Lineage-specific deletion of the gene indicated that the Rett-like phenotype is caused by Mecp2 deficiency in neurons. Biochemical evidence suggests that Mecp2 acts as a global transcriptional repressor, predicting that mutant mice should have genome-wide transcriptional deregulation. We tested this hypothesis by comparing global gene expression in wild-type and Mecp2 mutant mice. The results of numerous microarray analyses revealed no dramatic changes in transcription even in mice displaying overt disease symptoms, although statistical power analyses of the data indicated that even a small number of relatively subtle changes in transcription would have been detected if present. However, a classifier consisting of a combined small set of genes was able to distinguish between mutant and wild-type samples with high accuracy. This result suggests that Mecp2 deficiency leads to subtle gene expression changes in mutant brains which may be associated with the phenotypic changes observed.
Rett syndrome (RTT) is a common, severe mental retardation disorder affecting mainly females (incidence 1/20,000–1/15,000; Online Mendelian Inheritance in Man no. 312750; ref. 1). Behavioral findings include developmental stagnation after 7–18 months, ataxia, stereotyped hand-wringing motions, and autism (2). Microcephaly of affected girls has been reported (2–5), and histological findings include reduced neuron size (6).
Mutation of the MECP2 gene was found in most cases of RTT studied (7–10). The MECP2 gene encodes a methyl-CpG-binding protein which is thought to bind specifically to methylated CpG dinucleotides (11) and to act as a transcriptional repressor by virtue of its interaction with a histone deacetylase/Sin3 complex (12, 13). The involvement of the Mecp2 gene product in methylation-specific transcriptional repression suggests that RTT may be a result of misregulated gene expression. Because the prevailing model (based on biochemical evidence) suggests that Mecp2 acts as a global transcriptional repressor, it would predict that Mecp2 deficiency should result in widespread gene derepression.
Mice deficient for Mecp2 were generated by targeted mutagenesis (14, 15) and found to exhibit phenotypic similarities to RTT. Importantly, the specific deletion of Mecp2 in the brain (by using a Nestin-Cre;Mecp22lox conditional allele) mimicked the germline loss of Mecp2 (14, 15), indicating that Mecp2 is exclusively required for proper central nervous system function. Furthermore, it was shown that deletion of Mecp2 in postmitotic neurons (using a calmodulin kinase promoter-driven Cre recombinase) also mirrored the Rett phenotype, although with later onset (14). These results suggest that the Rett-like phenotype is not caused by a defect in early development, but rather is caused by dysfunction of postmitotic, differentiated neurons.
In this work, we tested the hypothesis that Mecp2 deficiency results in a widespread alteration of transcription by performing global transcriptional profiling of brain tissue from Mecp2 wild-type and mutant (14) mice by using oligonucleotide microarrays. The goal was to identify genes associated with the disease state, and, possibly, to identify genes whose expression may be regulated directly by Mecp2. Our results suggest that Mecp2 deficiency does not lead to global alterations in transcription but instead leads to subtle changes of gene expression that are only detectable by sensitive statistical analysis of relatively large datasets.
Materials and Methods
Mice.
The Mecp22lox allele (14) was bred to Nestin Cre (16) and Cam Kinase Cre 93 (Cre 93) transgenes (17). The former was used to recombine the Mecp22lox allele in the germline, thus producing Mecp21lox/y progeny for analysis of the germline-null mutants. The Mecp22lox/y;Cre93 mice were analyzed to assess the conditional phenotype. All mice were of mixed background (129/SVJae, BALB/c, C57BL/6).
RNA Samples and cRNA.
For forebrain samples, the forebrain was separated from the mid- and hindbrain by a coronal cut along the rostral border of the superior colliculi. For cerebral cortex and hippocampus samples, these structures were conservatively dissected: the cerebellum was removed and the brains were cut midsagittaly; under a dissecting microscope, the anterior border of the hippocampus was identified as the fimbria hippocampi; dorsally, the hippocampal white matter was carefully separated from overlying cerebral cortex along the alveus; and, finally, the subiculum was bisected in the middle to release the hippocampal gray matter. After removal of the hippocampus, the cerebral cortex was isolated; each hemisphere was cut coronally into blocks of ≈1.5-mm thickness, blocks between the interaural coordinates +5 mm and +1 mm, corresponding to the rostro-caudal extension of the corpus callosum, were flattened, and cortical gray matter was separated from underlying white matter. For each block, the band of cortical gray matter extending laterally from the perirhinal cortex to the cingulate and retrosplenial cortex medially, was collected. Samples were frozen on dry ice and subsequently extracted with Trizol (Life Technologies, Grand Island, NY). RNA samples were checked for integrity by gel electrophoresis and ethidium bromide staining. Targets for hybridization to Affymetrix arrays were produced from individual (i.e., not pooled) tissue samples, as described (18, 19). After hybridization and washing, arrays were scanned (18). Both Affymetrix Mu11k and MGU74A arrays were used in this study. Each array represents ≈10,000 genes (on two arrays and one array, respectively), with an overlap of ≈8,000 genes (although not necessarily represented by the same probes).
Data Scaling.
Mean response analysis (i.e., average difference of one sample's rank from those of all replicates) of the scaled data showed that even scaled data could be >20 SDs from zero sum variance within a genotype group. Analysis of the array images showed a mottled pattern of signal intensity, especially evident at the top of the array where bubbles were often observed during the fluidics protocol. This observation motivated a baseline equalization step: CEL files from the GENECHIP program (microarray suite 4, Affymetrix), were background subtracted by using the 25th percentile of a moving window (step 32 features, window 51 × 51 features) of the mismatch probes across the image of the array. The background-subtracted intensity values were then written into a CEL file for analysis.
Data Analysis.
Unmodified CEL files were used to obtain AD values by using the MICROARRAY SUITE 4.0 GENECHIP program, and background-subtracted CEL files were used to calculate modeled-expression values by using the DCHIP program (following DCHIP's invariant rank normalization; ref. 20). Both sets of data were analyzed and were qualitatively similar. Only data from the DCHIP program is presented. Analysis was limited to those genes showing a majority of “P” (present) calls as judged by the GENECHIP software. Although other filtering schemes were also explored, the majority P call criterion was chosen for its superior power (vs. considering all genes) and inclusivity (vs. considering genes with 100% P calls), although the differences between filters of 20–80% P calls were minor. Note also that “present” and “absent” are possibly misleading terms; “high-” and “low-confidence in the measured expression” are more evocative descriptions of the call algorithm used. It was found necessary to rescale this subset of data one last time by using a robust linear regression (robustfit routine of MATLAB) scaling of the majority P call data. Furthermore, gene expression values were floored to a low positive level (100 for Mu11K data and 10 for U74A data) to eliminate negative and zero values. At this point, the median correlation coefficient of the expression values from the arrays was determined, and those arrays falling below a value of 0.95 were discarded (six experiments were discarded for this reason, and five more were discarded because too few samples remained in an experiment to perform statistical analysis), yielding a final dataset of 100 profiles. Individual gene outliers [defined as absolute signal >2 × IQR (interquartile range) from median] were set to median ± 2 × IQR. For testing differential expression, several different methods were applied to the data (21–25). Data were processed and/or analysis results were compared by using EXCEL (Microsoft) and MATLAB (Mathworks).
Predictors.
The GENECLUSTER 2 (26) program [kindly provided in prerelease form by Pablo Tamayo and Keith Ohm (Whitehead Institute/MIT Center for Genome Research, Cambridge, MA)] was used to classify the data. Data from Mu11k and MG U74 arrays was combined by using a correspondence table from Affymetrix. In addition to the aforementioned P-call filter, a variation filter was also used whereby only genes showing a range of expression of 3-fold and 100 absolute units were analyzed. These three filters were applied to each experiment individually and all data points from genes which passed were used in the predictor. The predictors were constructed by crossvalidating and building on one experiment (of 9) or one “metaexperiment” (all whole forebrain, cortex, or hippocampus samples, after individual standardization), and testing on either the entire dataset or on one of the metaexperiments not included in the training set. Weighted voting and k-nearest neighbor predictors were constructed for a variety of features (1–20 features, 1–10 neighbors where applicable) and tested for accuracy on the validation sets.
RNase Protection Assays.
Candidate genes were chosen on the basis of a number of analyses including several successful classifiers (in addition to the optimal classifier presented) trained on different subsets of the data (prostaglandin D2 synthase, Rho GDI γ, parvalbumin, Cam kinase II, calcium channel β3, and serum-glucocorticoid related kinase) and consistent VERA/SAM or t test significance across several experiments and/or data processing methods (FABP7, α-synuclein). Probes were generated by PCR (except β-actin, which was supplied with the Ambion kits). Amplification primers added 10–15 nt of nonhomologous (T3 promoter sequence) to the 3′ end of probes. T7 RNA polymerase promoters were added to the PCR products by using the Lig'n Scribe kit (Ambion; the adapter adds another 10–12 nucleotides of nonhomologous sequence to the 5′ end of probes), and probes were transcribed by using the Maxiscript kit (Ambion) as described, except that reactions were carried out for 3 h. To equalize intensities of bands in the RNase protection assay, the probes were synthesized at different specific activities by diluting the UTPαP32 [3,000 Ci/mmol, 10 mCi/ml (1 Ci = 37 GBq), Amersham Pharmacia] with unlabeled nucleotide as follows: α-synuclein [amplified from an IMAGE consortium clone obtained from American Type Culture Collection (ATCC) catalog no. 3150394], 1:20 dilution; prostaglandin D2 synthase (21 kDa, brain; ATCC no. 6009404), 1:150; Rho GDP dissociation inhibitor (GDI) γ (ATCC no. 5664578), 1:1; parvalbumin (ATCC no. 5569832), 1:25; calcium/calmodulin-dependent protein kinase II α (ATCC no. 6483808), 1:250; calcium channel β-3 subunit (amplified directly from C57BL/6 first strand cDNA), 1:5; fatty acid-binding protein 7, brain (ATCC no. 1765841), 1:1; serum/glucocorticoid regulated kinase (amplified directly from C57BL/6 first strand cDNA), 1:10; Gapdh (ATCC no. 3897874), 1:50; 18s rRNA (ATCC no. 995394), 1:160,000; β-actin, 1:200. Probes were gel-purified, and 1,000 cpm each eluted probe was used for each assay. An amount of 10 μg of total RNA was used for each assay (using the Ambion RPA III kit), the samples were precipitated with the probes, resuspended in 9 μl of hybridization buffer, denatured, and hybridized overnight at 56°C. After digestion with 1:100 dilution of RNase A/T1 mix, the samples were separated on a 4% acrylamide (1:30 bisacrylamide) sequencing gel at 50 W, constant power. After drying, the gel was exposed to film and to a BAS 2000 phosphorimager screen (Fuji) for quantitation. For comparison, samples were scaled by using regression of the three control genes (actin, Gapdh, and 18s rRNA), as well as background measurements.
Results
Sample Collection.
The goal of our study was to compare gene expression in control and mutant mice before and after the onset of overt disease. Male mice mutant for Mecp2 are overtly normal until roughly one month of age [postnatal day 30 (P30)]. At this point, they begin to exhibit the phenotype characteristic of this gene disruption: tremors/seizures, lethargy, and variable weight gain. After deteriorating over the course of approximately another month, mutant mice generally die between P60 and P80. The course of disease progression is similar for mice which are Mecp22lox/y;Nestin-Cre+/o. These mice recombine the functional allele of Mecp2 mainly in neural progenitors and are, therefore, deficient for Mecp2 in the central nervous system. The disease progression in mice that are Mecp22lox/y;Cam Kinase Cre 93+/o (which delete the active Mecp2 allele only in postmitotic neurons; ref. 17) is delayed, with mice developing symptoms at ≈3 months of age and surviving to 8 or more months of age. We based our sample collection on these time courses (Table 1). Tissue samples were obtained from Mecp2 mutant mice (Mecp21lox/y; ref. 14) and wild-type sibling controls (Mecp22lox/y or Mecp2+/y). In initial experiments we isolated forebrains from P24, P35, and P56 mice. Thus, the mutant mice were asymptomatic, early symptomatic, and late symptomatic, respectively. The samples were hybridized to Affymetrix Mu11k arrays. We also analyzed dissected cerebral cortex and hippocampus at P35 and P63 and cerebral cortex and hippocampus of 4- to 6-month-old conditional mutant mice (Mecp22lox/y; Cam Kinase Cre 93+/o; ref. 14) and Mecp2+/y littermates by using MG U74Av1 arrays.
Table 1.
Samples analyzed
| Experiment | Age | Symptomatic? | Tissue | N, wt | N, mut | N, total |
|---|---|---|---|---|---|---|
| 1 | P24 | Pre | Forebrain | 4 | 6 | 10 |
| 2 | P35 | Early | Forebrain | 6 | 6 | 12 |
| 3 | Early | Cortex | 3 | 3 | 6 | |
| 4 | Early | Hippocampus | 3 | 3 | 6 | |
| 5 | P56 | Late | Forebrain | 4 | 4 | 8 |
| 6 | P63 | Late | Cortex | 6 | 6 | 12 |
| 7 | Late | Hippocampus | 7 | 6 | 13 | |
| 8 | P135–P180 | Late | Cortex | 8 | 7 | 15 |
| 9 | Late | Hippocampus | 9 | 9 | 18 | |
| Total | 100 | |||||
This table shows the sources and sizes of the samples used in the microarray experiments.
Mecp21lox/y vs. Mecp2+/y.
Mecp21lox/y or Mecp22lox/y; Nestin-Cre+/o vs. Mecp22lox/y or Mecp2+/y.
Mecp22lox/y; Cam Kinase Cre 93+/o vs. Mecp2+/y.
We note that the assayed regions of the brain are most likely to be involved in the Rett-like phenotype of Mecp2 mutant mice on the basis of the following observations. The Cam Kinase-Cre conditional mutants (which qualitatively recapitulate the germline-null phenotype) are deleted primarily in the neocortex, hippocampus, amygdala, and striatum (17). The cortex and hippocampus have been reported by various authors to be functionally or histologically abnormal in human RTT patients (6, 27–30), whereas there have not been reports of abnormalities in the striatum or amygdala. The observed histological abnormalities in the mutant mice (decreased neuron soma size) are most evident in the cerebral cortex and hippocampus (unpublished results; ref. 14). Finally, the structures were chosen for the practical reason that they can be reproducibly dissected.
Data Analysis and Statistical Tests.
Power analyses (see Figs. 3 and 4, Tables 3 and 4, and Supporting Text, which are published as supporting information on the PNAS web site, www.pnas.org) suggested that our data and statistical methods were of sufficient quality to detect even a small number of small-fold changes if they were present. Despite the apparent sensitivity of the experiments, the number of genes called statistically significant (t test, P < 0.05) that were also changed >1.5-fold (although none were changed more than 2-fold) varied between zero and three per experiment, close to the false-positive expectation. In addition, only one gene was called significantly changed when correcting for multiple testing in our Mecp2-mutant brain samples: in the Mecp22/y;CamK-Cre93+/o vs. Mecp2+/y cortex experiment, calcium channel β3-subunit, which was down 20% on average, adjusted P = 0.006. Because the Mecp2 allele we used (a deletion of exon 2; ref. 14) does not markedly affect the expression of the remainder of the transcript (not shown), we could not use differences in Mecp2 levels as a positive control for gene-expression changes in our studies. Genes that could tangentially be considered as candidates for expression changes based on published studies (31, 32) were individually inspected and confirmed to be unchanged in our experiments (not shown).
Supervised Learning.
Next, we explored the possibility that, although mutant and wild-type samples were not significantly different in expression of single genes, the behavior of multiple genes considered as a set may be correlated with Mecp2 deficiency. We constructed weighted voting (23) and k-nearest neighbor (33) classifiers. Surprisingly, predictors as small as five genes, trained on the hippocampus data, could successfully classify all of the samples in this study (100 samples from forebrain, cerebral cortex, as well as hippocampus) with >85% accuracy (proportional chance criterion (34) P < 0.05 for all experiments except P35 cerebral cortex samples: P = 0.2; P ≪ 0.001 for the classification of the entire dataset). The optimal predictor was trained on the hippocampus data, used 10 features (representing nine Unigene clusters; ref. 35), and compared query samples with the six nearest neighbors in the training set, weighted inversely to distance. This predictor was 93% accurate (93/100 correctly classified, P ≪ 0.001) in classifying the entire dataset and 89% accurate (56/63 correctly classified, P ≪ 0.001) in classifying samples not in its training set (i.e., whole forebrain and cortex samples). By using the proportional chance criterion, each of the nine experiments were successfully classified with P < 0.05. By using the more conservative Fisher's exact test, all experiments with sample size >8 were significantly correctly classified (P < 0.05).
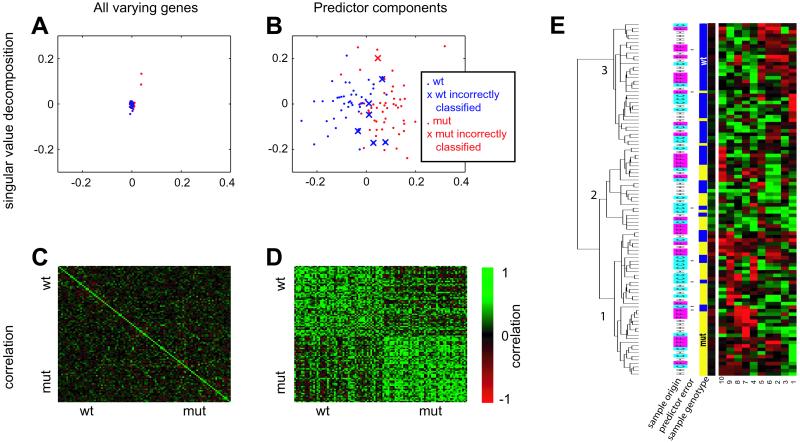
Fig. 1 A and B compare the singular value decomposition (SVD) of the predictor input and output genes. SVD describes multidimensional data (100 samples yields 100 dimensions in this case) in terms of the most significant variation in the data. Thus, distance between points on an SVD plot is related to the extent of difference of two expression profiles. Fig. 1A shows that mutant and wild-type samples are essentially superimposable when considering all genes passing a variation filter. Fig. 1B shows that, when restricting analysis to the genes comprising the optimal predictor, rough separation of mutant and wild-type samples is achieved, although the distinction is not perfect. Fig. 1 C and D show the inter-sample correlation matrix of expression levels for the same sets of genes. Again, Fig. 1C shows that there is relatively little correlation between individual samples, even between sibling replicates (not explicitly shown, but clustered together on each axis). However, note that these genes were chosen specifically for high variance to allow detection of differentially expressed genes, so this does not represent lack of correlation genome-wide. By comparison, Fig. 1D shows considerable correlation of all wild-type samples with all wild type and all mutant with all mutant samples when considering only the 10 features in the predictor. Taking these four plots together, it is apparent that the majority of the variation in the input data are not associated with the mutant/wild-type distinction, but a small subset of these genes, taken separately, does correlate with this division. Fig. 1E shows the result of hierarchical clustering (36) of the genes comprising the optimal predictor (columns, indexed along the bottom of the panel to the row numbers of Table 2). One observation which can be made is that the predictor is better able to classify the samples than is clustering analysis (or SVD or correlation matrices, for that matter). By using clustering analysis, three major clusters of experimental samples are formed (the dendrogram at the left of Fig. 1E) that respectively comprise mostly wild-type (branch no. 3, 35 samples in this category, including 3 mutant samples), mostly mutant (no. 1, 41 samples in this category, including 8 wild type), and a mixture of samples (no. 2, 24 in this category, 10 mutant, 14 wild type). The optimal predictor was able to unequivocally classify all of the samples and had a lower error rate, suggesting that supervised learning is better able to uncover class-specific expression differences than unsupervised approaches such as clustering. Conversely, it can be seen that the profiles of the majority of samples that were incorrectly classified by the predictor (denoted by asterisks in Fig. 1E) do indeed resemble those of the incorrect genotype more so than the correct one. Table 2 shows the average fold change and the P values associated with these genes across all samples. Note that none of the predictor components are individually significantly changed if a multiple-testing correction is applied.
Fig. 1.
Optimal predictor. A 10-feature, 6-nearest neighbor predictor was trained on the hippocampus data and was 93% accurate in classifying the entire 100-sample dataset. (A and B) Singular value decomposition (performed by using matlab svd function) of the variation-filtered data which was the input to the predictor training (A) (n = 95, see Materials and Methods for definition) and the components of optimal predictor (B). The first two principal components are shown (PC1 on x axis, PC2 on y axis), and the scale is the same. Samples that were misclassified by the optimal predictor are depicted as Xs. (C and D) The correlation matrix of the same sets of genes. High correlation is denoted by bright green. (E) A 10-feature, 6-nearest neighbor predictor was trained on the hippocampus data and was 93% accurate in classifying the entire 100-sample dataset. The 10 features (from nine Unigene clusters) comprising the predictor are shown with the columns corresponding to the numbered genes in Table 2, and with the expression of the individual experiments represented as rows. The yellow/blue bars represent mutant and wild-type samples, respectively. The cyan/magenta/white bars represent the cerebral cortex (C), whole forebrain (F), and hippocampus (H) samples. Asterisks denote samples incorrectly classified by the predictor. The data were clustered and visualized with the cluster and treeview programs (36). Black represents the median, red represents expression higher than the median (saturated at two interquartile ranges), and green represents expression lower than the median (saturated at two interquartile ranges).
Table 2.
Components of the optimal predictor
| Marker features | Mut/wt ratio | Wilcoxon test P value | Wilcoxon test, BY-corrected P value | |
|---|---|---|---|---|
| 1 | Rho GDI γ | 0.86 | 0.002 | 0.73 |
| 2 | Mm.204 | 0.93 | 0.32 | 1.00 |
| 3 | Prothymosin α | 0.95 | 0.74 | 1.00 |
| 4 | Serum/glucocorticoid-regulated kinase | 1.07 | 0.14 | 1.00 |
| 5 | Lipoprotein lipase | 0.92 | 0.08 | 1.00 |
| 6 | Mm.204 | 0.93 | 0.29 | 1.00 |
| 7 | Neurogenic differentiation 1 | 1.09 | 0.53 | 1.00 |
| 8 | Parvalbumin | 1.10 | 0.08 | 1.00 |
| 9 | Mm.2962 | 1.18 | 0.03 | 1.00 |
| 10 | Mm.22227 | 1.06 | 0.34 | 1.00 |
Shown are the genes that were components of the optimal predictor (10 marker features representing nine genes, six nearest-neighbor classifier). The following column shows the median mutant to wild-type ratios across all nine experiments (Table 1). Also shown are the Wilcoxon two-tailed P values for these comparisons, the Benjamini-Yekutieli (BY) multiple hypothesis testing-corrected Wilcoxon P values. Note that none of the genes is significantly changed if multiple-testing correction is applied.
Gene Expression Changes as Determined by RNase Protection Assays.
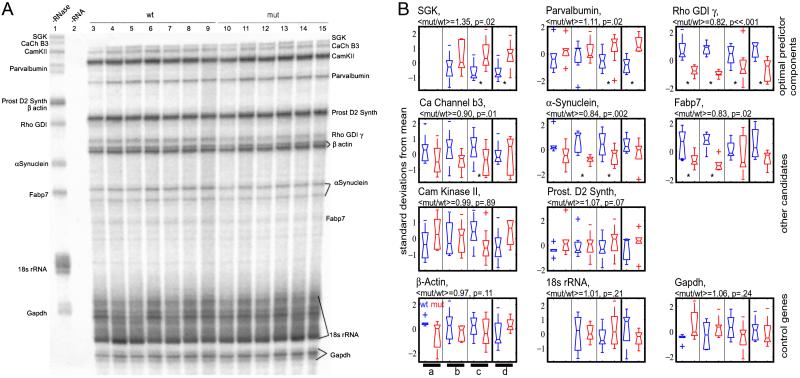
We set out to confirm some of the changes observed in our microarray experiments. By using a number of genes that were reported changed by different analyses, we developed an RNase protection assay to quantify simultaneously eight experimental and three control genes (Fig. 2). Three of these genes were components of the optimal predictor, whereas the remainder were candidates identified by other analyses (such as other successful predictors, consistent identification by t test, etc.). Assaying cerebral cortex and hippocampus of several sets of mutant and control animals, we were able to confirm a subset of the changes observed in our microarray experiments. Although the average fold-changes were small, as observed in our microarray data, they were nonetheless statistically significant (Fig. 2). In particular, we verified down-regulation of Rho GDI γ (also called “Rho GDI 5”) in all sample sets assayed. We also observed changes in α-synuclein, parvalbumin, fatty acid-binding protein 7, serum/glucocorticoid-regulated kinase, and voltage-gated calcium channel subunit-β3 in some sets but not in others, although aggregating the data of 43 cerebral cortex samples gave significant changes in all of these genes. We did not see convincing changes in calcium/calmodulin-dependent kinase II or prostaglandin D2 synthase. Note that Rho GDI γ, α-synuclein, and serum/glucocorticoid-regulated kinase were components of the optimal predictor.
Fig. 2.
(A) Representative RNase protection assay. Shown are the same samples used for the P63 Mecp21/y vs. Mecp2+/y, cerebral cortex microarray experiment (Table 1, experiment 6). Lane 1 is a 1/5 loading of the −RNase control. Lane 2 shows the −sample +RNase control. Lanes 3–9 are the wild-type cerebral cortex samples; lanes 10–15 are the mutant samples. (B) Bands from four separate RNase protection experiments were quantitated by phosphorimaging, the results were converted to z-scores (i.e., standardized), and the results are presented. Plots were generated by matlab boxplot function. Briefly, the median is the horizontal line inside each box, the interquartile range is represented by the box, the ranges of the data are represented by the whiskers, and any outliers are represented by crosses. Each pair of boxes represents an experiment; wild-type samples are in blue, mutants are in red. Experiment a: Mecp21lox/y (n = 6) vs. Mecp2+/y (n = 6), P40–60 cerebral cortex. Experiment b: Mecp21lox/y (n = 7) vs. Mecp2+/y (n = 6), P63 cerebral cortex (Table 1, experiment 6). Experiment c: Mecp22lox/y, Cam Kinase Cre 93+/o (n = 9) vs. Mecp2+/y (n = 9), P135–180 cerebral cortex (Table 1, experiment 8). Experiment d: Mecp21lox/y (n = 5) vs. Mecp2+/y (n = 6), P40–60 hippocampus. Asterisks denote comparisons that are statistically significant (P < 0.05 by two-tailed t test). Above each plot are shown the ratio of the means of mutant to wild type for all of the cerebral cortex samples (experiments a–c, n = 43) as well as the results of a two-tailed Wilcoxon signed rank test on the combined standardized data. The genes' data are organized according to their origin as candidates: the top three genes were components of the optimal predictor (Fig. 1) and the next five plots were candidates because of their appearance in other significantly successful predictors, or due to significant VERA/SAM or t test scores. The bottom three plots represent the loading controls used.
Discussion
Our results suggest that, despite striking physiological consequences including tremors, weight gain followed by wasting, death at 2 months, and the exclusive requirement of Mecp2 in the brain (deletion in the brain has the same phenotype as deletion throughout the body; refs. 14 and 15), the mutant brains have few, if any, genes that are significantly changed in expression level when considered singly. Nevertheless, a k-nearest neighbor classifier (33) was able to uncover a change in expression in all brain regions examined. Further examination of the components of this predictor shows that the average fold-changes of the genes comprising the predictor are small and generally well within the range of high false-positive rate seen when a t test is applied to all of the genes in the dataset. If the statistical tests are corrected for multiple hypothesis testing, no individual classifier component is found to be significantly changed. Our analysis does not indicate, however, whether these gene expression changes are a direct consequence of Mecp2 deficiency or a secondary result of physiological changes in the affected mutants.
Two simple explanations exist for our observations. First, it is possible that no gene is deterministically changed in expression in the absence of Mecp2. Alternatively, it is possible that small changes in expression are simply beyond the ability of the platform to detect reliably. This interpretation is bolstered by the ability of the RNase protection assay to detect significant (although low-fold) changes in expression where microarray analysis detected no changes with high confidence. Nevertheless, the significant accuracy of the predictor in classifying samples based on this small set of genes suggests that there are transcriptional differences in the brains of Mecp2 mutant vs. wild-type mice. Having uncovered a number of genes whose transcription seems to be changed in mutant mice, the question of the potential biological significance remains the subject of further study, although these genes are perhaps best thought of as markers for the phenotype rather than causative of the disease state.
The lack of more obvious changes in gene transcription in mutant mice is unexpected considering that Mecp2 has been proposed to function as a general transcription repressor. We consider the following possibilities to explain our observations. (i) Mecp2 is a member of a family of methyl-binding proteins that have similar and possibly redundant functions. For example, similar to Mecp2, Mbd2 and Mbd1 have both been implicated in methyl-CpG-specific DNA binding, recruitment of histone deacetylases, and transcriptional repression (37–39), and it is possible that expression of any of these proteins partially compensates for the loss of a member of the gene family. In this context, it is of interest that deficiency of Mbd2 or Mbd1 results in little or no overt phenotype (M.T., unpublished observations; ref. 40), possibly revealing redundancy in function between the different methyl-binding proteins. If functional redundancy is a factor in the methyl-cytosine-mediated transcriptional repression pathway and is responsible for the observed lack of global expression changes in Mecp2 mutants, the redundancy must be incomplete in view of the phenotype observed. To test this hypothesis, gene expression analyses need to be performed in mice that are deficient for several methyl-binding proteins. (ii) The Mecp2-null phenotype may be the result of transcriptional dysregulation in a small subset of cells which would not be detectable by our analysis. (iii) It is also possible that neurons are exquisitely sensitive to subtle changes in the dosage of many mRNAs, some of which we detected in our analysis, and that such subtle changes underlie the phenotype. (iv) Finally, we cannot rule out the possibility that the essential function of Mecp2 is not transcriptional, as has been suggested by the biochemical evidence. If this is the case, any changes in gene expression are secondary to another physiological role of the gene.
It should also be pointed out that Colantuoni et al. (32) have reported that samples from human Rett patients have more dramatic transcriptional changes than we observe. This may be caused by the differential sensitivity of humans and mice to loss of Mecp2 (note that male mutant mice are viable, whereas it is thought that many, although not all, human males with MECP2 mutations die perinatally; refs. 2 and 41). It is also possible that the relatively small sample size analyzed in the Colantuoni et al. study using disparate expression profiling techniques (which are likely to be inconsistent; ref. 42) and subsequent confirmation of array data with the same instead of independent samples (providing controls for the assay, not the biological variation) did not adequately control experimental noise and may have led to false-positive results. Interestingly, a study of human MECP2-mutant fibroblasts and lymphoblastoid cells suggests that loss of the Mecp2 protein does not cause reproducible changes in transcript levels (43).
It is possible that the subtle changes in gene expression found in our study are caused by the dysregulation of the most methylation-sensitive promoters, although the particular genes we identified may not be the primary targets of regulation and may instead be downstream of sensitive genes. If this were the case, one would expect that reduction of either additional MBD family members or of Dnmt1 (the maintenance methyltransferase) levels would aggravate the subtle molecular phenotype we see. To this end, the effect of compound mutations on the organismal and transcriptional phenotypes should be explored further.
Note Added in Proof.
In addition to the two mouse models cited (14, 15), another targeted disruption of the Mecp2 gene has recently been published (44).
Supplementary Material
Acknowledgments
We thank Todd Golub, Pablo Tamayo, and Keith Ohm for access to microarrays and prerelease software; Christine Ladd for help with microarray processing; Jessie Daussman and Ruth Flannery for assistance with mice; John Barnett, Trey Ideker, Nick Patterson, Jane Stauton, Vincent Carey, Fran Lewitter, and George Bell for computational help and discussion; and Uta Francke, Caroline Beard, Amir Eden, David Humpherys, Laura Lazzeroni, and Sara Cherry for critical reading of the manuscript. This work was supported by a grant from the Rett Syndrome Research Foundation and by National Institutes of Health/National Cancer Institute Grant 5RO1 CA87869 (to R.J.). This work was also supported in part by grants from Affymetrix and the Bristol-Myers Squibb Company. M.T. was supported by a predoctoral fellowship from the Howard Hughes Medical Institute; S.A. was supported by a Cure Autism Now fellowship; and R.Z.C. was supported by the Rett Syndrome Research Foundation.
Abbreviations
RTT, Rett syndrome
Pn, postnatal day n
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.McKusick-Nathans Institute for Genetic Medicine, (2000) Online Mendelian Inheritance in Man (National Center for Biotechnology Information, National Library of Medicine), www.ncbi.nlm.nih.gov/omin/.
- 2.Hagberg B., Aicardi, J., Dias, K. & Ramos, O. (1983) Ann. Neurol. 14 471-479. [DOI] [PubMed] [Google Scholar]
- 3.Villard L., Kpebe, A., Cardoso, C., Chelly, P. J., Tardieu, P. M. & Fontes, M. (2000) Neurology 55 1188-1193. [DOI] [PubMed] [Google Scholar]
- 4.Schanen N. C., Kurczynski, T. W., Brunelle, D., Woodcock, M. M., Dure, L. S., IV & Percy, A. K. (1998) J. Child Neurol. 13 229-231. [DOI] [PubMed] [Google Scholar]
- 5.Schanen C. & Francke, U. (1998) Am. J. Hum. Genet. 63 267-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauman M. L., Kemper, T. L. & Arin, D. M. (1995) Neurology 45 1581-1586. [DOI] [PubMed] [Google Scholar]
- 7.Amir R. E., Van den Veyver, I. B., Wan, M., Tran, C. Q., Francke, U. & Zoghbi, H. Y. (1999) Nat. Genet. 23 185-188. [DOI] [PubMed] [Google Scholar]
- 8.Dragich J., Houwink-Manville, I. & Schanen, C. (2000) Hum. Mol. Genet. 9 2365-2375. [DOI] [PubMed] [Google Scholar]
- 9.Van den Veyver I. B. & Zoghbi, H. Y. (2000) Curr. Opin. Genet. Dev. 10 275-279. [DOI] [PubMed] [Google Scholar]
- 10.Wan M., Lee, S. S., Zhang, X., Houwink-Manville, I., Song, H. R., Amir, R. E., Budden, S., Naidu, S., Pereira, J. L., Lo, I. F., et al. (1999) Am. J. Hum. Genet. 65 1520-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis J. D., Meehan, R. R., Henzel, W. J., Maurer-Fogy, I., Jeppesen, P., Klein, F. & Bird, A. (1992) Cell 69 905-914. [DOI] [PubMed] [Google Scholar]
- 12.Jones P. L., Veenstra, G. J., Wade, P. A., Vermaak, D., Kass, S. U., Landsberger, N., Strouboulis, J. & Wolffe, A. P. (1998) Nat. Genet. 19 187-191. [DOI] [PubMed] [Google Scholar]
- 13.Nan X., Ng, H. H., Johnson, C. A., Laherty, C. D., Turner, B. M., Eisenman, R. N. & Bird, A. (1998) Nature 393 386-389. [DOI] [PubMed] [Google Scholar]
- 14.Chen R. Z., Akbarian, S., Tudor, M. & Jaenisch, R. (2001) Nat. Genet. 27 327-331. [DOI] [PubMed] [Google Scholar]
- 15.Guy J., Hendrich, B., Holmes, M., Martin, J. E. & Bird, A. (2001) Nat. Genet. 27 322-326. [DOI] [PubMed] [Google Scholar]
- 16.Trumpp A., Depew, M. J., Rubenstein, J. L., Bishop, J. M. & Martin, G. R. (1999) Genes Dev. 13 3136-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minichiello L., Korte, M., Wolfer, D., Kuhn, R., Unsicker, K., Cestari, V., Rossi-Arnaud, C., Lipp, H. P., Bonhoeffer, T. & Klein, R. (1999) Neuron 24 401-414. [DOI] [PubMed] [Google Scholar]
- 18.Kikinis, Z. & Lee, P. (2000).
- 19.Tamayo P., Slonim, D., Mesirov, J., Zhu, Q., Kitareewan, S., Dmitrovsky, E., Lander, E. S. & Golub, T. R. (1999) Proc. Natl. Acad. Sci. USA 96 2907-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C. & Wong, W. H. (2001) Proc. Natl. Acad. Sci. USA 98 31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dudoit S., Yang, Y. H., Callo, M. & Speed, T., (2000) Technical Report #578 (Univ. of California Press, Berkeley).
- 22.Ideker T., Thorsson, V., Siegel, A. F. & Hood, L. E. (2000) J. Comput. Biol. 7 805-817. [DOI] [PubMed] [Google Scholar]
- 23.Golub T. R., Slonim, D. K., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J. P., Coller, H., Loh, M. L., Downing, J. R., Caligiuri, M. A., et al. (1999) Science 286 531-537. [DOI] [PubMed] [Google Scholar]
- 24.Benjamini Y. & Yekutieli, D. (2001) Ann. Stat. 29 1165-1188. [Google Scholar]
- 25.Manduchi E., Grant, G. R., McKenzie, S. E., Overton, G. C., Surrey, S. & Stoeckert, C. J., Jr. (2000) Bioinformatics 16 685-698. [DOI] [PubMed] [Google Scholar]
- 26.Tamayo P., Ohm, K., Subarmanian, A., Ross, K., Angelo, M. & Golub, T., (2002) GENECLUSTER VERSION 2 (Whitehead Institute/MIT Center for Genome Research, Cambridge, MA).
- 27.Bauman M. L., Kemper, T. L. & Arin, D. M. (1995) Neuropediatrics 26 105-108. [DOI] [PubMed] [Google Scholar]
- 28.Subramaniam B., Naidu, S. & Reiss, A. L. (1997) Neurology 48 399-407. [DOI] [PubMed] [Google Scholar]
- 29.Kaufmann W. E., Taylor, C. V., Hohmann, C. F., Sanwal, I. B. & Naidu, S. (1997) Eur. Child Adolesc. Psychiatry 6 75-77. [PubMed] [Google Scholar]
- 30.Armstrong D. D., Dunn, K. & Antalffy, B. (1998) J. Neuropathol. Exp. Neurol. 57 1013-1017. [DOI] [PubMed] [Google Scholar]
- 31.El-Osta A. & Wolffe, A. P. (2001) Biochem. Biophys. Res. Commun. 289 733-737. [DOI] [PubMed] [Google Scholar]
- 32.Colantuoni C., Jeon, O. H., Hyder, K., Chenchik, A., Khimani, A. H., Narayanan, V., Hoffman, E. P., Kaufmann, W. E., Naidu, S. & Pevsner, J. (2001) Neurobiol. Dis. 8 847-865. [DOI] [PubMed] [Google Scholar]
- 33.Pomeroy S. L., Tamayo, P., Gaasenbeek, M., Sturla, L. M., Angelo, M., McLaughlin, M. E., Kim, J. Y., Goumnerova, L. C., Black, P. M., Lau, C., et al. (2002) Nature 415 436-442. [DOI] [PubMed] [Google Scholar]
- 34.Shipp M. A., Ross, K. N., Tamayo, P., Weng, A. P., Kutok, J. L., Aguiar, R. C., Gaasenbeek, M., Angelo, M., Reich, M., Pinkus, G. S., et al. (2002) Nat. Med. 8 68-74. [DOI] [PubMed] [Google Scholar]
- 35.Schuler G. D., Boguski, M. S., Stewart, E. A., Stein, L. D., Gyapay, G., Rice, K., White, R. E., Rodriguez-Tome, P., Aggarwal, A., Bajorek, E., et al. (1996) Science 274 540-546. [PubMed] [Google Scholar]
- 36.Eisen M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng H. H., Zhang, Y., Hendrich, B., Johnson, C. A., Turner, B. M., Erdjument-Bromage, H., Tempst, P., Reinberg, D. & Bird, A. (1999) Nat. Genet. 23 58-61. [DOI] [PubMed] [Google Scholar]
- 38.Ng H. H., Jeppesen, P. & Bird, A. (2000) Mol. Cell. Biol. 20 1394-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hendrich B. & Bird, A. (1998) Mol. Cell. Biol. 18 6538-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hendrich B., Guy, J., Ramsahoye, B., Wilson, V. A. & Bird, A. (2001) Genes Dev. 15 710-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dotti M. T., Orrico, A., De Stefano, N., Battisti, C., Sicurelli, F., Severi, S., Lam, C. W., Galli, L., Sorrentino, V. & Federico, A. (2002) Neurology 58 226-230. [DOI] [PubMed] [Google Scholar]
- 42.Kuo W. P., Jenssen, T. K., Butte, A. J., Ohno-Machado, L. & Kohane, I. S. (2002) Bioinformatics 18 405-412. [DOI] [PubMed] [Google Scholar]
- 43.Traynor, J., Agarwal, P., Lazzeroni, L. & Francke, U. (2002) BMC Med. Genet., in press. [DOI] [PMC free article] [PubMed]
- 44.Shahbazian M., Young, J., Yuva-Paylor, L., Spencer, C., Antalffy, B., Noebels, J., Armstrong, D., Paylor, R. & Zoghbi, H. (2002) Neuron 35 243-254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.