Abstract
Mutations are a hallmark of cancer. Normal cells minimize spontaneous mutations through the combined actions of polymerase base selectivity, 3′ → 5′ exonucleolytic proofreading, mismatch correction, and DNA damage repair. To determine the consequences of defective proofreading in mammals, we created mice with a point mutation (D400A) in the proofreading domain of DNA polymerase δ (polδ, encoded by the Pold1 gene). We show that this mutation inactivates the 3′ → 5′ exonuclease of polδ and causes a mutator and cancer phenotype in a recessive manner. By 18 months of age, 94% of homozygous Pold1D400A/D400A mice developed cancer and died (median survival = 10 months). In contrast, only 3–4% of Pold1+/D400A and Pold1+/+ mice developed cancer in this time frame. Of the 66 tumors arising in 49 Pold1D400A/D400A mice, 40 were epithelial in origin (carcinomas), 24 were mesenchymal (lymphomas and sarcomas), and two were composite (teratomas); one-third of these animals developed tumors in more than one tissue. Skin squamous cell carcinoma was the most common tumor type, occurring in 60% of all Pold1D400A/D400A mice and in 90% of those surviving beyond 8 months of age. These data show that polδ proofreading suppresses spontaneous tumor development and strongly suggest that unrepaired DNA polymerase errors contribute to carcinogenesis. Mice deficient in polδ proofreading provide a tractable model to study mechanisms of epithelial tumorigenesis initiated by a mutator phenotype.
Cancer is characterized by an accumulation of mutations in genes that regulate somatic cell homeostasis (1). The multitude of mutations required for neoplastic transformation exceeds the number predicted for a normal cell, suggesting that an accelerated mutation rate (“mutator phenotype”) may be an early event in tumorigenesis (2). Consistent with this hypothesis, at least two cancer predisposition syndromes in humans result from defects in DNA repair pathways that affect mutagenesis: xeroderma pigmentosum (caused by defective processing of UV DNA damage; ref. 3) and hereditary nonpolyposis colon cancer [a consequence of defective DNA mismatch repair (MMR); ref. 4]. MMR functions, together with exonucleolytic proofreading and polymerase base selection, to ensure that DNA replication proceeds with high fidelity (5). Although defective MMR leads to cancer in humans (4) and mice (6, 7), the role of exonucleolytic proofreading in preventing cancer is unknown.
DNA polymerase δ (polδ) possesses intrinsic proofreading activity and is essential for nuclear DNA replication (8). The polymerase and 3′ → 5′ exonucleolytic proofreading active sites of polδ reside in the p125 subunit of this multimeric protein and are encoded by the Pold1 gene (9). The exonuclease active site is composed of three highly conserved motifs (Exos I, II, and III) that include four absolutely conserved carboxylate residues that coordinate exonucleolytic catalysis (Fig. 1A; refs. 10 and 11). Mutations of these residues in Saccharomyces cerevisiae reduce proofreading and cause a strong mutator phenotype (12, 13). As predicted from homology alignments (10), equivalent amino acid substitutions in phage and bacterial polymerases disrupt 3′ → 5′ exonuclease activity with little impact on DNA polymerase activity (10, 11). Thus, these substitutions in mammalian polδ are also expected to selectively disrupt proofreading.
Fig 1.
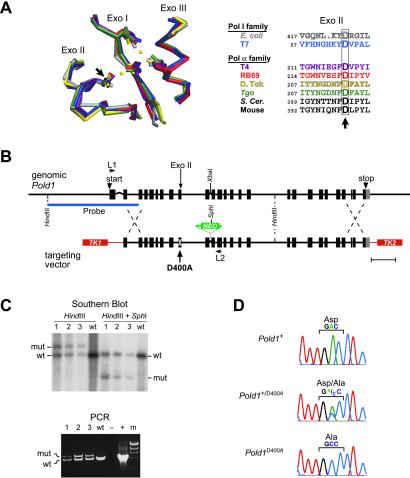
Generation of Pold1D400A mice. (A Left) Structural conservation of the Exo I, II, and III motifs from Klenow (gray), T7 (blue), T4 (purple), RB69 (red), Tgo (green), and D. Tok (yellow) DNA polymerases (Protein Data Bank ID codes , , , , , and , respectively). Cα atoms were superimposed by using SWISS-PDBVIEWER 3.5 (www.expasy.ch/spdbv/mainpage.htm). Catalytic carboxylate side chains and metal ions (yellow dots) are shown. (Right) Alignment of Exo II amino acid sequences. Arrows, conserved aspartic acid residue mutated in this report. (B) Mouse Pold1 gene (Upper) and the vector used for targeted homologous recombination (Lower). (Scale bar = 1 kb.) Boxes, exons; lines, introns; white X, D400A point mutation; NEO, neomycin-resistance gene; TK1 and TK2, thymidine kinase genes; L1 and L2, PCR primers; Probe, Southern blot probe. (C) Confirmation of gene targeting by Southern blot (Upper) and PCR (Lower) analyses. Lanes 1–3, targeted ES cell clones; wt, WT ES clone; −, no DNA control; +, WT Pold1 plasmid control; m, λ HindIII marker; mut, Pold1D400A mutant allele. (D) Sequencing of RT-PCR products from heterozygous Pold1+/D400A ES cells. (Middle) Direct sequencing of unfractionated RT-PCR products. (Top and Bottom) Sequencing of WT and Pold1D400A alleles after subcloning.
In a brief letter, we previously reported that mice homozygous for the polδ Exo II mutation D400A develop lymphomas early in life (14). Here we demonstrate that this mutation effectively eliminates polδ exonuclease activity in vitro and increases the spontaneous mutation rate of mouse cells in culture. Moreover, we show that Pold1D400A/D400A mice exhibit a high incidence of epithelial cancers later in life. These data indicate that unrepaired DNA polymerase errors contribute to carcinogenesis.
Experimental Procedures
Generation and Housing of Mutant Mice.
A 129/SvJ λ Fix II phage library (Stratagene) was screened by using oligonucleotide probes, and three clones spanning the entire coding sequence of mouse Pold1 were isolated (ref. 15; GenBank accession no. AF024570). To generate the targeting vector (Fig. 1B), Pold1 genomic fragments from sequence-verified λ clones were digested with XbaI and subcloned. The 5′ XbaI fragment, extending from exon 3 to the intron between exons 11 and 12, was subcloned into pALTER (Promega), and the 3′ XbaI fragment was subcloned into pBluescript II SK (Stratagene). A mutation was introduced to change the D400 codon to an alanine codon by using the oligonucleotide 5′-GAGGTATGGGAGGGCAAAGTTCTGAATGTTGTAGCC-3′ (alanine anticodon is underlined) and the Altered Sites II Mutagenesis System (Promega). The resultant mutated 5′ Pold1 XbaI fragment was ligated to the 3′ XbaI genomic fragment to generate a contiguous Pold1 clone extending from exon 3 to the end of the gene. A neomycin selection marker (provided by Kirk Thomas, University of Utah) was introduced into the intronic XbaI site between exons 11 and 12. This construct was subcloned into a targeting vector (TK1-TK2C, also provided by Kirk Thomas), which was then linearized by cutting between the TK1 and TK2 genes (Fig. 1B) and electroporated into R1 embryonic stem (ES) cells (16). Candidate recombinant clones were isolated by growth in the presence of G418 and gancyclovir (17). Southern blotting, PCR, and DNA sequencing confirmed gene targeting (see Supporting Materials and Methods, which are published as supporting information on the PNAS web site, www.pnas.org, for details). Chimeric male mice were generated by aggregation of these recombinant ES cell clones with C57BL/6J morulas (18). Chimeras were crossed with C57BL/6J mice, and germ-line transmission of the mutant allele was verified by PCR and DNA sequencing.
Mice were cared for in accordance with Institutional Animal Care and Use Committee guidelines in plastic cages with microisolator filter covers and corncob bedding (GreenTru, Green Products, Conrad, IA). Mice of each genotype were in cages distributed on multitiered racks in a specific pathogen-free room with 12-h on/12-h off lighting from conventional ceiling fluorescent lamps. Food (Capecchi Diet 7080, Harlan Teklad, Madison, WI) and water were provided ad libitum.
Histology and Immunophenotyping.
Tumor samples were fixed in buffered formalin, embedded in paraffin, sectioned, stained with hematoxylin and eosin, and examined by light microscopy. Lesions were scored as tumors only if they were macroscopically visible at necropsy and confirmed by histology. Thymic lymphomas from killed moribund animals were immunophenotyped by three-color flow cytometry as described (19).
Determination of Mutation Rates.
Mouse embryonic fibroblasts were cultured from day 14–16 embryos and allowed to spontaneously immortalize by continuous passage in αMEM (GIBCO/Invitrogen). Immortalized fibrosarcoma cell lines were similarly derived starting from two spontaneous sarcomas (confirmed by histology): one arising in a Pold1+/+ mouse and the other in a Pold1D400A/D400A mouse. Pold1 genotypes were confirmed by PCR and sequencing for each cell line after immortalization. Rates of spontaneous mutation to ouabain resistance were determined by fluctuation analyses (20) using the method of the mean (21). For each analysis, 10–18 replica cultures were initiated with 300–1,000 cells, and each replica was expanded to 1–3 × 106 cells. Cells were then plated into 2 mM ouabain (Fisher Scientific). After 10–14 days of growth, resistant colonies (≥50 cells) were stained with methylene blue and counted. Plating efficiencies were determined by scoring colonies that grew in the absence of ouabain after plating a known number of cells. Two or three independently derived polyclonal cell lines of each genotype were analyzed.
Polymerase and Exonuclease Assays.
A baculovirus system (Invitrogen/Life Technologies, Grand Island, NY) was used to express and partially purify (>80%) recombinant WT and D400A polδ p125 proteins. Polymerase activities were determined by measuring incorporation of [3H]dTTP into activated poly(dA-dT) by using a procedure adapted from Lee et al. (22). One unit of polymerase activity is the amount of enzyme required to incorporate 1 pmol of dNTP per min in this assay.
Exonuclease activities were determined on single- and double-stranded DNAs. Activities on single-stranded DNA were measured by quantifying the release of radioactivity from a 5′-(dT)33[3H]dT1–3-3′ substrate. Activities on double-stranded DNA were determined by a gel assay using a 5′ 32P-labeled DNA 20-mer (5′-TGATAGCACTGATATACCGA-3′) hybridized to a 40-mer DNA template (5′-TCATGGGTCGTCGGTATATCAGTGCTATCACATTAGTGTA-3′) as substrate (underline indicates the residue base paired with the 3′ terminus of the 20-mer); products were visualized by PhosphorImaging after electrophoresis through 7 M urea/12% polyacrylamide gels.
See Supporting Materials and Methods for details about recombinant polδ p125 purification and polymerase and exonuclease assays.
Results
Generation of Pold1D400A Mice.
A point mutation (D400A) was created in the exonuclease-encoding domain of mouse Pold1 (Fig. 1A), and the mutant allele was introduced into SvJ-129 mouse ES cells by using targeted homologous recombination (Fig. 1 B and C). RT-PCR and sequencing of transcripts from heterozygous Pold1+/D400A ES cells showed that the mutant and WT alleles were expressed at similar levels (Fig. 1D). Both alleles were evident in the unfractionated RT-PCR product (Fig. 1D Middle), and subcloning revealed a near equal distribution of WT and Pold1D400A sequences (six and four of 10 subclones, respectively; Fig. 1D Top and Bottom). Western blot analyses of fibroblasts cultured from Pold1D400A/D400A, Pold1+/D400A, and Pold1+/+ embryos also demonstrated similar levels of total polδ p125 protein (14), further supporting the conclusion that the WT and mutant alleles are equally expressed.
Two male chimeras were crossed with C57BL/6J females to obtain heterozygous F1 mice. Eleven F1 Pold1+/D400A × Pold1+/D400A breeding pairs were then used to create F2 animals. Two cohorts of animals were generated to assess phenotypes conferred by the Pold1D400A allele: 180 agouti F1 mice (100 Pold1+/+ WT and 80 Pold1+/D400A heterozygotes) and 199 F2 mice (53 Pold1+/+ WT, 97 Pold1+/D400A heterozygous, and 49 Pold1D400A/D400A homozygous mutants). A normal Mendelian distribution of genotypes was observed, indicating that the mutant allele does not affect embryo viability. Subsequent F2 × F2 breeding experiments showed that both male and female homozygous mutant mice are fertile.
Survival of Pold1D400A Mice.
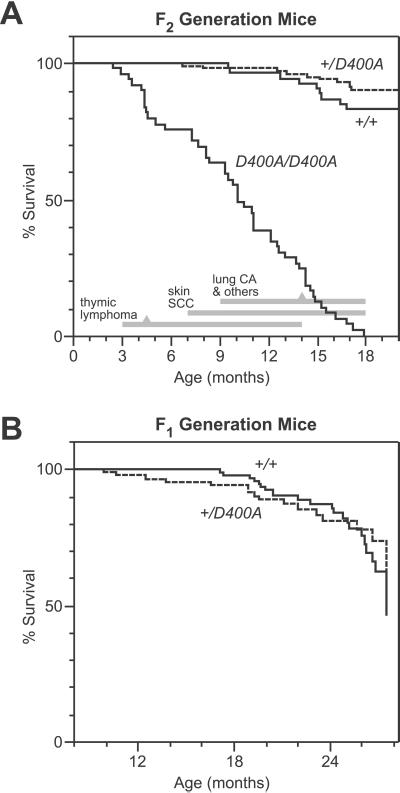
Homozygous Pold1D400A/D400A mice from the F2 generation developed into adulthood without visible abnormalities. However, after 3 months of age they exhibited significantly higher mortality than Pold1+/+ and Pold1+/D400A mice (Fig. 2A). By 18 months, all 49 of the Pold1D400A/D400A mice died spontaneously or were moribund and killed; the median age of death was 10 months. No significant difference in survival was observed between Pold1+/+ and Pold1+/D400A mice (Fig. 2A).
Fig 2.
Survival of Pold1D400A mice. (A) Kaplan–Meier survival estimates for F2 generation mice (53 Pold1+/+, 97 Pold1+/D400A, and 49 Pold1D400A/D400A). Pold1D400A/D400A mouse survival was significantly reduced (P < 0.0001 by log-rank and Wilcoxon tests) compared with Pold1+/+ and Pold1+/D400A mice, which were statistically indistinguishable from each other (P > 0.2). The major types of tumors observed in Pold1D400A/D400A mice and their time of appearance are indicated at the bottom. Gray triangles mark the median age of death caused by each tumor type (except skin tumors that were not lethal). Lung CA, lung adenocarcinomas; others include diffuse abdominal lymphomas, sarcomas, and teratomas (see Table 1). (B) Kaplan–Meier survival estimates for F1 generation mice (100 Pold1+/+ and 80 Pold1+/D400A). The two curves are statistically equivalent (P > 0.2). Statistical analyses were performed by using JMP IN 3.2.1 software (SAS Institute, Cary, NC).
To determine whether the Pold1D400A allele has a subtle effect in heterozygotes, we examined an additional 180 mice from the F1 generation for an extended period (up to 27 months of age). No significant difference in survival was observed in this cohort at a median age of 2 years (Fig. 2B). Similarly, no difference in survival was seen when the data from the F1 and F2 cohorts were combined (comparing a total of 153 WT and 177 heterozygous mice). Thus, the Pold1D400A allele acts in a recessive manner with respect to survival.
Increased Tumor Incidence in Pold1D400A/D400A Mice.
Most Pold1D400A/D400A mice (94%) harbored one or more forms of cancer at death (Table 1). Sixteen animals (33%) had two or three different primary tumors. Most of the tumors were of epithelial origin. In contrast, relatively few Pold1+/+ and Pold1+/D400A mice developed tumors (five tumors among 150 animals in the F2 generation), and all were solitary primary lesions. Thus, homozygous mutation of the polδ proofreading domain substantially accelerates tumorigenesis, particularly epithelial tumors, in mice.
Table 1.
Tumor incidence in Pold1D400A mice
| Tumor
|
Incidence | ||
|---|---|---|---|
| Pold1+/+ | Pold1+/D400A | Pold1D400A/D400A | |
| Carcinoma | |||
| Skin squamous | 0 | 0 | 31 |
| Lung adeno | 0 | 0 | 7 |
| Other adeno | 1 | 1 | 2 |
| Total | 1 (2%) | 1 (1%) | 40 (63%) |
| Lymphoma | |||
| Thymic | 0 | 0 | 15 |
| Diffuse abdominal | 0 | 2 | 4 |
| Total | 0 | 2 (2%) | 19 (39%) |
| Sarcoma | |||
| Histiocytic | 0 | 0 | 3 |
| Fibro | 0 | 0 | 1 |
| Hemangio | 1 | 0 | 0 |
| Rhabdomyo | 0 | 0 | 1 |
| Total | 1 (1%) | 0 (0%) | 5 (10%) |
| Teratoma | 0 (0%) | 0 (0%) | 2 (4%) |
| No. of mice with tumors/ total no. of mice | 2/53 (4%) | 3/97 (3%) | 46/49 (94%) |
Values represent the number (and percentage) of animals with the indicated tumor. Seven, six, and three Pold1+/+, Pold1+/D400A, and Pold1D400A/D400A mice, respectively, had no visible tumors at autopsy. There were no significant differences in cancer incidence by gender.
Sixteen Pold1D400A/D400A mice (33%) developed tumors in more than one tissue (12 with two tumors and four with three tumors); all of these had skin squamous cell carcinoma (SCC) as one of the tumor types. A total of 66 different tumors were observed in 49 Pold1D400A/D400A mice. Mice with multiple SCCs were scored as having a single tumor of this type.
Includes 27 mice with tail tumors, two with both tail and ear tumors, one with tail tumors and a hind paw tumor, and one with an isolated ear tumor.
Includes three myoepitheliomas (one from each genotype) derived from either breast or salivary glands and a Pold1D400A/D400A adenocarcinoma of the uterus with metastases to the lung.
All nine mice with adenocarcinomas also had skin SCCs. Thus, 31 of the 49 Pold1D400A/D400A mice (63%) developed carcinomas in one or more tissues.
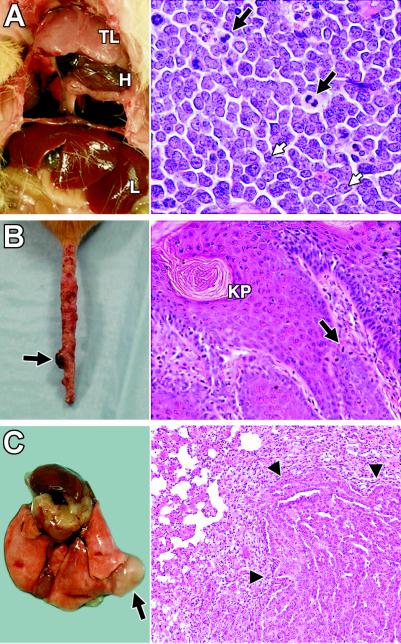
The tumors arising in Pold1D400A/D400A mice were derived from multiple cell lineages (Table 1) and developed in a distinct temporal order (Fig. 2A). Initially, thymic lymphomas predominated, developing between 3 and 14 months of age (median age = 4.5 months). These tumors often filled the chest cavity, displacing and constraining the lungs and heart, and were fatal (Fig. 3A Left). All were composed of a homogeneous population of cells with high nuclear-to-cytoplasmic ratio, fine chromatin, prominent nucleoli, and frequent mitoses, consistent with a morphological classification of lymphoblastic lymphoma (Fig. 3A Right). Immunophenotyping of three thymic lymphomas showed that they were all of T cell origin expressing CD8, but not CD4 or CD3, on the cell surface.
Fig 3.
Tumors in Pold1D400A/D400A mice. (A) Mediastinal thymic lymphoma (TL). H, heart; L, liver. Lungs are collapsed and not visible. (Right) Hematoxylin and eosin (H&E)-stained section of thymic lymphoma (original magnification, ×400). White arrows, nucleoli; black arrows, mitotic figures. (B) Tail with marked irregularities throughout its length and a focal skin tumor (arrow). (Right) Section of skin tumor stained with H&E (original magnification, ×200). KP, keratin pearl; arrow, mitotic figure. (C) Lung adenocarcinoma (arrow). (Right) H&E-stained section showing the tumor (arrowheads) compressing the normal lung parenchyma (original magnification, ×100).
Skin tumors began to emerge at 7 months of age (Fig. 3B) and continued to develop through the end of the study. These were the most frequent tumors observed in the Pold1D400A/D400A mice, developing in 30 of the 34 animals (88%) who survived beyond 8 months of age. They occurred in areas of exposed skin, predominantly on the tail but also occasionally on the ears and hind limbs. Pold1D400A/D400A mice exhibited a series of observable changes in the skin. Initially, the tail skin became dry and scaly over its length. This was followed by the development of isolated small nodules (<3 mm) with progression to larger nodules (up to 10 mm) on a background of lesions involving most of the tail (Fig. 3B Left). Classic pedunculated papillomas usually were not observed as part of this progression. Histologic examination of the small nodules revealed features of carcinoma in situ. The larger nodules demonstrated marked epithelial hyperplasia, numerous mitotic figures, intercellular bridges, and keratosis with keratin pearls typical of SCCs (Fig. 3B Right). These lesions appeared restricted to the dermis with no evidence of metastases. Most animals developed multiple tail tumors before succumbing to a second primary tumor in an internal organ. This last wave of tumors occurred at 9–18 months of age (median = 14 months) and included adenomas and adenocarcinomas (predominantly lung; Fig. 3C), diffuse abdominal lymphomas, sarcomas, and teratomas.
Increased Spontaneous Mutation Rate of Cultured Pold1D400A/D400A Cells.
To assess whether the Pold1D400A allele causes a mutator phenotype, fibroblast cell lines derived from WT, heterozygous, and homozygous mutant embryos were assayed for rates of spontaneous mutation to ouabain resistance (Table 2). The mutation rates of the WT and heterozygous cell lines were at or below the background level of detection. In contrast, the Pold1D400A/D400A cells consistently exhibited a rate that was about 10 times above background. Cells derived from a Pold1D400A/D400A fibrosarcoma exhibited a similar increase in mutation rate when compared with WT fibrosarcoma cells. Thus, homozygous expression of the Pold1D400A allele significantly increases the spontaneous mutation rate of mouse cells in culture.
Table 2.
Mutation rates of cultured cells derived from Pold1D400A mice
| Cell line
|
Mutation rate, × 10−7 | ||
|---|---|---|---|
| Pold1+/+ | Pold1+/D400A | Pold1D400A/D400A | |
| Embryonic fibroblast | <5 (3) | 5 ± 2 (3) | 72 ± 29 (4) |
| Fibrosarcoma | 6 ± 1 (2) | NA | 34 ± 14 (3) |
Mutation rates were determined by fluctuation assays using the method of the mean and are expressed as ouabain-resistant mutants/cell generation after correction for plating efficiency. Embryonic fibroblast values are the means ± SE of at least three fluctuation experiments (number indicated in parentheses) conducted on two independently derived, polyclonal cell lines for each genotype. Fibrosarcoma values are the means ± SE of cell lines derived from two spontaneous fibrosarcomas, one arising in a Pold1+/+ mouse and the other in a Pold1D400A/D400A mouse. NA, cell line not available.
D400A Mutation Disrupts Polδ p125 Exonuclease Activity.
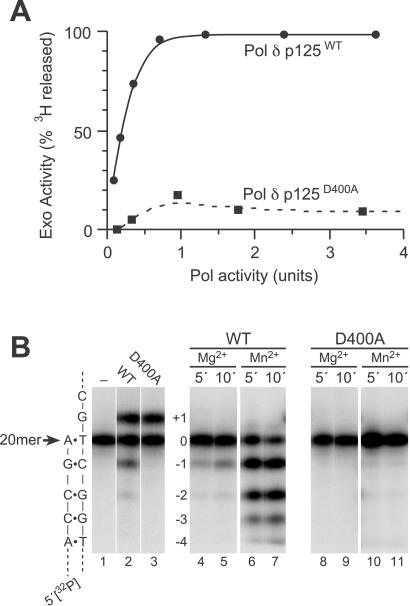
To determine the effect of the D400A mutation on polδ 3′ → 5′ exonuclease activity, we partially purified recombinant WT and mutant catalytic subunits (p125) of polδ and assayed their polymerase and exonuclease activities in vitro (Fig. 4). On a single-stranded 5′-(dT)33[3H]dT1–3-3′ substrate, the WT enzyme catalyzed release of 3′-terminal [3H]dTMP residues in proportion to the amount of polymerase added to the assay (Fig. 4A). In contrast, the D400A mutant exhibited no significant single-stranded exonuclease activity above the background of the assay even at high polymerase concentrations. Similar results were observed on a double-stranded [5′-32P]20-mer⋅40-mer substrate (Fig. 4B). When WT polδ p125 was incubated with this duplex in the presence of the next correct nucleotide dCTP and Mg2+ as divalent metal activator, products both longer and shorter than the 20-mer primer were observed (Fig. 4B, lane 2). The +1 product results from templated incorporation of dCMP by the polymerase, whereas the −1 and −2 products reflect 3′ → 5′ exonuclease activity. As expected, incubations lacking dCTP generated only the −1 and −2 exonuclease products (Fig. 4B, lanes 4 and 5). Substituting Mn2+ for Mg2+ in these reactions increased both the activity and processivity of the exonuclease, yielding a “ladder” of product bands down to the −4 position (Fig. 4B, lanes 6 and 7). In contrast, the mutant D400A polymerase yielded no detectable exonuclease products with Mg2+ as divalent metal activator (Fig. 4B, lanes 3, 8, and 9) and only a trace of product in the presence of Mn2+ (Fig. 4B, lanes 10 and 11). Together, these data show that the D400A mutation substantially reduces polδ p125 3′ → 5′ exonuclease activity on both single- and double-stranded substrates.
Fig 4.
3′ → 5′ exonuclease activities of polδ p125 proteins in vitro. (A) Activities on single-stranded DNA. Recombinant WT (•) or D400A mutant (▪) polδ p125 proteins were incubated with single-stranded (dT)33[3H]dT1–3 substrate, and the release of [3H]dTMP from 3′ termini was measured and plotted as a function of polymerase activity. Polymerase units were determined on poly(dA-dT) as described in Materials and Methods. (B) Activities on double-stranded DNA. Recombinant WT (lanes 2 and 4–7) or D400A mutant (lanes 3 and 8–11) polδ p125 proteins were incubated for 5 min (lanes 1–4, 6, 8, and 10) or 10 min (lanes 5, 7, 9, and 11) with double-stranded [5′-32P]20-mer⋅40-mer substrate, and products were analyzed by urea-PAGE and PhosphorImaging. Incubations contained comparable amounts of WT and mutant enzymes as indicated by the similar yields of +1 product after 5-min incubations in the presence of 500 μM dCTP (lanes 2 and 3). Either Mg2+ (lanes 1–5, 8, and 9) or Mn2+ (lanes 6, 7, 10, and 11) was included as divalent metal activator. A schematic of the [5′-32P]20-mer⋅40-mer substrate is shown (Left). 3′ → 5′ exonucleolytic cleavage products are indicated as −1, −2, −3, and −4. Products down to −6 were also observed in the incubations with WT enzyme and Mn2+ (data not shown). Lane 1, control incubation (5 min) lacking polδ p125 protein.
Discussion
DNA polymerase proofreading is a highly conserved mechanism that corrects spontaneous replication errors (5, 10, 11). Here we show that mice homozygous for an inactivating point mutation in the proofreading domain of polδ (D400A; Fig. 1) are prone to cancer. These mice developed lymphomas at a young age and a high incidence of epithelial tumors later in life (Figs. 2 and 3 and Table 1). The D400A mutation abolished polδ 3′ → 5′ exonuclease activity in vitro (Fig. 4) and conferred a mutator phenotype in homozygous, but not heterozygous, mutant cell lines (Table 2). Correspondingly, heterozygous mice did not exhibit a cancer phenotype (Fig. 2). These studies demonstrate the importance of polδ proofreading for mutation avoidance in mammalian cells and show that epithelial tissues are particularly susceptible to polymerase error-induced cancers in mice.
DNA polymerase errors are corrected by both proofreading and MMR (13), and several studies show that loss of MMR also predisposes mice to cancer (reviewed in refs. 6 and 7). However, the cancer phenotypes of MMR- and proofreading-deficient mice are significantly different (see below), and it is not clear that replication errors alone account for the cancer susceptibility of MMR-deficient mice (23), as MMR proteins also function in DNA damage recognition and apoptosis (24, 25). Thus, cancers in MMR-deficient mice may result from a combination of cellular events that disrupt homeostasis and drive oncogenic transformation. Our data indicate that polδ errors are sufficient to accelerate tumorigenesis as predicted by the “mutator” hypothesis of carcinogenesis (2).
The cancer phenotypes of MMR- and proofreading-deficient mice are similar in some ways but significantly different in others. Both mutant mice develop thymic lymphomas early in life. However, these tumors appear to originate from different cell populations. Flow cytometric analyses of Pold1D400A/D400A thymic lymphomas showed that they were of T cell origin with a CD3−/CD4−/CD8+ surface antigen expression pattern. In contrast, CD3+ lymphomas predominate in MMR-deficient mice as determined by immunohistochemistry (26). Although surface antigens were not evaluated in the MMR tumors, these data suggest that loss of polδ proofreading and MMR affect different pathways of lymphomagenesis. The thymic lymphomas in Pold1D400A/D400A mice may have originated from a CD3−/CD4−/CD8+ intermediate during T cell maturation (27). Alternatively, these lymphomas may exhibit aberrant expression of the CD8 antigen. Polδ has been implicated in V(D)J recombination during B cell lymphocyte maturation (28), and polδ exonuclease function may affect analogous T cell receptor rearrangements.
MMR- and polδ proofreading-deficient mice also differ in their distribution of epithelial cancers: intestinal adenomas/adenocarcinomas in MMR-deficient mice (6, 7) and skin SCCs in Pold1D400A/D400A mice (Fig. 3 and Table 1). Thus, MMR is rate-limiting for tumor formation in intestinal epithelium, whereas polδ proofreading is rate-limiting in skin epidermis. This finding may reflect tissue- or cell-specific differences in the levels of MMR or polδ proofreading, DNA damage and repair, lesion bypass, oncogene and tumor suppressor expression, mutation types and available target sequences, mutagenic processing of polδ errors (29), and/or MMR-mediated apoptosis (24, 25). Additional studies are required to characterize the contribution of these and other processes to tissue-specific tumorigenesis in mutator mice.
Tumors developed in homozygous, but not heterozygous, Pold1D400A mice (Fig. 2). The low and high cancer susceptibilities of Pold1+/D400A and Pold1D400A/D400A animals parallel the respective low and high mutator phenotypes of cultured fibroblasts derived from these animals (Table 2). Heterozygous yeast with analogous polδ proofreading mutations also show a limited mutator effect (12, 13). Thus, defects in polδ proofreading are recessive in diverse eukaryotes. The low mutation rate of heterozygotes suggests that other pathways such as MMR (13) and/or intermolecular proofreading (30, 31) correct errors introduced by mutant polδ molecules that lack proofreading but exhibit normal polymerase activity (Fig. 4) and thus presumably participate in cellular DNA synthesis. Although tumors might arise in heterozygotes by loss of the WT allele, this was not observed in our Pold1+/D400A animals (Fig. 2). Mice carrying heterozygous defects in MMR genes also rarely develop tumors (6, 7). Thus, loss of heterozygosity is not a common pathway for tumor initiation in these mouse model systems. The reason for this is unknown. In the case of Pold1, hemizygosity may confer a growth disadvantage if polδ levels in the cell are rate-limiting for normal DNA synthesis (8). Maintenance of high replication fidelity after loss of one proofreading or MMR allele presumably benefits the organism by minimizing spontaneous mutations and cancers.
The high incidence of skin SCCs in Pold1D400A/D400A mice is a striking and unusual phenotype (Fig. 3B). These tumors were clearly carcinomas of squamous cell origin and did not resemble the keratoacanthomas or sebaceous gland tumors that occasionally arise in the skin of MMR-deficient mice (ref. 32; L.E.H. and B.D.P., unpublished results). Spontaneous skin tumors are extremely rare in both 129 and C57BL/6 mice (33, 34), indicating that SCC tumorigenesis is controlled largely by the Pold1D400A allele and not genetic background in this model system. The SCCs in Pold1D400A/D400A mice arose exclusively on exposed skin (predominantly tail), suggesting that environmental factors, such as contact with irritants in the cage and/or UV light exposure, may contribute to tumorigenesis. The tumors did not appear to result from wounds, as none were observed on the snout, face, or flanks where self- and fight-inflicted wounds are most common. There was also no association between skin tumor and lymphoma development (lymphomas were observed in only 10% of mice bearing SCCs). The skin lesions progressed through a series of morphologic changes (initial epidermal thickening and cracking followed by the growth of small and then large nodules) that appear similar to human cutaneous SCCs (35), UV-induced mouse skin cancers (36), and SCCs arising in K14-HPV16 transgenic mice (37). In contrast, the carcinomas in Pold1D400A/D400A mice differ from chemical-induced skin tumors, which are predominantly benign papillomas (38). The spectrum and timing of mutations in ras, p53, and other target genes should be determined to better characterize the molecular events driving epidermal tumor development in Pold1D400A/D400A mice. The high incidence of carcinomas in a readily accessible tissue make this a tractable model for studying genetic and molecular mechanisms of epithelial tumorigenesis initiated by a mutator.
Epithelial cells serve as the first line of defense against exogenous agents, including carcinogens that generate mutagenic and polymerase-blocking DNA lesions. Thus, DNA damage, repair, and lesion bypass may influence the cancer phenotype in Pold1D400A/D400A mice. Perhaps similar to Pold1D400A/D400A mice, humans with defects in nucleotide excision repair (NER) or the lesion bypass polymerase η (polη) also develop skin cancer (3). Polδ is responsible for DNA synthesis during NER as well as long-patch base excision repair (8). Thus loss of polδ proofreading would be expected to increase the error rates of these repair pathways, including the repair of UV-induced DNA damage. Polδ may also work together with polη or related polymerases (39) to affect lesion bypass and faithful DNA synthesis (31, 40, 41). Pold1D400A/D400A mice and derivative cell lines provide tools for examining interactions of these pathways with polδ proofreading and how they influence mutagenesis and tumorigenesis.
Although our study demonstrates that defective polδ proofreading can lead to cancer in mammals, it is uncertain whether this represents a mechanism for human carcinogenesis. Several nucleotide alterations have been identified in the human polδ gene (refs. 42–44; http://egp.gs.washington.edu/data/pold1). da Costa et al. (42) found two colon cancer cell lines with mutations in or near the exonuclease domain of polδ. One of these cell lines was subsequently noted to have a defect in the MMR protein Msh6, and the mutator phenotype was mostly corrected by introduction of WT Msh6 into these cells (45). We have also shown that polδ mutations homologous to those observed by da Costa et al. (42) in humans do not confer a mutator effect in yeast (M. Singh, N. Lawrence, and B.D.P., unpublished results). The phenotypes of other polδ variants are unknown.
In summary, we show that a single point mutation in the highly conserved exonuclease domain of mouse polδ impairs exonucleolytic activity and results in a recessive mutator and cancer phenotype. These mice, along with MMR-defective mice, demonstrate the importance of DNA replication fidelity in preventing cancer. There are numerous genetic alterations that lead to mutator phenotypes in bacteria and yeast (46), suggesting that a large number of mutator genes, singly or in combination, may contribute to carcinogenesis (47). It is likely that mutator proteins affecting DNA replication fidelity will also interact with DNA damage and repair pathways, particularly in the skin and other epithelial tissues that are exposed to exogenous agents.
Supplementary Material
Acknowledgments
We thank Kirk Thomas for advice and reagents used in the gene targeting experiments and Peggy Lewis and Peggy O'Neil of the Gene Targeting Core Facility for their help in performing the gene targeting experiments. The Gene Targeting Core Facility is part of the National Institutes of Health Center of Excellence in Molecular Hematology at the University of Utah. We are indebted to Cheng-Keat Tan and Antero So for help and reagents used for the purification and analyses of mouse polδ and Aym Berges for preparation of gel-purified oligonucleotides. We thank Jamie Bugni, Robert Smith, Mark Meuth, Mario Capecchi, William Carroll, George Klarmann, Jim Kushner, Kirk Thomas, and David Virshup for valuable discussions and critical reading of the manuscript, Diana Lim for assistance with graphic design, Aurelia Meloni-Ehrig of the Cytogenetics Core Facility for assistance with karyotype analysis, Ken Boucher of the Huntsman Cancer Institute Biostatistics Shared Resource for help with statistical analyses, and Donovan Anderson, Michelle Carroll, and Jing Xu for technical assistance. This work was supported by grants from the Primary Children's Medical Center Foundation, the University of Utah, the Leukemia Society of America (Fellowship 5119-97), the Hope Street Kids Foundation, the American Cancer Society (RPG-00-299-01-GMC), and the National Institutes of Health (K08 CA72731, R01 ES09927, R01 ES09927-S1, and U01 ES11045).
Abbreviations
ES, embryonic stem
MMR, mismatch repair
polδ, DNA polymerase δ
SCC, squamous cell carcinoma
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hanahan D. & Weinberg, R. A. (2000) Cell 100 57-70. [DOI] [PubMed] [Google Scholar]
- 2.Loeb L. A. (2001) Cancer Res. 61 3230-3239. [PubMed] [Google Scholar]
- 3.Cleaver J. E. (2000) J. Dermatol. Sci. 23 1-11. [DOI] [PubMed] [Google Scholar]
- 4.Peltomäki P. (2001) Hum. Mol. Genet. 10 735-740. [DOI] [PubMed] [Google Scholar]
- 5.Kunkel T. A. & Bebenek, K. (2000) Annu. Rev. Biochem. 69 497-529. [DOI] [PubMed] [Google Scholar]
- 6.Buermeyer A. B., Deschenes, S. M., Baker, S. M. & Liskay, R. M. (1999) Annu. Rev. Genet. 33 533-564. [DOI] [PubMed] [Google Scholar]
- 7.Heyer J., Yang, K., Lipkin, M., Edelmann, W. & Kucherlapati, R. (1999) Oncogene 18 5325-5333. [DOI] [PubMed] [Google Scholar]
- 8.Hindges R. & Hübscher, U. (1997) Biol. Chem. 378 345-362. [DOI] [PubMed] [Google Scholar]
- 9.Lee M. Y., Jiang, Y. Q., Zhang, S. J. & Toomey, N. L. (1991) J. Biol. Chem. 266 2423-2429. [PubMed] [Google Scholar]
- 10.Bernad A., Blanco, L., Lázaro, J. M., Martin, G. & Salas, M. (1989) Cell 59 219-228. [DOI] [PubMed] [Google Scholar]
- 11.Derbyshire V., Pinsonneault, J. K. & Joyce, C. M. (1995) Methods Enzymol. 262 363-385. [DOI] [PubMed] [Google Scholar]
- 12.Simon M., Giot, L. & Faye, G. (1991) EMBO J. 10 2163-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrison A., Johnson, A. L., Johnston, L. H. & Sugino, A. (1993) EMBO J. 12 1467-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldsby R. E., Lawrence, N. A., Hays, L. E., Olmsted, E. A., Chen, X., Singh, M. & Preston, B. D. (2001) Nat. Med. 7 638-639. [DOI] [PubMed] [Google Scholar]
- 15.Goldsby R. E., Singh, M. & Preston, B. D. (1998) Mamm. Genome 9 92-93. [DOI] [PubMed] [Google Scholar]
- 16.Nagy A., Rossant, J., Nagy, R., Abramow-Newerly, W. & Roder, J. C. (1993) Proc. Natl. Acad. Sci. USA 90 8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansour S. L., Thomas, K. R. & Capecchi, M. R. (1988) Nature 336 348-352. [DOI] [PubMed] [Google Scholar]
- 18.Wood S. A., Allen, N. D., Rossant, J., Auerbach, A. & Nagy, A. (1993) Nature 365 87-89. [DOI] [PubMed] [Google Scholar]
- 19.Kim M., Cooper, D. D., Hayes, S. F. & Spangrude, G. J. (1998) Blood 91 4106-4117. [PubMed] [Google Scholar]
- 20.Reitmair A. H., Risley, R., Bristow, R. G., Wilson, T., Ganesh, A., Jang, A., Peacock, J., Benchimol, S., Hill, R. P., Mak, T. W., et al. (1997) Cancer Res. 57 3765-3771. [PubMed] [Google Scholar]
- 21.Capizzi R. L. & Jameson, J. W. (1973) Mutat. Res. 17 147-148. [DOI] [PubMed] [Google Scholar]
- 22.Lee M. Y. W. T., Tan, C.-K., Downey, K. M. & So, A. G. (1984) Biochemistry 23 1906-1913. [DOI] [PubMed] [Google Scholar]
- 23.Tomlinson I. P., Novelli, M. R. & Bodmer, W. F. (1996) Proc. Natl. Acad. Sci. USA 93 14800-14803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li G. M. (1999) Oncol. Res. 11 393-400. [PubMed] [Google Scholar]
- 25.Fishel R. (2001) Cancer Res. 61 7369-7374. [PubMed] [Google Scholar]
- 26.Lowsky R., DeCoteau, J. F., Reitmair, A. H., Ichinohasama, R., Dong, W. F., Xu, Y., Mak, T. W., Kadin, M. E. & Minden, M. D. (1997) Blood 89 2276-2282. [PubMed] [Google Scholar]
- 27.Guidos C. J., Weissman, I. L. & Adkins, B. (1989) Proc. Natl. Acad. Sci. USA 86 7542-7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jessberger R., Schar, P., Robins, P., Ferrari, E., Riwar, B. & Hübscher, U. (1997) Nucleic Acids Res. 25 289-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Datta A., Schmeits, J. L., Amin, N. S., Lau, P. J., Myung, K. & Kolodner, R. D. (2000) Mol. Cell 6 593-603. [DOI] [PubMed] [Google Scholar]
- 30.Perrino F. W. & Loeb, L. A. (1990) Biochemistry 29 5226-5231. [DOI] [PubMed] [Google Scholar]
- 31.Bebenek K., Matsuda, T., Masutani, C., Hanaoka, F. & Kunkel, T. A. (2001) J. Biol. Chem. 276 2317-2320. [DOI] [PubMed] [Google Scholar]
- 32.Reitmair A. H., Redston, M., Cai, J. C., Chuang, T. C., Bjerknes, M., Cheng, H., Hay, K., Gallinger, S., Bapat, B. & Mak, T. W. (1996) Cancer Res. 56 3842-3849. [PubMed] [Google Scholar]
- 33.Smith G. S., Walford, R. L. & Mickey, M. R. (1973) J. Natl. Cancer Inst. 50 1195-1213. [DOI] [PubMed] [Google Scholar]
- 34.Blackwell B. N., Bucci, T. J., Hart, R. W. & Turturro, A. (1995) Toxicol. Pathol. 23 570-582. [DOI] [PubMed] [Google Scholar]
- 35.Alam M. & Ratner, D. (2001) N. Engl. J. Med. 344 975-983. [DOI] [PubMed] [Google Scholar]
- 36.Brash D. E. & Ponten, J. (1998) Cancer Surv. 32 69-113. [PubMed] [Google Scholar]
- 37.Coussens L. M., Hanahan, D. & Arbeit, J. M. (1996) Am. J. Pathol. 149 1899-1917. [PMC free article] [PubMed] [Google Scholar]
- 38.Frame S., Crombie, R., Liddell, J., Stuart, D., Linardopoulos, S., Nagase, H., Portella, G., Brown, K., Street, A., Akhurst, R. & Balmain, A. (1998) Philos. Trans. R. Soc. London B 353 839-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodman M. F. (2002) Annu. Rev. Biochem. 71 17-50. [DOI] [PubMed] [Google Scholar]
- 40.Kannouche P., Broughton, B. C., Volker, M., Hanaoka, F., Mullenders, L. H. & Lehmann, A. R. (2001) Genes Dev. 15 158-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thakur M., Wernick, M., Collins, C., Limoli, C. L., Crowley, E. & Cleaver, J. E. (2001) Genes Chromosomes Cancer 32 222-235. [DOI] [PubMed] [Google Scholar]
- 42.da Costa L. T., Liu, B., el-Deiry, W., Hamilton, S. R., Kinzler, K. W., Vogelstein, B., Markowitz, S., Willson, J. K., de la Chapelle, A., Downey, K. M., et al. (1995) Nat. Genet. 9 10-11. [DOI] [PubMed] [Google Scholar]
- 43.Chung D. W., Zhang, J. A., Tan, C. K., Davie, E. W., So, A. G. & Downey, K. M. (1991) Proc. Natl. Acad. Sci. USA 88 11197-11201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flohr T., Dai, J. C., Buttner, J., Popanda, O., Hagmuller, E. & Thielmann, H. W. (1999) Int. J. Cancer 80 919-929. [DOI] [PubMed] [Google Scholar]
- 45.Umar A., Koi, M., Risinger, J. I., Glaab, W. E., Tindall, K. R., Kolodner, R. D., Boland, C. R., Barrett, J. C. & Kunkel, T. A. (1997) Cancer Res. 57 3949-3955. [PubMed] [Google Scholar]
- 46.Miller J. H. (1998) Mutat. Res. 409 99-106. [DOI] [PubMed] [Google Scholar]
- 47.Schar P. (2001) Cell 104 329-332. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.