Abstract
The prevalence of hepatitis C virus (HCV)-related hepatocellular carcinoma (HCC) is considerably lower in the U.S. than in Japan. To elucidate this difference, we determined the time origin of the HCV epidemic in each country by using molecularly clocked long-term serial samples obtained from HCV carriers of genotypes 1a and 1b. The molecular clock estimated that HCV genotype 1 first appeared in Japan in around 1882, whereas emergence in the U.S. was delayed until around 1910. In addition, by statistical analysis using coalescent theory, the major spread time for HCV infection in Japan occurred in the 1930s, whereas widespread dissemination of HCV in the U.S. occurred in the 1960s. These estimates of viral spread time are consistent with epidemiologic observations and predict that the burden of HCC in the U.S. will increase in the next two to three decades, possibly to equal that currently experienced in Japan.
Chronic hepatitis C virus (HCV) infection is an endemic disease affecting millions of individuals globally (1, 2). HCV infection is usually clinically mild, but ≈20% of patients progress to severe chronic hepatitis and cirrhosis, occasionally culminating in hepatocellular carcinoma (HCC) over the course of 2–3 decades, the latter especially in Japan, Italy, and Spain (3–7).
In the United States (U.S.) from 1988 to 1994, the overall prevalence of antibody to HCV was 1.8%, and the calculated prevalence of viremia was 1.3%. These figures led to the estimate that ≈2.7 million Americans were chronically infected with HCV (8), with an associated ≈10,000 deaths per year from cirrhosis and HCC (9). However, the prevalence of HCC in the U.S. is well below that reported in Japan (3–5). The difference in the prevalence of HCC between Japan and the U.S. may because of various genetic and/or environmental influences, but an important factor is postulated to be the time of exposure to HCV. The HCV epidemic has been present in Japan for a longer time than in the U.S. To investigate the hypothesis that a longer duration of the HCV epidemic accounts for the differing HCC burden between the U.S. and Japan, the constant evolutionary rate of the virus over time, a molecular clock of HCV evolution, can be determined by retrospective extrapolation, and it provides an insight into both the time when a virus enters a population (“divergence time”) and the time when it expands within that population (“spread time”).
Previous studies (10–14) have estimated the rate of synonymous and nonsynonymous substitutions in the HCV genome. However, these earlier estimates must be regarded as preliminary because most were based on relatively short observation times, analyzed only short regions of the HCV genome, examined a small number of samples, and/or used simplified models of nucleotide evolution. Additionally, the extrapolation of substitution rates based on pairwise comparisons can give misleading estimates. To address these issues, this study used serial samples obtained from individuals under long-term medical follow-up for up to 20 years (15); additionally, by sequencing long segments of the HCV genome, the accuracy of any extrapolation is enhanced. The assumption underlying this study is that an accurate molecular clock of HCV can lead to a more valid extrapolation of the divergence time and spread time of HCV infection in the U.S. and Japan and, hence, establish the relative influx of HCV infection in these populations. Differences in viral divergence and spread time then may be compared with differences in the clinical burden of HCV infection, in particular, to ascertain whether the higher rate of HCC in Japan is primarily a function of the total time of viral exposure within these populations.
Methods
Long-Term Serial Samples.
Thirty-six serial samples (varying from two to seven serial samples for each subject) from 10 U.S. subjects with HCV genotype 1a were used, supplemented by single samples from 11 additional U.S. genotype 1a subjects. All samples were obtained from patients enrolled in prospective studies of transfusion-associated hepatitis conducted at National Institutes of Health since 1972 (15) and from anti-HCV positive blood donors whose exposure history suggested infection of >20 years' duration (16). The interval between the first and last sample in the 10 subjects for whom serial samples were available ranged from 7 to 21.6 years (mean ± SD:15.1 ± 5.4). Some of the samples were obtained within the first 6 months of infection. The clinical data for each subject are shown in Table 1. For Japanese subjects, 14 serial samples from six subjects with HCV genotype 1b in Nagoya City University Graduate School of Medical Sciences and 18 HCV genotype 1b sequences from the GenBank database (accession nos. D13558, D90208, D14484, AF165046, AF165048, AF165050, AF165052, AF207752–AF207761, and AF208024) were used. All U.S. and Japanese participants provided informed consent.
Table 1.
Clinical data of subjects used in our study
| Subjects
|
Sample intervals, years
|
Sample number
|
Age, years
|
Age at infection, years
|
Risk factors
|
Gender
|
Race
|
HBc Ab
|
Degree of chronic hepatitis, HAI | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mild | Moderate | Bridging | Cirrhosis | |||||||||
| CF | 15.6 | 4 | 71 | 48 | BTF (1977) | M | B | Neg | X | |||
| FM | 20.6 | 5 | 55 | 32 | BTF (1978) | M | W | Neg | X | |||
| JL | 21.6 | 4 | 52 | 24 | BTF (1973) | F | B | Neg | X | |||
| HW | 17.7 | 7 | 84 | 60 | BTF (1977) | M | W | Neg | X | |||
| PR | 15.5 | 4 | 62 | 39 | BTF (1978) | M | W | Neg | X | |||
| PEN | 17.8 | 4 | 53 | 29 | BTF (1977) | M | W | Neg | X | |||
| KM | 19 | 2 | 76 | 46 | BTF (1971) | M | W | Neg | X | |||
| C107 | 8 | 2 | 41 | <21 | IDU (1972) | F | W | Neg | X | |||
| C115 | 8.1 | 2 | 44 | <24 | Unknown | M | W | Neg | X | |||
| C159 | 7 | 2 | 49 | 29–33 | OPE (1989) | F | B | Neg | X | |||
The first seven subjects were enrolled in prospective studies of transfusion-associated hepatitis conducted at the National Institutes of Health starting in 1972 (15) and the final three subjects were from anti-HCV positive blood donors whose exposure history suggested infection of >20 years' duration (16).
BTF, blood transfusion; IDU, injection drug user; OPE, surgical operation. The putative infection time appears in parentheses.
B, African American; W, Caucasian; HBcAb, antibody to HBV core antigen; Neg, negative.
RNA Extraction, PCR, and Sequencing.
To extract total RNA from human sera, the RNA isolator (Genosys, The Woodlands, TX) was used, with slight modification from the manufacturer's instructions; Superscript II RNase H− reverse transcriptase (GIBCO/BRL) was used for reverse transcription. To reduce the number of artifactual substitutions occurring during PCR, Platinum Pfx DNA Polymerase (GIBCO/BRL) with very high fidelity was used. For the PCR in HCV core, E1, and E2 regions, long PCR fragments (core to NS2 regions) were amplified by HCV1- and 2-core-s1 (346–373): GCACGAATCCTAAACCTCAAAGAAAAAC and HCV1ab-ns2-as (2540–2565): CTATCAGCAGCATCATCCACAAGCAG. The first products were nested by using four primers sets: (i) HCV1&2-core-s1 and HCV1&2-core-as (865–885): GCARRGCCARRAGGAAGATAG [R = A or G]; (ii) HCV1ab-core-s2 (808–828): GGGTTCTGGARGACRGCGTGA and 45AT-as2 (1299–1321): GACCARTTCATCATCATRTCCCA; (iii) HCVE1-s1 (834–859): GCAACAGGGAACCTTCCTGGTTGCTC and HCVE2-as1 (1600–1624): TTCAAGGCMGTSCTRTTGATGTGCC [M = A or C, S = C or G]; (iv) 45AT-s1 (1290–1310): CGCATGGCNTGGGAYATGATG [n = any, Y = C or T] and HCVE2-as2 (1848–1868): GAAGCAATAYACYGGRCCACA. For PCR in NS5B region, NS5B 8278S (8258–8278): TGATACCCGCTGYTTTGACTC and NS5B 8618AS (8618–8639): GTACCTGGTCATAGCCTCCGTG were used. As recommended by Simmonds (17), to increase stringency, the PCR products of a single target molecule were directly sequenced rather than initially cloned.
Molecular Evolutionary Analyses.
Twenty-two sequences within combined long regions (1,778 nt; core, E1, E2, NS5B) were generated from samples obtained over the course of around 20 years from four subjects (duration: 15.6–21.6 years) and were aligned with CLUSTAL W V.1.8. Nine types of evolutionary distance models were used: Kimura's two-parameter method (K2P), Tamura-Nei (TN) model, K2P compensated by γ-distribution, Tamura-Nei compensated by γ-distribution, Nei-Gojobori (NG) model, modified Nei-Gojobori model, Li-Wu-Luo model, Pamilo-Bianchi-Li (P-B-L) model, and the Kumar model. MEGA V.2.1 (18) was used to determine the number of nucleotide substitutions per site (evolutionary distance) between the strains. The γ-shape parameter (α) was presumed in using the Yang-Kumar model, resulting in 0.46 for combined regions and 0.64 for the NS5B region. Based on these values by nine distance models, phylogenetic trees were constructed by the neighbor-joining (NJ) tree method, and then the distances from the ancestral sequence of the most recent common ancestor to each strain were estimated. To confirm the reliability of the phylogenetic tree, bootstrap resampling tests were performed 1,000 times. ML trees under Hasegawa-Kishino-Yano model (γ) implemented in PAUP V.4.0B8. The ancestral sequence of the most recent common ancestor was parsimoniously estimated from ML tree reconstructed by heuristic search in PAML (19), and then the distances from the ancestral sequence to each strain were estimated by the above nine distance models in MEGA V.2.1 (18).
Effective Population Size.
By using 17 U.S. genotype 1a sequences from 1998 to 1999 and 16 Japanese genotype 1b sequences in 1999 within the NS5B region, a phylogenetic tree was constructed by dnamlk, DNA maximum likelihood with molecular clock, by using PHYLIP V.3.6, and a parametric maximum-likelihood estimate of demographic history was conducted with the computer software GENIE V.2.0 (20). In brief, demographic history is represented mathematically by the function N(t), which represents the effective population size of the viral epidemic at t years before the present. For a parametric model to estimate the growth of HCV in a population, a logistic growth model was derived from a basic epidemiological model (21).
Results
Molecular Clock of HCV.
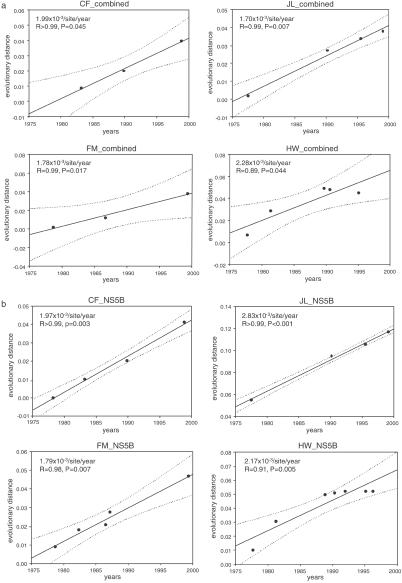
HCV is characterized by a high degree of genetic heterogeneity (22), resulting from the fact that the viral RNA polymerase lacks a proof-reading 3′–5′ exonuclease activity (23), an important repair mechanism. The rate of nucleotide misincorporation by the viral RNA polymerase of HCV has been calculated to be on the order of 10−3 per site per year (10, 11, 13), which is similar to the other RNA viruses (24, 25). However, the accuracy of any estimated evolutionary rate depends not only on a reliable time frame but also on a precise estimation of evolutionary distances among taxa and a correct depiction of the tree topology. To increase the precision in the estimation of the evolutionary rate, 10 different methods of phylogenetic tree analysis (nine NJ trees and one ML tree) with long-term serial sequences were used for each data set. Two basic approaches determined the divergence time, each measuring long sequences (1,778 nt) in combined genomic regions (Fig. 1a) as well as in the shorter NS5B region and testing multiple long-term serial samples from most patients (Fig. 1b). Based on these robust phylogenetic trees, the evolutionary distances from the most recent common ancestor to each strain were deduced. Linear regression showed that although the figures for molecular clocks differed somewhat from patient to patient, for each individual patient the molecular clock remained within a well defined range (P < 0.05, Fig. 1), and that the mean figure for the molecular clocks for all patients was also within well defined parameters (P < 0.01, Fig. 2 a and b). Specifically for the U.S. cohort, the mean evolutionary rate of all codon substitutions within the combined nucleotide sequences was 0.67 (0.53–0.79) × 10−3 per site per year, and the rate of synonymous substitutions was 1.32 (1.00–1.74) × 10×3 per site per year (Fig. 2c). Of note, these results are slower than previously reported for large genomic regions [0.9 (12), 1.44 (13) and 1.92 × 10−3 (10), respectively] and may reflect differences between patients, viral quasi-species, or the fact that prior studies, using only two time points per patient, may have overestimated the rate of codon substitutions. However, this rate of genetic change is similar to the figure obtained from a cohort infected with HCV after administration of HCV-contaminated anti-D Ig 17 years earlier (14).
Fig. 1.
The molecular clock within each individual obtained from National Institutes of Health prospective studies. (a) The regression analyses within combined regions were based on the evolutionary distances using Pamilo-Bianchi-Li model (P-B-L; P < 0.05). Note that each molecular clock within each single host was different [(1.70–2.28) × 10−3 nucleotide substitutions per site per year]. (b) Based on a phylogenetic tree within the NS5B region constructed by the NJ method, the figures for the four patients in A became statistically more significant with the inclusion of more data points (P < 0.01).
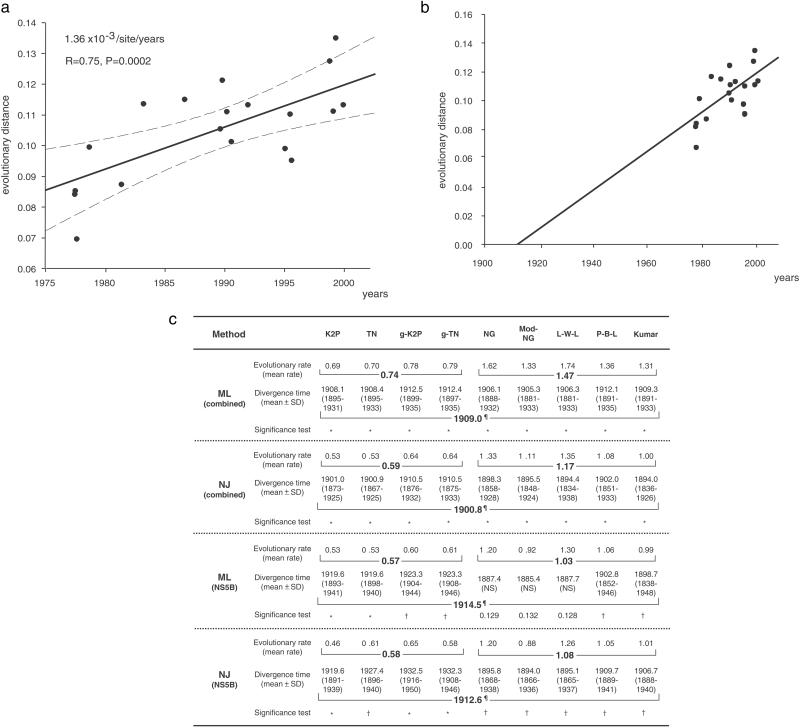
Fig. 2.
Estimating the most recent common ancestor of genotype 1a in the U.S. based on data collected over the last three decades. (a) A regression analysis within combined regions was performed to estimate a mean molecular clock. The mean evolutionary rates (solid regression line) of the P-B-L model indicated 1.36 × 10−3 per site per year (P = 0.0002). The 95% confidence intervals of the regression line are indicated by broken lines. (b) The divergence time of the most recent common ancestor of U.S. genotype 1a was estimated to have occurred around 1910, which is supported by most evolutionary models (c). (c) By using the mean molecular clock derived from regression analyses of serially determined phylogenetic trees, the divergence time of the most recent common ancestor of genotype 1a in the U.S. was estimated. Models for evolutionary analyses are explained in Methods. Phylogenetic trees were analyzed by the maximum likelihood and the NJ methods. To estimate the divergence time (mean ± SD), 500 bootstrap replicates were generated by random-with-replacement resampling of the data points to determine the molecular origin of ancestral sequences. Mean and SD of the divergence times also were determined. Standard significance testing was conducted. *, P < 0.01; †, P < 0.05; ¶, mean of 95% significance divergence time (year); NS, not significant. Evolutionary rate = number of nucleotide substitutions × 10−3 per site per year.
Divergence Time of HCV.
Given a phylogenetic tree, one can plot the total branch length from the tips of the branches to the ancestral node against the year of sampling. To estimate the time when the branch length was zero, i.e., the time of the ancestral sequence, a retrospective regression backward in time (Fig. 2 a and b) was made. It was deduced that the divergence time of the most recent common ancestor of genotype 1a in the U.S. was ≈1910. This estimate lies well within the 95% significance data (1894–1912) of the divergence time estimated by most models, even taking into account the bias toward synonymous transition substitutions (Fig. 2c).
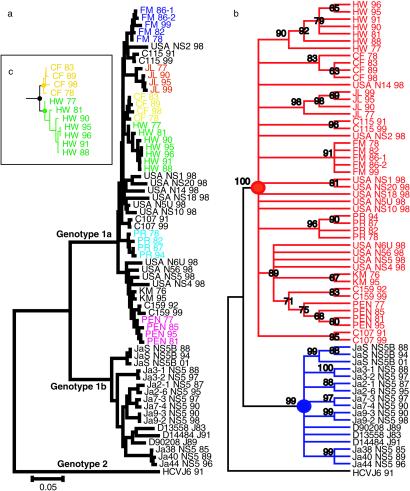
To confirm the divergence time of U.S. genotype 1a and compare it with genotype 1b strains in Japan, a total of 47 sequences of HCV genotype 1a from 21 U.S. subjects and 17 sequences of genotype 1b from 9 Japanese subjects were analyzed for mutational changes in the NS5B region, which was chosen as it has the highest phylogenetic signal and a relatively low-noise content (26). The same tree topologies and consensus for the most recent common ancestor point of U.S. genotype 1a were obtained in the NS5B region as in the larger genomic segments (Fig. 3 a and b). By using the NS5B region for the entire U.S. cohort, the divergence time of the most recent common ancestor of U.S. genotype 1a was again around 1910, with the 95% significant divergence time ranging from 1894 to 1932 (Fig. 2c). Similarly, the divergence time of the most recent common ancestor of HCV genotype 1b in Japan was estimated to be before 1882, and thus ≈30 years earlier than in the U.S. When more samples from each patient were analyzed by using the short NS5B sequences, the statistical significance increased (P < 0.01, Fig. 1b), thus providing evidence of the validity of this analysis.
Fig. 3.
A phylogenetic tree within NS5B region constructed by NJ method. (a) HCV genotype 2a (HCV J6 strain: D00944) was used as an outgroup. Each subject with serial strains in the National Institutes of Health study that had a significantly independent cluster is indicated by color. (b) By using an outgroup (HCV J6), U.S. genotype 1a strains (red) and Japanese genotype 1b strains (blue) were shown to diverge from each ancestral point (solid circle). The numbers indicate bootstrap reliability (1,000×). Most individual clusters were reliable at >80%. (c) The timing of an individual's HCV infection was estimated as the interval between an individual's most recent ancestral point and the divergence point of each individual's cluster. For example, a solid yellow circle was the most recent ancestral point of patient “CF.” A solid green circle was the most recent ancestral point of patient “HW.” A solid black circle was the divergence point of both “CF” cluster and “HW” cluster. In this phylogenetic tree, HW's HCV infection timing was thought to be the period between the divergence time of the solid green circle and the divergence time of the solid black circle.
Our figures agree with a report that used a maximum likelihood approach under the assumption of a constant rate of nucleotide substitutions within E1 and NS5 gene sequences that estimated the ancestral point of genotype 1a originated ≈100 years ago (21), thus disagreeing with Smith et al. (14), who have a figure of 58–65 years ago.
It is possible that HCV was introduced into the U.S. population during the Spanish-American war in 1898–1900, when soldiers were infected with the virus overseas and returned to the U.S. Additionally, the divergence time of the most recent common ancestor of this blood-borne virus in the U.S. is also consistent with the introduction of modern blood transfusion practice after the discovery of blood types by Landsteiner in 1900.
Spread Time of HCV Strains Between the U.S. and Japan.
Based on the various figures obtained from each individual and plotting them into phylogenetic trees within combined long regions or the NS5B region, the mean spread time of HCV genotype 1a in the U.S., defined as the period between an individual's most recent ancestral point and the divergence point of each individual's cluster (Fig. 3c), was estimated to have occurred between 1954 and 1978. At this time, injection drug use, sharing needles, and transfusion of unscreened blood and blood products were widespread (8). Thus, our molecular analysis is congruent with epidemiological data (8, 27) regarding the spread of HCV in the U.S.
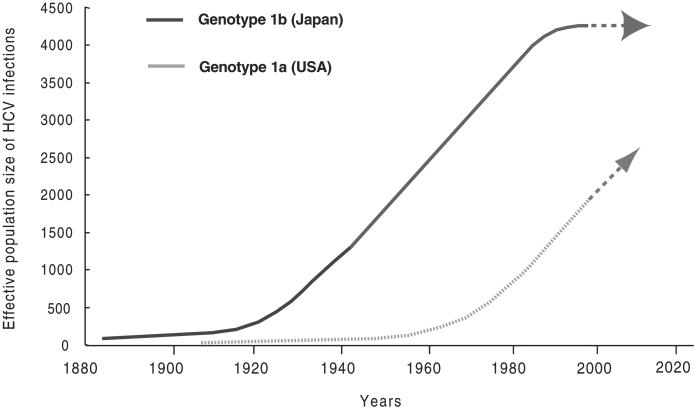
Coalescent theory examines the history of changes in virus effective population size, proportional to the diversity among a population, inferred from a phylogenetic tree reconstructed by nucleotide sequences of the virus genome in the context of an accurate molecular clock. Application of this approach suggests the growth of the U.S. HCV genotype 1a-infected population occurred around 1960, at least 30 years later than the widespread introduction of genotype 1b into the Japanese population (Fig. 4). Of note, the spread of genotype 1b in the Japanese population started to decrease around 1995, whereas HCV genotype 1a in the U.S. is still growing exponentially (Fig. 4). These data were also supported by analyses of longer sequences, such as combined sequences (1,778 nt) for genotype 1a and nearly full-genome sequences for genotype 1b. Although the effective HCV genotype 1a-infected population size of the U.S. at present is smaller than that of Japan (Fig. 4), the actual prevalence of HCV infection in the U.S. is almost the same as Japan because HCV-infected patients in the U.S. have expanded during the last 2–3 decades because of injection drug use, sharing needles, and transfusion of unscreened blood.
Fig. 4.
The effective population size of HCV in the U.S. relative to the population size in Japan over the past century. The growth (spread time) of the U.S. HCV genotype 1a population was estimated to have occurred around 1960, at least 30 years later than the spread time of genotype 1b population in Japan. The HCV genotype 1b population in Japan has already reached an equilibrium prevalence while U.S. genotype 1a is still growing exponentially. Future prediction is indicated by arrows.
Discussion
These analyses indicate that both the divergence time and the spread time for HCV in Japan occurred decades before these same events in the U.S. When the differing HCC prevalence in the U.S. and Japan simply reflects the longer duration of the epidemic in Japan, then it is predicted there will be an increase in the disease burden of HCC in the U.S. over the next few decades.
Indeed, an increase in the HCC prevalence affecting mainly younger age groups (28) has been reported to have occurred in the United States over the past 2 decades. El-Serag et al. (29) reported a 3-fold increase in the age-adjusted incidence of HCV-associated HCC (2.3 per 100,000 between 1993 and 1995 to 7.0 per 100,000 between 1996 and 1998), whereas age-adjusted rates for primary liver cancer caused by HBV and alcoholic cirrhosis remained stable. More recently, Nair et al. (30) reported a marked increase in U.S. deaths related to HCC over the past 3 decades. Such data support the hypothesis that HCC incidence is related to HCV exposure time within a population. However, contrary data from a follow-up study of young men who were infected with HCV at the time they were recruited into the U.S. Air Force showed that none of them developed HCC over an approximately 50-year interval (31).
In Japan, our data support that the spread of HCV is likely to be linked to two distinct occurrences: the widespread treatment of schistosomiasis with i.v. antimony sodium tartrate since 1923 (32), and the use of i.v. stimulants during and after World War II. Additionally, the spread of genotype 1b in the Japanese population started to decrease around 1995, while HCV genotype 1a in the U.S. is still growing exponentially (Fig. 4). These conclusions are congruent with current epidemiological reports (27, 33). Thus, this study also demonstrates that viral gene sequences constitute a potentially significant source of information for predicting the course of an epidemic. The combination of a classical epidemiological approach and a molecular evolutionary analyses could be equally useful in the study of other endemic infectious diseases.
In summary, long-term serial HCV-samples from the U.S. and Japan were molecularly clocked, and the analysis suggests that the HCV was introduced into the U.S. population around 100 years ago and widely disseminated in the 1960s. In contrast, HCV was introduced into Japan >100 years ago and widely disseminated in the 1930s and 1940s. Because Japan has a much higher HCC prevalence than does the U.S. (3–5), it is predicted that an increased HCC prevalence will occur in the U.S. over the next two to three decades. To date, no environmental or genetic predisposition factors that account for the high incidence of HCC in Japan have been identified. If there are few nonviral predeterminants of HCC incidence in Japan, or if they play a minor role, then the spread time of HCV in the U.S. relative to Japan will assume greater importance, and it is probable that the incidence of HCC in the U.S. will increase in subsequent years as predicted by the molecular clock.
Acknowledgments
We thank O. G. Pybus and A. Rambaut for helping us to use their software.
Abbreviations
HCV, hepatitis C virus
HCC, hepatocellular carcinoma
NJ, neighbor-joining
References
- 1.Alter M. J. (1995) Semin. Liver Dis. 15 5-14. [DOI] [PubMed] [Google Scholar]
- 2.Mansell C. J. & Locarnini, S. A. (1995) Semin. Liver Dis. 15 15-32. [DOI] [PubMed] [Google Scholar]
- 3.Nishioka K., Watanabe, J., Furuta, S., Tanaka, E., Iino, S., Suzuki, H., Tsuji, T., Yano, M., Kuo, G., Choo, Q. L., et al. (1991) Cancer 67 429-433. [DOI] [PubMed] [Google Scholar]
- 4.Shiratori Y., Shiina, S., Imamura, M., Kato, N., Kanai, F., Okudaira, T., Teratani, T., Tohgo, G., Toda, N., Ohashi, M., et al. (1995) Hepatology 22 1027-1033. [DOI] [PubMed] [Google Scholar]
- 5.Kiyosawa K., Sodeyama, T., Tanaka, E., Gibo, Y., Yoshizawa, K., Nakano, Y., Furuta, S., Akahane, Y., Nishioka, K., Purcell, R. H., et al. (1990) Hepatology 12 671-675. [DOI] [PubMed] [Google Scholar]
- 6.Colombo M., de Franchis, R., Del Ninno, E., Sangiovanni, A., De Fazio, C., Tommasini, M., Donato, M. F., Piva, A., Di Carlo, V. & Dioguardi, N. (1991) N. Engl. J. Med. 325 675-680. [DOI] [PubMed] [Google Scholar]
- 7.Lopez-Labrador F. X., Ampurdanes, S., Forns, X., Castells, A., Saiz, J. C., Costa, J., Bruix, J., Sanchez Tapias, J. M., Jimenez de Anta, M. T. & Rodes, J. (1997) J. Hepatol. 27 959-965. [DOI] [PubMed] [Google Scholar]
- 8.Alter M. J., Kruszon-Moran, D., Nainan, O. V., McQuillan, G. M., Gao, F., Moyer, L. A., Kaslow, R. A. & Margolis, H. S. (1999) N. Engl. J. Med. 341 556-562. [DOI] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention (1998) Morbid. Mortal. Wkly. Rep. 47 RR-19., 1–39. [Google Scholar]
- 10.Ogata N., Alter, H. J., Miller, R. H. & Purcell, R. H. (1991) Proc. Natl. Acad. Sci. USA 88 3392-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ina Y., Mizokami, M., Ohba, K. & Gojobori, T. (1994) J. Mol. Evol. 38 50-56. [DOI] [PubMed] [Google Scholar]
- 12.Abe K., Inchauspe, G. & Fujisawa, K. (1992) J. Gen. Virol. 73 2725-2729. [DOI] [PubMed] [Google Scholar]
- 13.Okamoto H., Kojima, M., Okada, S., Yoshizawa, H., Iizuka, H., Tanaka, T., Muchmore, E. E., Peterson, D. A., Ito, Y. & Mishiro, S. (1992) Virology 190 894-899. [DOI] [PubMed] [Google Scholar]
- 14.Smith D. B., McAllister, J., Casino, C. & Simmonds, P. (1997) J. Gen. Virol. 78 1511-1519. [DOI] [PubMed] [Google Scholar]
- 15.Alter H. J., Purcell, R. H., Shih, J. W., Melpolder, J. C., Houghton, M., Choo, Q. L. & Kuo, G. (1989) N. Engl. J. Med. 321 1494-1500. [DOI] [PubMed] [Google Scholar]
- 16.Conry-Cantilena C., VanRaden, M., Gibble, J., Melpolder, J., Shakil, A. O., Viladomiu, L., Cheung, L., DiBisceglie, A., Hoofnagle, J., Shih, J. W., et al. (1996) N. Engl. J. Med. 334 1691-1696. [DOI] [PubMed] [Google Scholar]
- 17.Simmonds P., Balfe, P., Peutherer, J. F., Ludlam, C. A., Bishop, J. O. & Brown, A. J. (1990) J. Virol. 64 864-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S., Tamura, K. & Nei, M. (1994) Comput. Appl. Biosci. 10 189-191. [DOI] [PubMed] [Google Scholar]
- 19.Yang Z. (1997) Comput. Appl. Biosci. 13 555-556. [DOI] [PubMed] [Google Scholar]
- 20.Pybus O. G., Rambaut, A. & Harvey, P. H. (2000) Genetics 155 1429-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pybus O. G., Charleston, M. A., Gupta, S., Rambaut, A., Holmes, E. C. & Harvey, P. H. (2001) Science 292 2323-2325. [DOI] [PubMed] [Google Scholar]
- 22.Choo Q. L., Richman, K. H., Han, J. H., Berger, K., Lee, C., Dong, C., Gallegos, C., Coit, D., Medina-Selby, R., Barr, P. J., et al. (1991) Proc. Natl. Acad. Sci. USA 88 2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holland J., Spindler, K., Horodyski, F., Grabau, E., Nichol, S. & VandePol, S. (1982) Science 215 1577-1585. [DOI] [PubMed] [Google Scholar]
- 24.Fitch W. M., Leiter, J. M., Li, X. Q. & Palese, P. (1991) Proc. Natl. Acad. Sci. USA 88 4270-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wain-Hobson S. (1993) Curr. Opin. Genet. Dev. 3 878-883. [DOI] [PubMed] [Google Scholar]
- 26.Salemi M. & Vandamme, A. M. (2002) J. Mol. Evol. 54 62-70. [DOI] [PubMed] [Google Scholar]
- 27.Armstrong G. L., Alter, M. J., McQuillan, G. M. & Margolis, H. S. (2000) Hepatology 31 777-782. [DOI] [PubMed] [Google Scholar]
- 28.El-Serag H. B. & Mason, A. C. (1999) N. Engl. J. Med. 340 745-750. [DOI] [PubMed] [Google Scholar]
- 29.El-Serag H. B. & Mason, A. C. (2000) Arch. Intern. Med. 160 3227-3230. [DOI] [PubMed] [Google Scholar]
- 30.Nair S., Shivakumar, K. S. & Thuluvath, P. J. (2002) Am. J. Gastroenterol. 97 167-171. [DOI] [PubMed] [Google Scholar]
- 31.Seeff L. B., Miller, R. N., Rabkin, C. S., Buskell-Bales, Z., Straley-Eason, K. D., Smoak, B. L., Johnson, L. D., Lee, S. R. & Kaplan, E. L. (2000) Ann. Intern. Med. 132 105-111. [DOI] [PubMed] [Google Scholar]
- 32.Iida F., Iida, R., Kamijo, H., Takaso, K., Miyazaki, Y., Funabashi, W., Tsuchiya, K. & Matsumoto, Y. (1999) Bull. W. H. O. 77 573-581. [PMC free article] [PubMed] [Google Scholar]
- 33.Moriya T., Koyama, T., Tanaka, J., Mishiro, S. & Yoshizawa, H. (1999) Intervirology 42 153-158. [DOI] [PubMed] [Google Scholar]