Abstract
We have recently reported that the mean number of CCR5 coreceptors at the surface of CD4+ T cells (CCR5 density) correlates with viral load and disease progression in HIV-1-infected persons. Here, we definitively establish that CCR5 density determines the level of virus production and identify the stages of HIV-1 replicative cycle modulated by this effect. We show, by transducing the CCR5 gene into CCR5+ cells, that CCR5 overexpression resulted in an HIV-1 overinfectability. We sorted HOS-CD4+-CCR5+ cells into two subpopulations, HOShigh and HOSlow, the former expressing seven times more cell surface CCR5 molecules than the latter. Virus production was 30–80 times higher in HOShigh cells than in HOSlow cells after a single round of infection. In contrast, only twice as many viral particles entered the cytosol of HOShigh cells as compared with the cytosol of HOSlow cells. Yet, seven times as many early, and 24 times as many late, reverse transcription products were found in HOShigh cells as compared with HOSlow cells. Moreover, a 24- to 30-fold difference in the number of copies of integrated HIV-1 DNA was observed. No difference in HIV-1 LTR activation between the two cell lines was evident. Finally, we show that the higher virus production observed in HOShigh cells is inhibited by pertussis toxin, a Gαi protein inhibitor. Thus, CCR5 density mainly modulates postentry steps of the virus life cycle, particularly the reverse transcription. These data explain why CCR5 density influences HIV-1 disease progression and underline the therapeutic interest of lowering CCR5 expression.
Keywords: coreceptor, chemokine, activation, retrovirus life cycle, AIDS
There is a large interindividual variability in viral load among HIV-1-infected persons. As HIV RNA plasma level is a major predictive indicator of bioclinical outcome (1), it is important to identify the host factors responsible for this variability. The C-C chemokine receptor CCR5 plays a key role during HIV-1 infection. It is used in addition to CD4 as a coreceptor by HIV-1 strains isolated from the blood of infected persons from the beginning of the infection at least until AIDS develops (2). The importance of CCR5 in the physiopathology of HIV-1 infection is illustrated by the effect of a 32-bp deletion in the CCR5 gene, resulting in a mutant gene, Δ32CCR5, encoding a truncated CCR5 molecule that is not expressed at the cell surface. Homozygotes for this Δ32 deletion are usually resistant to the infection, and heterozygotes progress slowly in the infection (3, 4). We have recently shown that one of the factors determining the level of viral load in HIV-1-infected persons is the density of CCR5 coreceptors at the single CD4+ T cell level (5). Thus, persons exhibiting high CCR5 expression exhibit high viral loads and persons exhibiting low CCR5 expression exhibit low viral loads. Interestingly, the correlation between CCR5 expression and viremia is logarithmic, a small difference in CCR5 density resulting in a marked difference in HIV RNA plasma levels. As a consequence, we have also established that in infected persons CCR5 cell surface density correlates with disease progression (6). The aim of the present study was to analyze the molecular mechanisms responsible for this correlation. In vitro, CCR5 cell surface density has already been involved in the infectability and/or productivity of macrophages (7–9), thymocytes (10, 11), and cell lines (12). By contrast, Vicenzi et al. (13) have shown similar R5 replication in Th1 and Th2 cell lines, although these cell lines express different CCR5 cell surface densities. The mechanisms accounting for the correlation between CCR5 cell surface density and HIV infectability, and particularly the reasons for its logarithmic nature, have not been addressed. The simplest explanation would be that CCR5 density determines viral entry. Here, we show that the facilitation of viral entry is in fact a minor effect of high CCR5 expression, whereas a major effect is exerted postentry.
Materials and Methods
Cells.
HOS-CD4+-CCR5+ cells (AIDS Reagent Program, Rockville, MD) and simian virus 40 T antigen-transformed human embryonic kidney 293T cells (Genethon) were grown in DMEM supplemented with 10% FCS and antibiotics.
Flow Cytometry.
For antibody staining, 105 cells were incubated with the anti-CCR5 mAb 2D7 (PharMingen), the anti-CD4 mAb 13B8-2 (Beckman Coulter), or an isotype control (Beckman Coulter) for 1 h on ice at final concentration of 10 μg/ml. After washing, cells were incubated with a 1:50 dilution of FITC-conjugated F(ab′)2 fragment goat anti-mouse IgG (H+L, Jackson ImmunoResearch) for 1 h on ice. Cells were then washed, fixed in paraformaldehyde, and analyzed on a FACSCalibur flow cytometer (Becton Dickinson). For quantitative determination of the mean number of CCR5 molecules at the surface of each CD4+ T cell, fluorescence intensity was converted into antibody-binding capacity by using populations of standard microbeads coated with different quantities of mAb molecules (Dako QIFIKIT) as described (5).
Vector Construction.
The BamHI–XhoI fragment of the pHR-CMVlacZ plasmid (14) containing the lacZ gene was replaced with the linker gatccgtcgacacgcgtcctaggactagtc, creating SalI and SpeI cloning sites 3′ of the cytomegalovirus promoter (plasmid pHR-BX). The NotI site of pEGFP-N1 (CLONTECH) was destroyed by a linker introducing an SpeI site (plasmid pEGFP-N1S). The WT CCR5 gene and the Δ32CCR5 gene were obtained by PCR amplification of the cDNA with the oligomers 5′-CGTCGACTCTCCCCGGGTGGAACAA-3′ and 5′-TGGATCCAAGCCCACAGATATTTCCTGC-3′ or 5′-TGGATCCCTGTATGGAAAATGAGAGCT-3′, by using Expand High Fidelity polymerase (Roche Molecular Biochemicals). The PCR products were cloned in pGEMT-easy (Promega). The plasmids were sequenced, and as only a silent mutation was detected at amino acid 163 (GGA for GGG) for CCR5, the SalI–BamHI fragments were cloned into pEGFP-N1S to generate the CCR5-EGFP (enhanced GFP) and Δ32-EGFP fusion genes. These fusion genes were inserted into the lentiviral vector pHR-BX after a SalI–SpeI digestion (plasmids VCCR5/EGFP and VΔ32/EGFP). We checked that the CCR5/EGFP fusion protein was functional as an HIV-1 coreceptor by transducing CD4+CCR5− cells with VCCR5/EGFP and infecting them with an R5 HIV-1 strain (data not shown). The SalI–SpeI fragment from pEGFP-N1S was introduced into pHR-BX to generate the VEGFP control vector.
Transduction.
Four million 293T cells were plated into 75-cm2 flasks. To produce HIV vectors, VCCR5/EGFP, VΔ32/EGFP, or VEGFP were cotransfected with pMD.G, encoding the vesicular stomatitis virus (VSV) envelope glycoprotein and the HIV packaging plasmid p8.2 (14) by the calcium phosphate method. Supernatants were collected at day 2 and concentrated 100 times by ultracentrifugation at 17,000 rpm for 2 h. HOSlow cells (8 × 104 per well) were plated in 24-well plates. They were transduced with 300 ng of p24 equivalents of VCCR5/EGFP, VΔ32/EGFP, or VEGFP virions in 8 μg/ml polybrene for 16 h. The cells were amplified and controlled for CCR5/EGFP expression.
Infection Assays.
HOS cells were plated in 24-well plates at 8 × 104 cells per well. They were infected in duplicate with 2 ng of p24 equivalent of the primary R5 strain CON overnight, washed, and cultured for 14 days. Virus production was monitored by measuring p24 gag concentration in the cell supernatant by using a commercial ELISA kit (Beckman).
One-Round Infection Assay.
To produce replication-defective HIV virions, 293T cells were cotransfected with the pNL4.3-env−-luc (AIDS Reagent Program) plasmid on one hand, and with the pCMV.AD8-Env plasmid encoding the R5 envelope of HIV-1 prototype AD8 (15) or the pMD.G plasmid (14) at a molecular ratio of 2:1 on the other hand. For the one-round infection assay, 8 × 104 HOShigh or HOSlow cells were plated per well in a 24-well plate. They were exposed to AD8-pseudotyped or VSV-pseudotyped defective virions for 18 h, washed, and cultured for 72 h. Luciferase activity was measured by luminometry at the end of the culture, by using the Luciferase Assay System (Promega). To study the effect of pertussis toxin (PTX) on HIV-1 cell cycle, the toxin was added to the cells 18 h before the infection and throughout the assay.
Viral Entry Assay.
Ten million HOS cells in a 75-cm2 flask were exposed to an HIV-1 suspension (AD8, NL4.3, or the primary HIV-1 R5 strain CON) containing 200 ng of p24 gag antigen in culture medium. After 3 h at 37°C, cells were washed three times in ice-cold Dulbecco's PBS and treated for 10 min at 4°C with 1 ml of ice-cold DMEM containing pronase (7 mg/ml, Sigma). Cells were washed once in DMEM supplemented with 10% FCS and three times in ice-cold PBS to eliminate pronase and resuspended in 2 ml of swelling buffer (10 mM Tris⋅HCl, pH 8/10 mM KCl/1 mM EDTA) for 15 min at 4°C. Cells were disrupted by Dounce homogenization (15 strokes, 7-ml B pestles), and nuclei and cell debris were pelleted by centrifugation (3,000 rpm for 3 min). The resulting postnuclear extracts were ultracentrifuged at 60,000 rpm for 10 min at 4°C in a Beckman TL100 centrifuge. The supernatant representing the cytosolic fraction was adjusted to 0.5% Triton X-100, while the pellet was resuspended in lysis buffer (20 mM Hepes/0.5% Triton X-100/150 mM NaCl) to obtain the vesicular fraction. The amount of p24 was measured in both fractions.
Real-Time PCR.
Virions were treated with 100 units/ml RNase-free DNase 1 h at 37°C with 10 mM MgCl2. HOSlow and HOShigh cells were cultured at 106 cells per 100-mm dish and infected with or without (first negative control) various quantities of virus in 3 ml of fresh medium under agitation at 37 or 4°C (second negative control). Two hours later, cells were rinsed three times with cold PBS, collected with a cell lifter, and centrifuged at 2,000 rpm to remove the cell supernatant. Cells were then resuspended in lysis buffer (10 mM Tris, pH 8/0.5 mM EDTA/0.0001% SDS/0.001% Triton/100 μg/ml Proteinase K), incubated 3 h at 50°C and 10 min at 95°C, and frozen/thawed three times. Cellular debris were removed by centrifugation, and DNA concentration was normalized by dilution in lysis buffer. DNA was amplified with the oligomers 5′-GCTCTCTGGCTAACTAGGGAAC-3′ and 5′-TGACTAAAAGGGTCTGAGGGAT-3′ (strong stop) or 5′-GCTCTCTGGCTAACTAGGGAAC-3′ and 5′-CTCTGGCTTTACTTTCGCTTTC-3′ at 65°C in a LightCycler (Roche Diagnostics) with SYBR green following the manufacturer's recommendations. Amplified products were analyzed by denaturation/renaturation to verify the specific Tm, analyzed on agarose gel and sequenced. Standard curve was established by analyzing serial dilution of a positive control plasmid. The yield was ≈1. The PCR cycle at which the amplification signal entered the exponential range was used to quantify the cellular DNA.
LTR Activation.
HOShigh cells were seeded at 5 × 105 cells/well in 6-well plates. They were transfected with 2 μg/well of (Igκ)3-conaluc plasmid or HIV-1 LTR-luc by using the calcium-phosphate method. (Igκ)3-conaluc contains three copies of the Ig κ chain enhancer κB site upstream of the minimal conalbumin promoter fused to the firefly luciferase reporter gene (16). The TK-RL plasmid (Promega), encoding the Renilla luciferase reporter gene, was cotransfected in each well to normalize the transfection efficiency. Two micrograms of pCMV-tat was cotransfected with HIV-1 LTR-luc in one experimental group as a positive control. Eighteen hours posttransfection, the culture media were changed to fresh DMEM with 10% FCS. The transfected cells were then treated with or without 100 ng/ml tumor necrosis factor α in (Igκ)3-conaluc-transfected cells or 100 ng p24 of Ada-M virus in both groups for 6 h. Luciferase activity was then analyzed immediately after the treatment by using the Dual luciferase assay kit (Promega).
Results
CCR5 Cell Surface Density Determines HIV-1 Production.
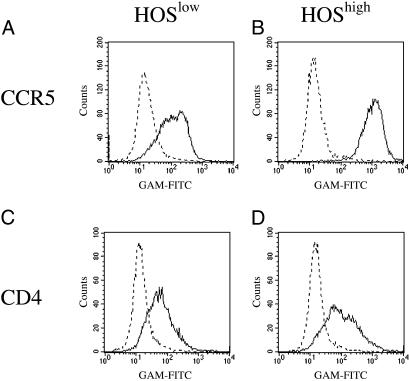
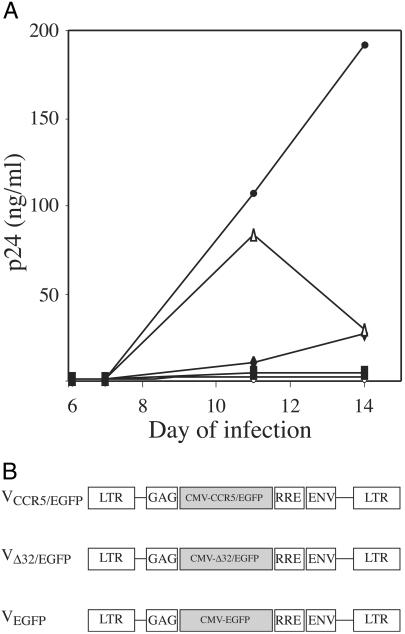
To demonstrate that CCR5 cell surface density determines viral production, we derived two cell sublines expressing different membrane CCR5 densities from a single cell line. For this purpose, we sorted HOS-CD4+-CCR5+ cells to obtain two sublines, HOShigh and HOSlow, that display a similar number of surface CD4 molecules but different numbers of CCR5 molecules. Indeed, HOShigh cells express seven times more CCR5, as illustrated in Fig. 1. When we exposed both cell lines to the R5 HIV-1 strain AD8, we observed that virus production was earlier and higher in HOShigh cells than in HOSlow cells (Fig. 2A). To definitively establish that the difference in CCR5 density between both cell lines was responsible for this difference in infectability, we prepared HIV-1 vectors harboring the WT CCR5 gene fused to the EGFP gene (VCCR5/EGFP) and negative control HIV-1 vectors harboring either the Δ32-CCR5 mutant gene, encoding a nonfunctional coreceptor, fused to EGFP (VΔ32/EGFP), or the EGFP gene alone (VEGFP) as represented in Fig. 2B, and transduced them into HOSlow cells. CCR5 density at the surface of VCCR5/EGFP-transduced HOSlow cells was similar to CCR5 density at the surface of nontransduced HOShigh cells. In contrast, transduction of HOSlow cells with VΔ32/EGFP or VEGFP did not modify their CCR5 expression (data not shown). These different cell lines were exposed to the HIV-1 strain AD8, and viral production was monitored over time. Fig. 2A shows that the increase in CCR5 expression in HOSlow cells transduced with the CCR5 gene resulted in an increase in the capacity of these cells to produce virions. On the other hand, HOSlow cells transduced with the negative control vectors VΔ32/EGFP and VEGFP displayed no increase in their viral productivity, which remained below 2.5 and 5 ng of p24 antigen per ml, respectively. Thus, the level of CCR5 expression at the surface of a target cell determines its potential for HIV-1 production.
Fig 1.
CCR5 (A and B) and CD4 (C and D) expression on HOSlow (A and C) and HOShigh (B and D) cells. Cells were exposed to an anti-CCR5 or anti-CD4 antibody (full line) or to a negative control antibody (dotted line), labeled with an FITC-conjugated anti-mouse Ig probe, and analyzed by flow cytometry.
Fig 2.
(A) Effect of CCR5 overexpression on HIV-1 R5 production. HOShigh cells (▵), HOSlow cells either nontransduced (♦) or transduced with VCCR5/EGFP (•), VΔ32/EGFP (○), or VEGFP (▪), were exposed to the HIV-1 R5 strain AD8 and cultured. Infection was monitored by measuring p24 concentration in the cell supernatant. (B) Structure of the HIV-1 vectors used for gene transfer.
CCR5 Cell Surface Density Determines the Efficiency of HIV-1 Infection at a Postentry Step.
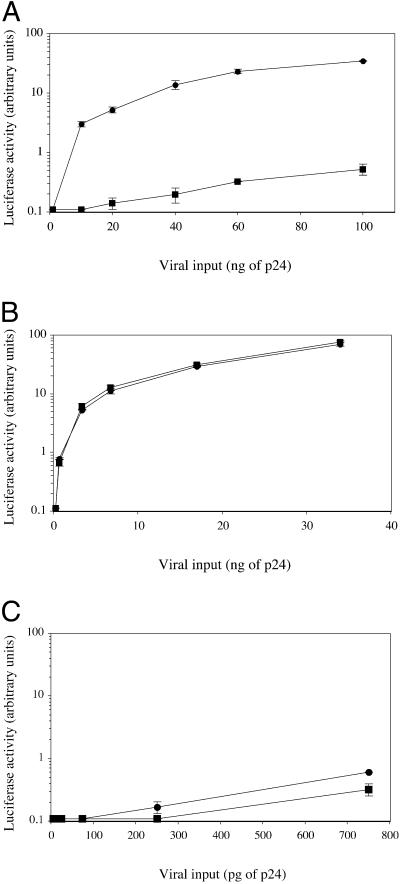
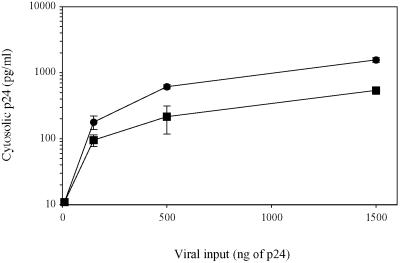
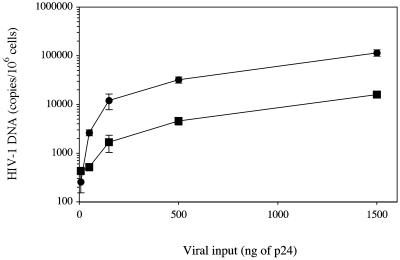
Next, we analyzed the stages of the HIV-1 life cycle that may be modulated by CCR5 density. First, we wanted to determine the effect of membrane CCR5 density on viral production after a single round of infection. For this purpose, we exposed HOShigh and HOSlow cells to defective virions obtained by cotransfecting 293T cells with a plasmid encoding the R5 envelope of AD8 and another plasmid containing the entire HIV-1 genome deleted in the envelope gene and harboring a luciferase reporter gene fused to nef. Fig. 3A shows that after a single replication cycle the amount of virus produced, as evaluated by measuring luciferase activity, is 30–80 times lower in HOSlow cells than in HOShigh cells. We also compared the efficiency of one round of infection in HOShigh and HOSlow cells with Δenv-defective viruses containing the luciferase gene as described above, pseudotyped with the G protein of the VSV. Contrary to AD8-enveloped virions, G protein-enveloped virions were as infectious in HOSlow cells as in HOShigh cells. This finding was observed either by using the same viral inocula (Fig. 3B) or 150-fold lower viral inocula (Fig. 3C) to normalize viral copy number entering the cells, because HIV entry was 150-fold more efficient with a G protein envelope as compared with an R5 envelope (data not shown). Thus, the difference in the infectability of HOSlow and HOShigh cells with R5 viruses is R5 envelope-dependent. The simplest hypothesis to account for this phenomenon is that high CCR5 density facilitates virus entry, and thereby virus production. To test this hypothesis, we measured the quantity of viral particles that productively entered HOSlow and HOShigh cells after 3 h of exposure to the virus. Maréchal et al. (17) have shown that cytosolic uptake leads to productive infection, whereas a nonspecific vesicular uptake of HIV-1 particles is a dead end. Therefore, we measured the cytosolic and endosomic uptake of p24 from the R5 HIV-1 strain AD8 in both cell lines. Table 1 shows a representative experiment where 40 and 78 pg of AD8 p24 entered the cytosol of HOSlow and HOShigh cells, respectively. In contrast, the amount of p24 found in the nonproductive vesicular pathway was roughly the same (163 and 191 pg of p24, respectively). The same result was obtained with the primary R5 strain CON with a 50% reduction in the amount of p24 found in the cytosol of HOSlow cells as compared with the cytosol of HOShigh cells. As a negative control we used the X4 strain NL4–3 that exclusively entered the vesicular pathway. Preincubation with the anti-CCR5 mAb 2D7, which maps at the gp20 binding site, inhibited cytosol uptake (data not shown). Fig. 4 shows a dose-dependent experiment with the R5 HIV-1 strain Ada-M. The amount of virus entering the cytosol of HOS cells increased linearly with viral input, and the difference in efficiency between the entry in HOShigh and HOSlow cells (twice to thrice) remained constant. Thus, the ratio between the productive entry of R5 HIV-1 strains into HOSlow cells and HOShigh cells is only 1:2 to 1:3 and cannot account for the 1:30 to 1:80 ratio between the viral production of HOSlow cells and HOShigh cells observed after a single round of replication.
Fig 3.
Efficiency of one round of HIV-1 R5 infection correlates with CCR5 density on the target cell. HOSlow (▪) and HOShigh (•) cells were exposed to env-defective HIV-1 R5 virus harboring the luciferase marker gene and pseudotyped with an R5 envelope at various quantities (A) or pseudotyped with the G protein of the VSV at the same quantities (B) or at 150-fold lower quantities to result in equal viral copy number entering the cells (C). Luciferase activity was measured in cell lysates 72 h later.
Table 1.
Correlation between HIV-1 R5 entry and CCR5 density on the target cell surface
| Virus | Cell | Endosomic p24, pg/ml | Cytosolic p24, pg/ml | Cytosolic fraction, % |
|---|---|---|---|---|
| CON (primary R5) | HOShigh | 3,750 | 1,140 | 23 |
| HOSlow | 4,895 | 555 | 10 | |
| AD8 (laboratory R5) | HOShigh | 191 | 78 | 29 |
| HOSlow | 163 | 40 | 20 | |
| NL4-3 (laboratory X4) | HOShigh | 386 | 0 | 0 |
| HOSlow | 380 | 0 | 0 |
HOSlow and HOShigh cells were exposed to R5 or X4 HIV-1 strains for 3 h. Cytosolic and endosomic fractions were then prepared, and p24 concentration was measured in each fraction. Comparable results were obtained in three independent experiments.
Fig 4.
Efficiency of HIV-1 R5 entry into HOSlow (▪) and HOShigh (•) cells. HOSlow and HOShigh cells were exposed to various amounts of R5 HIV-1 strains for 3 h. Cytosolic and endosomic fractions were then prepared, and p24 concentration was measured in each fraction.
CCR5 Cell Surface Density Influences the Efficiency of Early Postentry Stages of HIV-1 Replicative Cycle.
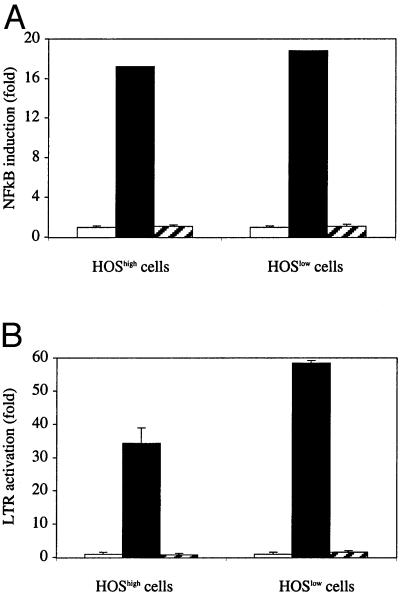
Our data suggest that CCR5 cell surface density influences postentry stages of the HIV replicative cycle. Therefore, we compared the efficiency of HIV-1 RNA reverse transcription, HIV-1 DNA integration, and LTR activation in HOSlow and HOShigh cells exposed to Δenv-defective viruses containing the luciferase gene. First, we quantified the amount of early products of reverse transcription in HOS cells after 3 h of exposure to the virus by quantitative PCR, by using LTR R/U5 primers. We detected six to eight times as many early reverse transcripts in HOShigh cells as compared with HOSlow cells at various viral inputs (Fig. 5). Thus, the difference in early reverse transcription efficiency between HOSlow and HOShigh cells is higher than the difference in the amount of virions that entered the cytosol, suggesting that cell surface CCR5 density modulated early reverse transcription efficiency. We also measured the amount of late products of reverse transcription in HOS cells 48 h after exposure to the virus, by using LTR and gag primers. We detected 21–26 times as many late reverse transcripts in HOShigh cells as compared with HOSlow cells. A representative experiment is shown in Table 2. Second, we measured with the LTR R/U5 primers the number of HIV-1 DNA copies in HOS cells 15 days after viral exposure, a stage at which all HIV-1 DNA copies are integrated (18). In independent experiments, we observed a 1:24 to 1:30 ratio between the number of HIV-1 DNA copies integrated in HOSlow cells and the number of HIV-1 DNA copies integrated in HOShigh cells. One of these experiments is represented in Table 2. In this experiment, virus production was also evaluated at day 15 by measuring luciferase activity. The ratio between virus production in HOShigh cells and virus production in HOSlow cells was 1:33. Thus, the difference in HIV-1 DNA integration (1:24) accounted almost entirely for the difference in virus production (1:33). Consequently, cell surface CCR5 density appeared to modulate HIV production mostly by interfering with early stages of the virus replicative cycle, rather than with late stages of the virus replicative cycle, including LTR activation. To confirm this observation, we looked for a potential effect of virus binding to the target cell on LTR activation. As NF-κB plays a key role in LTR activation, we first tested the hypothesis that virus-cell interaction could modulate NF-κB-mediated gene activation in the course of the virus life cycle. For this purpose, we transfected HOShigh cells with the luciferase marker gene driven by three NF-κB sites upstream of a minimal conalbumin promoter and analyzed the effect of HIV-1 R5 exposure on the marker gene expression. As a positive control, we used tumor necrosis factor α exposure. Fig. 6A shows that tumor necrosis factor α, but not virus exposure, induced NF-κB-dependent luciferase gene expression. Because additional factors are involved in LTR activation, apart from NF-κB, we repeated the experiment with the luciferase gene under the control of the HIV-1 LTR. A tat-expressing plasmid was cotransfected as a positive control. Fig. 6B shows that virus exposure also had no effect on the LTR-driven marker gene expression.
Fig 5.
Efficiency of HIV-1 R5 early reverse transcription in HOSlow (▪) and HOShigh (•) cells. HOSlow and HOShigh cells were exposed to various amounts of HIV-1 R5 for 3 h, and the quantity of early transcripts was determined by quantitative PCR.
Table 2.
Efficiency of HIV-1 R5 reverse transcription and integration in HOSlow and HOShigh cells
| Cells | Late transcripts day 2, copies/106 cells | Integration day 15, copies/106 cells | Virus production day 15, arbitrary units of luciferase activity |
|---|---|---|---|
| HOSlow | 238 ± 65 | 189 ± 133 | 3,154 ± 1,276 |
| HOShigh | 6,292 ± 1,879 | 4,464 ± 997 | 103,220 ± 24,043 |
| HOShigh/HOSlow | 26 ± 7 | 24 ± 5 | 33 ± 7 |
HOSlow and HOShigh cells were exposed to 500 ng of p24 of env-defective HIV-1 R5 virus harboring the luciferase marker gene and analyzed at different times. The number of late transcripts was determined by LTR-gag quantitative PCR at day 2. The number of integrated proviral DNA copies was determined by LTR-LTR quantitative PCR at day 15. Virus production was also quantitated at day 15 by measuring luciferase activity.
Fig 6.
Effect of exposure of HOSlow and HOShigh cells to HIV-1 R5 on LTR expression. (A) HOSlow and HOShigh cells were transfected with a luciferase gene driven by a minimal promoter containing three NF-κB boxes, and exposed or not (empty bars) to 100 ng/ml tumor necrosis factor α (filled bars), or to 100 ng of p24 of the Ada-M strain (hashed bars). (B) HOSlow and HOShigh cells were transfected with an HIV-1 LTR-driven luciferase gene and exposed or not (empty bars) to 100 ng of p24 of the Ada-M strain (hatched bars); as a positive control, cells were cotransfected with a tat-expressing plasmid (filled bars). Luciferase activity was measured, and the induction of gene expression was calculated.
CCR5 Signaling Modulates the HIV-1 Replicative Cycle.
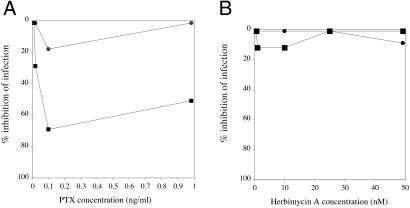
Finally, we tested the hypothesis that the facilitation of postentry steps of HIV replicative cycle at high cell surface CCR5 densities could be mediated by CCR5 signaling, secondary to the binding of the virus to its coreceptor. For this purpose, we analyzed the antiviral effects of various inhibitors of activation pathways potentially involved in CCR5 signaling, screening for compounds able to inhibit the infection of WT R5 virions AD8 but not of VSV-G pseudotyped virions in HOShigh cells. Among the various inhibitors tested (wortmannin and LY294002, phosphatidylinositol 3-kinase inhibitors; SB203580, p38-mitogen-activated protein kinase inhibitor; lovastatin, isoprenylation inhibitor; genistein, tyrosine kinase inhibitor; okadaic acid, phosphatases 1A and 1B inhibitor; HA1004 and H7, phosphokinases inhibitors; and PTX, Gαi protein inhibitor), only PTX induced a dose-dependent reduction of AD8 infection without reducing the infection by VSV-G pseudotyped viruses (Fig. 7A). In contrast, the other inhibitors tested, such as herbimycin A, a tyrosine kinase inhibitor, had no effect (Fig. 7B).
Fig 7.
PTX reduces the efficiency of a single round of R5-enveloped, but not VSV-pseudotyped, HIV-1 virions. HOShigh cells treated with PTX (A) or herbimycin A (B) were exposed to R5-enveloped (▪) or VSV-pseudotyped (•) env-defective HIV-1 R5 virions harboring the luciferase marker gene, and luciferase activity was measured in cell lysates 72 h later. Comparable results were obtained in other experiments (n = 4).
Discussion
In the present study, we provide evidence that cell surface CCR5 density determines HIV-1 production. Various authors, but not all of them (13), had correlated CCR5 expression in vitro (12) and ex vivo (7–9, 19) with HIV-1 infectability, but the causality was not demonstrated. Interestingly, in our study a moderate difference in CCR5 density (1:7) resulted in a strong difference in the amount of virus produced after a single round of replication (1:30 to 1:80). This observation is consistent with the data reported by Platt et al. (12) that below 20,000 CCR5 molecules per cell the correlation between CCR5 expression and cell infectability is logarithmic. Moreover, this result explains the logarithmic link we previously established in HIV-1-infected persons between CCR5 density on one hand and viral load (5) or disease progression (6) on the other hand.
The simplest explanation for this phenomenon would have been that CCR5 density determined cell infectability by facilitating virus entry. We show here that the difference in CCR5 density results in a small difference in virus entry that cannot account for the strong difference in virus production observed. Moreover, the fact that the differential between early reverse transcription efficiency (1:7) is higher than the differential between entry (1:2 to 1:3) suggests that CCR5 density influences the efficiency of reverse transcription. This observation is reinforced by the differential (1:21 to 1:26) between late reverse transcription efficiency observed at day 2. Yet, at day 2, reverse transcription and integration are mostly carried out (18), and this differential may reflect differences in reverse transcription as well as integration efficiency. Interestingly, the difference in the number of copies of virus DNA integrated in the genome of HOSlow and HOShigh cells almost entirely accounts for the difference in virus production observed. This finding is consistent with the fact that we observed no effect of the exposure of the cells to the virus on LTR activation. Therefore, CCR5 density does not seem to modulate HIV gene expression. Thus, CCR5-dependent virus-cell interaction seems to modulate HIV-1 entry, reverse transcription, and possibly integration, but not HIV-1 gene expression.
Our observation that CCR5 density influences a postentry step of HIV-1 replicative cycle is consistent with previous reports. In 1993, Mori et al. (20) already noticed that the replication of the SIV strain 239 in macrophages was restricted by env, although at a postentry step. Likewise, the infection of human macrophages by X4 HIV-1 strains has been shown to be restricted at a postentry level (21). Interestingly, Schmidtmayerova et al. (22) localized the block at the reverse transcription stage. Moreover, Chackerian et al. (23) reported that HIV-1 infection of macaques cells expressing human CD4 was blocked at a preintegration level and that this block was removed if the cells additionally expressed CCR5. Finally, Fauci's group (24) has shown a correlation between the capacities of R5 HIV-1 strains to signal through CCR5 and replicate in macrophages. They even showed that the productive infection of macrophages by a low signaling R5 strain can be rescued by activating the macrophages with macrophage inflammatory protein-1α. One explanation for theses observations might be that the signaling induced by gp120-CCR5 interaction (24–29) facilitates various stages of the HIV replicative cycle known to depend on the state of cell activation, especially reverse transcription and integration. Recent reports by Alfano et al. (30, 31) are in agreement with this hypothesis. Those authors showed that the B-oligomer of the PTX specifically inhibits various stages of R5 HIV-1 replication. A signal induced by this B-oligomer was proposed by those authors to be responsible for this effect, through heterogeneous desensitization of CCR5. The reduction of HIV production by PTX we observed in cells expressing high surface CCR5 densities is an argument for the involvement of Gαi activation pathway in optimal virus production.
The model we propose might explain why infection of macrophages by some R5 viruses is not productive. Furthermore, our results might explain why persons exhibiting low CCR5 densities, including persons heterozygous for the Δ32 deletion in the CCR5 gene, exhibit low viral loads and progress slowly in the disease. This study also emphasizes the potential interest of CCR5 density as a pronostic tool and strengthens the idea that lowering this density may have a therapeutic relevance.
Acknowledgments
We thank M. Segondy for providing the HIV-1 CON strain and M. Mathieu-Mahul for providing the (Igκ)3-conaluc plasmid. We are grateful to J. Estaquier for fruitful discussions and J. Corbeil, A. Gervaix, and N. Taylor for critical revision of the manuscript. HOS-CCR5 cells were obtained through the National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, from N. Landau. The study was supported by the Délégation à la Recherche Clinique du Centre Hospitalier Universitaire de Montpellier, the Agence Nationale de Recherche sur le SIDA, SIDACTION, and the Fondation pour la Recherche Médicale.
Abbreviations
EGFP, enhanced GFP
VSV, vesicular stomatitis virus
PTX, pertussis toxin
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mellors J. W., Rinaldo, C. R., Jr., Gupta, P., White, R. M., Todd, J. A. & Kingsley, L. A. (1996) Science 272 1167-1170. [DOI] [PubMed] [Google Scholar]
- 2.Connor R. I., Sheridan, K. E., Ceradini, D., Choe, S. & Landau, N. R. (1997) J. Exp. Med. 185 621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu R., Paxton, W. A., Choe, S., Ceradini, D., Martin, S. R., Horuk, R., MacDonald, M. E., Stuhlmann, H., Koup, R. A. & Landau, N. R. (1996) Cell 86 367-377. [DOI] [PubMed] [Google Scholar]
- 4.Mummidi S., Ahuja, S. S., Gonzalez, E., Anderson, S. A., Santiago, E. N., Stephan, K. T., Craig, F. E., O'Connell, P., Tryon, V., Clark, R. A., et al. (1998) Nat. Med. 4 786-793. [DOI] [PubMed] [Google Scholar]
- 5.Reynes J., Portales, P., Segondy, M., Baillat, V., André, P., Réant, B., Avinens, O., Couderc, G., Benkirane, M., Clot, J., et al. (2000) J. Infect. Dis. 181 927-932. [DOI] [PubMed] [Google Scholar]
- 6.Reynes J., Portales, P., Segondy, M., Baillat, V., André, P., Avinens, O., Picot, M.-C., Clot, J., Eliaou, J.-F. & Corbeau, P. (2001) AIDS 15 1627-1634. [DOI] [PubMed] [Google Scholar]
- 7.Fear W. R., Kesson, A. M., Naif, H., Lynch, G. W. & Cunningham, A. L. (1998) J. Virol. 72 1334-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naif H. M., Li, S., Alali, M., Sloane, A., Wu, L., Kelly, M., Lynch, G., Lloyd, A. & Cunningham, A. L. (1998) J. Virol. 72 830-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuttle D. T., Harrison, J. K., Anders, C., Sleasman, J. W. & Goodenow, M. M. (1998) J. Virol. 72 4962-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaitseva M., Lee, S., Rabin, R. L., Tiffany, H. L., Farber, J. M., Peden, K. W., Murphy, P. M. & Golding, H. (1998) J. Immunol. 161 3103-3113. [PubMed] [Google Scholar]
- 11.Pedroza-Martins L., Gurney, K. B., Torbett, B. E. & Uittenbogaart, C. H. (1998) J. Virol. 72 9441-9452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Platt E. J., Wehrly, K., Kuhman, S. E., Chesebro, B. & Kabat, D. (1998) J. Virol. 72 2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vicenzi E., Bordignon, P. P., Biwas, P., Brambilla, A., Bovolenta, C., Cota, M., Sinigaglia, F. & Poli, G. (1999) J. Virol. 73 7515-7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naldini L., Blomer, U., Gallay, P., Ory, D., Mulligan, R., Gage, F. H., Verma, I. M. & Trono, D. (1996) Science 272 263-267. [DOI] [PubMed] [Google Scholar]
- 15.Cho M. W., Shibata, R. & Martin, M. A. (1996) J. Virol. 70 7318-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrier R., Nougarede, R., Doucet, S., Kahn-Perles, B., Imbert, J. & Mathieu-Mahul, D. (1999) Oncogene 18 995-1005. [DOI] [PubMed] [Google Scholar]
- 17.Maréchal V., Clavel, F., Heard, J.-M. & Schwartz, O. (1998) J. Virol. 72 2208-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler S. L., Hansen, M. S. T. & Bushman, F. D. (2001) Nat. Med. 7 631-634. [DOI] [PubMed] [Google Scholar]
- 19.Wu L., Paxton, W. A., Kassam, N., Ruffing, N., Rottman, J. B., Sullivan, N., Choe, H., Sodroski, J., Newman, W., Koup, R. A. & Mackay, C. R. (1997) J. Exp. Med. 185 1681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mori K., Ringler, D. J. & Desrosiers, R. C. (1993) J. Virol. 67 2807-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verani A., Pesenti, E., Polo, S., Tresoldi, E., Scarlatti, G., Lusso, P., Siccardi, A. G. & Vercelli, D. (1998) J. Immunol. 161 2084-2088. [PubMed] [Google Scholar]
- 22.Schmidtmayerova H., Alfano, M., Nuovo, G. & Bukrinsky, M. (1998) J. Virol. 72 4633-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chackerian B., Long, E. M., Luciw, P. A. & Overbaugh, J. (1997) J. Virol. 71 3932-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arthos J., Rubbert, A., Rabin, R. L., Cicala, C., Machado, E., Wildt, K., Hanbach, M., Steenbeke, T. D., Swofford, R., Farber, J. M. & Fauci, A. S. (2000) J. Virol. 74 6418-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis C. B., Dikic, I., Unumatz, D., Hill, C. M., Arthos, J., Siani, A., Thompson, D. A., Schlessinger, J. & Littman, D. (1997) J. Exp. Med. 186 1793-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cicala C., Arthos, J., Ruiz, M., Vaccarezza, M., Rubbert, A., Riva, A., Wildt, K., Cohen, O. & Fauci, A. S. (1999) J. Immunol. 163 420-426. [PubMed] [Google Scholar]
- 27.Popik W., Hesselgesser, J. E. & Pitha, P. M. (1998) J. Virol. 72 6406-6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Q.-H., Williams, D. A., McManus, C., Baribaud, F., Doms, R. W., Schols, D., De Clerq, E., Kotlikoff, M. I., Collman, R. G. & Freedman, B. D. (2000) Proc. Natl. Acad. Sci. USA 97 4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weissman D., Rabin, R. L., Arthos, J., Rubbert, A., Dybul, M., Swofford, R., Venkatesan, S., Farber, J. M. & Fauci, A. S. (1997) Nature 389 981-985. [DOI] [PubMed] [Google Scholar]
- 30.Alfano M., Schmidtmayerova, M., Amella, C. A., Pushkarsky, T. & Bukrinsky, M. (1999) J. Exp. Med. 190 597-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alfano M., Pushkarsky, T., Poli, G. & Bukrinsky, M. (2000) J. Virol. 74 8767-8770. [DOI] [PMC free article] [PubMed] [Google Scholar]