Abstract
Hereditary hemochromatosis (HH) is a disorder of iron metabolism caused by common mutations in the gene HFE. The HFE protein binds to transferrin receptor-1 (TfR1) in competition with transferrin, and in vitro, reduces cellular iron by reducing iron uptake. However, in vivo, HFE is strongly expressed by liver macrophages and intestinal crypt cells, which behave as though they are relatively iron-deficient in HH. These latter observations suggest, paradoxically, that expression of wild-type HFE may lead to iron accumulation in these specialized cell types. Here we show that wild-type HFE protein raises cellular iron by inhibiting iron efflux from the monocyte/macrophage cell line THP-1, and extend these results to macrophages derived from healthy individuals and HH patients. In addition, we find that the HH-associated mutant H41D has lost the ability to inhibit iron release despite binding to TfR1 as well as wild-type HFE. Finally, we show that the ability of HFE to block iron release is not competitively inhibited by transferrin. We conclude that HFE has two mutually exclusive functions, binding to TfR1 in competition with Tf, or inhibition of iron release.
Hereditary hemochromatosis (HH) is a common inherited disease (1, 2) characterized by chronic excessive intestinal absorption of dietary iron that is subsequently deposited, with associated toxicity, in a variety of parenchymal tissues. Approximately 80% of HH patients have mutations in the gene HFE that encodes a β2-microglobulin-associated protein with structural resemblance to MHC class I proteins (3, 4). The most common disease-associated mutation of HFE protein is C260Y† (4), which disrupts a disulfide bond leading to misfolding of the heavy chain and failure to associate with β2-microglobulin (3). A second mutation or polymorphism linked with HH is H41D (3), but its mechanism of action is unknown. WT and H41D bind to transferrin-receptor 1 (TfR1) with similar nanomolar affinity constants (5). HFE has been shown to compete with transferrin (Tf) for binding to TfR1 (6), and several cell lines engineered to express HFE take up less Tf-bound iron and demonstrate reduced ferritin and enhanced TfR1 levels (7–11).
These observations led to the view that HFE inhibits Tf-bound iron uptake in vivo, with the prediction that cells expressing dysfunctional HFE should take up excess iron, as occurs in many tissues of patients with HH. However, in vivo HFE is expressed strongly by Kupffer cells and intestinal crypt cells (12, 13); and in HH these cells paradoxically behave as though they are relatively iron-deficient (14–17). This finding suggests that the normal function of HFE in these cell types may be to raise iron levels, not to reduce them (12, 18, 19), a concept that is consistent with the finding that expression of WT HFE in HH macrophages leads to iron accumulation (20).
This study confirms that exposure to WT HFE does lead to the accumulation of iron in a monocyte/macrophage cell line THP-1. The mechanism of enhanced cellular iron accumulation is shown to result from the inhibition of iron efflux. Analysis of HFE mutants and experiments with transferrin indicate that the ability of HFE to inhibit iron release is independent of binding to TfR1. We propose a scheme by which loss of the ability to inhibit iron release, caused by mutations in HFE, can lead to HH.
Materials and Methods
Proteins, Viruses, and Cells.
Soluble WT and mutant HFE proteins produced in Chinese hamster ovary cells were gifts from J. Lebrón and P. Björkman, or purified in our laboratory by using their protocol (4). Highly purified human apo-transferrin was a gift from A. Bomford. We obtained human Fe-Tf (holo-Tf) from Sigma. The monoclonal anti-HFE antibody 10G4 was a gift from Y. Yang (9); the monoclonal anti-HFE antibody 8C10 and recombinant vaccinia encoding HFE were gifts from R. Ehrlich (21). Recombinant vaccinia encoding H-2 Kb was a gift from J. Yewdell (National Institutes of Health, Bethesda). The anti H2-Kb monoclonal antibody Y3 was obtained from American Type Culture Collection; anti-CD68 was obtained from Dako; U937 and THP-1 cells were obtained from American Type Culture Collection. HeLa cells expressing HFE under the control of tetracycline were a gift from C. Enns (7). Ex vivo macrophages were grown from peripheral blood monocytes (taken from healthy volunteers in the Weatherall Institute of Molecular Medicine and consenting HH patients undergoing treatment at the John Radcliffe Hospital, with the help of C. Mackintosh, Weatherall Institute of Molecular Medicine) as described (20); HFE genotyping was kindly performed by K. Livesey, Weatherall Institute of Molecular Medicine.
Vaccinia Infection of Cells.
Stationary-phase THP-1 cells at 2 × 106 cells per ml were infected with HFE-vacc or Kb-vacc at 5 plaque-forming units per cell for 24 h, then washed and double stained for HFE or Kb, and TfR1 or ferritin expression. Infection levels were reproducibly above 40%. The anti-HFE monoclonal antibody 8C10 was used to detect HFE; the Y3 antibody was used to detect Kb. A second-layer goat anti-mouse IgG-phycoerythrin conjugate (Sigma) was used to detect first-layer binding. TfR1 expression was analyzed by using a directly conjugated anti-TfR1-FITC antibody (Dako); ferritin expression was analyzed as below.
Analysis of HFE, Ferritin, and TfR1 Expression.
HeLa–HFE cells, grown as described (7), were cultured for 2 days in media plus 10% FCS with given amounts of human cold Fe-Tf or in media plus 5% human serum (Sigma) in the presence or absence of tetracycline to control HFE expression. Log-phase U937 cells, or THP-1 cells grown to stationary phase (these growth stages were found to be optimal; data are available on request), were incubated in media plus 10% FCS at 5 × 105 cells per ml for 16 h in the presence of 333 nM (20 μg/ml) WT or mutant HFE proteins or 50 μM desferrioxamine (DFO; Sigma) or 250 μg/ml ferric ammonium citrate (FAC; Sigma) or 333 nM HFE proteins plus given amounts of Fe-Tf. Cells were then washed and stained with anti-HFE antibody 10G4 or anti-TfR1 antibody BerT-9 (Dako) or permeabilized with permeafix reagent (Ortho Diagnostics) and stained with polyclonal anti-ferritin antibodies (Dako). Mouse anti-rabbit monoclonal antibody MR12 (Dako) and rabbit anti-mouse polyclonal antibody (Dako) were used as control Ig for nonspecific staining. Cells were then washed and second-layer antibodies (goat anti-mouse IgG-FITC or goat anti-rabbit IgG-FITC, Sigma) added. After washing and fixing, cells were analyzed for HFE, TfR1, and ferritin expression levels by using a Becton Dickinson FACSCalibur and cellquest software. Results by using this method of ferritin detection were confirmed by Western blotting (data available on request).
59Fe-Tf Uptake Studies.
Apo-transferrin was loaded with 59Fe (NEN) to 66% saturation as described (22). Log-phase U937 or THP-1 cells at 106 cells per ml were exposed to given amounts of 59Fe-Tf for the times indicated, with soluble HFE proteins or cold Fe-Tf also present. Experiments were performed in triplicate. At the end of the uptake time, cells were washed four times at 4°C in PBS/0.1%NaN3 and incorporated cellular radioactivity was measured by using a Packard Cobra γ-counter.
59Fe Release Studies.
59Fe-Tf [53 nM (4 μg/ml)] was added to growing THP-1 or U937 cells for 2 days. At the end of this time, cells were washed four times in Hanks' buffered saline solution and resuspended at 106 cells per ml in serum-free RPMI medium 1640 with the addition of HFE proteins and cold human Tf as shown. At the time points indicated cells were spun at 4°C and, taking care not to disturb the cell pellet, half of the cell supernatant was removed and assayed for released radioactivity. For ex vivo macrophages, 1 μM 59Fe-Tf was added to each well of growing cells on day 4 of culture. Release assays from adherent CD68 positive cells on day 7 of culture were performed as for THP-1 cells; release after 30 min was measured.
Results
Effect of HFE on Iron Content of THP-1 Cells Compared with U937 and HeLa Cells.
To investigate how HFE might increase cellular iron, we measured the effect of HFE on two monocyte cell lines, THP-1 (23) and U937, of which THP-1 is the more macrophage-like. THP-1, but not U937 cells react weakly with the antibody to HFE 10G4 (Fig. 1 A and B); THP-1 reacts strongly and U937 weakly with our anti-HFE antibody JB-1. We found that THP-1 cells infected with recombinant vaccinia encoding full-length HFE (Fig. 1 C and D), and THP-1 cells exposed to soluble truncated HFE/β2-microglobulin heterodimer (Fig. 1 G and H), increase intracellular ferritin (Fig. 1 C and G) and decrease TfR1 levels (Fig. 1 D and H) relative to control cells. THP-1 cells infected with a control vaccinia encoding the mouse MHC class I protein H-2 Kb do not alter their ferritin or TfR1 levels (Fig. 1 E and F). This response of THP-1 cells is the opposite to that of the immature monocytic cell line U937 exposed to soluble HFE (Fig. 1 I and J) or of HeLa cells engineered to express full-length HFE (7, 8) (Fig. 1 K and L), which both reduce ferritin and increase TfR1 levels. Control conditions show that U937 and THP-1 respond as expected to iron loading (with 250 μg/ml FAC) by increasing ferritin and decreasing TfR1, and to iron deprivation (chelation with 50 μM DFO) by decreasing ferritin and increasing TfR1. These results indicate that THP-1 cells, like macrophages, accumulate iron in response to HFE, whereas HeLa and U937 reduce their iron content.
Fig 1.
HFE increases ferritin and reduces TfR1 expression by the reticuloendothelial cell line THP-1. (A and B) Expression of HFE by THP-1 and U937 cells. (A) THP-1 endogenously express low levels of HFE (green, 10G4 antibody). (B) HFE is not detected on U937 cells with this antibody. TfR1 expression at log phase is shown in red. (C–H) Effect of HFE on ferritin and TfR1 expression in THP-1 cells. Full-length HFE was expressed by infection with recombinant vaccinia virus (C and D), or cells were exposed for 16 h to soluble HFE at 300 nM (20 μg/ml) (G and H). Both forms of HFE increased ferritin expression (C and G, green histograms) and decreased TfR1 (D and H, green histograms) compared with untreated THP-1 cells (filled gray histograms). Controls: expression of mouse H2-Kb by vaccinia infection (E and F, green line) does not alter ferritin or TfR1 levels in THP-1 cells. (I and J) In contrast, a 16-h incubation of U937 cells with 300 nM soluble HFE (or infection of U937 cells with HFE-vacc, not shown) reduces ferritin expression (I, green) and raises TfR1 (J, green) compared with untreated U937 cells (filled gray). (G–J) In both THP-1 and U937, iron loading with 250 μg/ml FAC (blue) raises ferritin and reduces TfR1, whereas chelation of iron with 50 μM DFO (red) reduces ferritin and raises TfR1. (K Inset) HeLa-HFE cells grown in the absence of tetracycline express full-length HFE (red), whereas cells grown with tetracycline are HFE negative (blue). (K and L) HeLa-HFE cells positive for full-length HFE (green histograms) express less ferritin (K) and more TfR1 (L) than HFE-negative HeLa-HFE cells (filled gray). Filled purple histograms represent staining with isotype control antibodies.
HFE Inhibits Iron Release from THP-1 Cells.
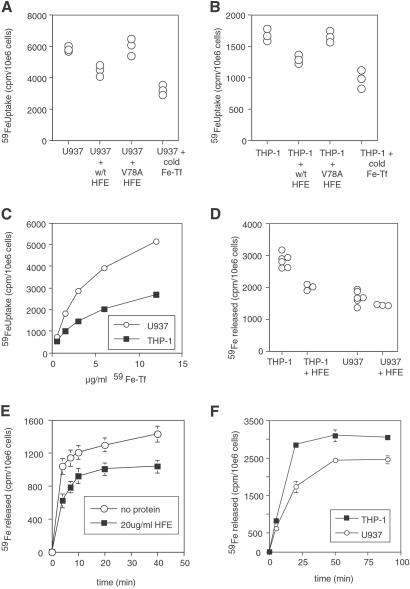
HFE could raise iron in THP-1 cells by either increasing uptake or inhibiting release of iron. Fig. 2 A and B shows that for cells in log-phase growth, a 30-fold molar excess of sHFE reduces Tf-iron uptake into both U937 and THP-1 cells, although the slower growing THP-1 cells take up much less Tf-iron overall (and express less TfR1) than U937 cells (Fig. 2C). The effect is specific because the mutant V78A HFE, which no longer binds TfR1 (5), is unable to inhibit iron uptake. Fig. 2 D and E show that sHFE inhibits release of iron from stationary phase THP-1 cells but not significantly from U937 cells; again the effect is lost with the V78A mutant of HFE (see Fig. 3B, discussed below). Fig. 2F shows that THP-1 cells release more iron per cell than U937 cells. These data show that HFE is able to block both iron uptake and iron release. As HFE increases ferritin, and decreases TfR1, on THP-1 cells it suggests that the dominant function of HFE on these cells is to block iron release. However, on the faster growing U937, HeLa, and other cell types investigated (HL-60, K562, KG-1, data not shown) in vitro, the dominant effect of HFE is to lower Tf-iron uptake, reducing cellular iron and ferritin.
Fig 2.
Effect of HFE on iron uptake and release. (A and B) WT HFE blocks iron uptake into THP-1 and U937 cells. Log-phase U937 (A) and THP-1 (B) cells were grown with 27 nM 59Fe-Tf for 6 h in the presence of 270 nM cold human holo-Tf, 300 nM soluble HFE, or 300 nM soluble V78A HFE as indicated, and then cellular 59Fe was measured. Each point represents the uptake into one aliquot of 106 cells. (C) Comparison of iron uptake by U937 and THP-1 cells. Log phase U937 and THP-1 cells at 106 cells per ml were incubated with given doses (10 μg/ml = 133 nM) of 59Fe-Tf for 4 h, and then cellular 59Fe was measured. (D) Effect of HFE on iron release by U937 and THP-1 cells. Cells were grown in 53 nM 59Fe-Tf for 2 days, then washed and allowed to export iron into media for 45 min in the presence of 300 nM HFE protein as shown. Each point represents the release of one aliquot of 106 cells. (E) Time course of iron release from THP-1 cells in the presence or absence of HFE. THP-1 cells were loaded with iron, as in C, and allowed to export iron for the times indicated with or without addition of soluble HFE (300 nM) to the medium. Error bars indicate the range of values in the triplicate samples for each condition. (F) Time course of iron release from U937 and THP-1 cells. Cells were grown in 53 nM 59Fe-Tf for 2 days, then washed and allowed to export iron into media for times shown. Error bars indicate the range of values in the triplicate samples for each condition.
Fig 3.
Effects of mutations in HFE on iron handling by U937 cells, THP-1 cells, and ex vivo macrophages. (A) Log-phase U937 (left two columns) and stationary phase THP-1 (right two columns) cells were exposed to 300 nM WT HFE, S43C HFE, V78A HFE, H41D HFE, R44E HFE, or W141A HFE soluble proteins for 16 h, and TfR1 (first and third columns) and ferritin expression (second and fourth columns) were measured (green line histograms) relative to untreated cells (gray filled histogram). Controls: Cells exposed to 50 μM DFO (red) or 250 μg/ml FAC (blue) are also shown. Staining with isotype control antibodies are shown as filled purple histograms. (B and C) Effect of mutations in HFE on iron release by THP-1 cells; cells were loaded with iron as in Fig. 2D and allowed to export iron in the presence of WT or mutant HFE proteins at 300 nM for the times shown (B) or for 30 min (C). Error bars in B indicate the range of values in the triplicate samples for each condition; each point in C represents 59Fe released by one aliquot of 106 cells. (D) Effect of HFE on iron release by ex vivo macrophages (homozygous WT HFE). (E) Effect of HFE on iron release by ex vivo macrophages from a treated HH patient (homozygous C260Y). (F) Summary of percent inhibition of iron release by WT HFE from macrophages from controls (open bars) and treated HH patients (black bars). The HFE genotype of each individual is shown. Error bars represent the range of inhibition when multiple experiments were performed on cells from the same individual (each experiment was performed on triplicate wells). (Inset) Adherent macrophages stained with anti-CD68 antibody (red histogram) or isotype control (purple). (G) Location of amino acids on HFE (α1 and α2 domains depicted by green ribbons) required for inhibition of iron release (red spheres). The HFE-contacting regions of TfR1 are shown by white strands. The figure was derived from the crystal structure of HFE:TfR1 (24).
Effects of Various HFE Mutants on THP-1 Cells, U937 Cells, and ex Vivo Macrophages.
To investigate further we examined the effect of various HFE mutants (whose binding affinities for TfR1 have been determined; ref. 5) on the U937 and THP-1 cell lines, and ex vivo macrophages. Fig. 3A shows that WT HFE reduces ferritin and increases TfR1 on U937 cells, but increases ferritin and reduces TfR1 on THP-1 cells, and that the S43C mutant which binds TfR1 as WT (5) behaves as WT HFE. The V78A mutant of HFE, that does not bind TfR1 (5), alters neither ferritin nor TfR1 on either U937 or THP-1 cells. Three HFE mutants, H41D (which is associated with HH; ref. 3), R44E and W141A, which all bind TfR1 with WT affinity (5), retain the ability to make U937 cells iron-deficient, but lose the ability to increase ferritin and reduce TfR1 on THP-1 cells. Fig. 3 B and C show that H41D and V78A have lost the ability to block iron release, and W141A and R44E have reduced ability to block iron release from THP-1 cells compared with WT HFE.
We then extended these experiments to CD68-positive macrophages (Fig. 3 D–F) derived ex vivo from peripheral blood monocytes. Fig. 3D shows that WT HFE inhibits iron release from macrophages from a healthy donor, V78A has no inhibitory effect, and R44E has a reduced effect, consistent with the results on THP-1. Fig. 3E shows that WT but not V78A HFE inhibits iron release from a C260Y homozygous individual. Fig. 3F summarizes experiments using macrophages derived from healthy volunteers (open bars) or HH patients (filled bars) whose HFE genotype is shown. The percentage inhibition of iron release by WT HFE is shown. We did not find a significant difference in the effect of HFE on the macrophages from healthy volunteers versus HH patients.
The results with the HFE mutants show that V78A abolishes both binding to TfR1 and inhibition of iron release, whereas H41D, R44E, and W141A have no effect on binding to TfR1, but abolish or reduce the inhibitory effect on iron release. The S43C mutant behaves as WT. The positions of mutated amino acids that influence iron release are displayed in Fig. 3G. The amino acid residues W141, H41, and R44 (Fig. 3G, red spheres) do not contact TfR1 (white strands) (24).
Effect of Transferrin on the Ability of HFE to Inhibit Iron Uptake and Iron Release.
These results left open the question of whether binding of HFE to TfR1 was required for the observed effect on iron release. To address this question, we looked at the effect of increasing levels of iron-loaded transferrin on iron uptake and release. We reasoned that if the effect of HFE on iron release required HFE to bind TfR1, competition for binding by high levels of Fe-Tf should abolish the effect. Fig. 4A shows that when human Tf is titrated into cultures of HeLa cells expressing HFE, the ability of HFE to reduce ferritin is abolished at levels of Tf <100 μg/ml (1.3 μM; well below levels in human serum). Fig. 4B shows that, similarly, sHFE does not reduce ferritin in U937 cells cultured in media supplemented with 1.3 μM human Tf. Fig. 4 C and D show that sHFE can reduce 59Fe-Tf uptake into U937 cells when HFE is at a 30× molar excess over Tf, but that when Tf is present at 2 μM, 400 nM HFE does not affect iron uptake. These results are consistent with HFE and Fe-Tf competing for binding to TfR1 (6).
Fig 4.
Human Fe-Tf inhibits the effect of HFE on iron uptake but not release. (A) Human Fe-Tf was titrated into cultures of HFE-positive (open squares) or HFE-negative (filled circles) HeLa-HFE cells. With no added Tf, HFE-negative cells contain more ferritin than HFE-positive cells, but as human Fe-Tf increases toward 100 μg/ml (1.33 μM), the difference between the ferritin expression in HFE-positive and -negative cells disappears. Points plotted are mean fluorescence of 25,000 cells. (B) U937 cells grown in media containing 670 nM (50 μg/ml) human Fe-Tf express the same level of ferritin in the presence (green line histogram) or absence (gray filled histogram) of added 300 nM WT HFE. (C and D) The effect of HFE on iron uptake is dose-dependent: 400 nM HFE reduces the uptake of iron by U937 cells exposed to 26.6 nM 59Fe-Tf (C) but does not have a detectable effect on cells exposed to 2 μM 59Fe-Tf (D). (E–G) By contrast Fe-Tf does not prevent the effect of HFE on THP-1 cells. HFE expressed by HFE-vacc infection still reduces TfR1 (green histogram, E) and soluble HFE raises ferritin (green line, G) in THP-1 cells grown in media containing 4 μM (300 μg/ml) Fe-Tf. (H) Unlabeled Fe-Tf (133 nM) slightly enhanced the inhibitory effect of HFE (at given doses) on iron release over 20 min from THP-1 cells. Controls: Expression of H2-Kb by vacc-Kb infection did not alter TfR1 (F) or ferritin expression (not shown) in THP-1 cells cultured in media containing 4 μM Fe-Tf. U937 cells (B) and THP-1 cells (G) responded to DFO (red) and FAC (blue) as in Fig. 1.
On the other hand, Fig. 4 E and G show that full-length HFE and sHFE reduce TfR1 and increase ferritin in THP-1 cells even when cells are cultured with 4 μM human Tf. Fig. 4H shows a dose response of sHFE blocking iron release from THP-1 cells; when sHFE is coincubated with human Tf, the effect of sHFE on blocking iron release is, if anything, slightly enhanced. These results show that the effect of HFE on iron release is not inhibited by Fe-Tf. Together with the results in Fig. 3, these data suggest that binding of HFE to TfR1 is not required for the effect of HFE on iron release.
Discussion
The major features of HH are increased dietary iron absorption and poor iron storage in reticuloendothelial cells (2). HFE protein is strongly expressed by intestinal crypt cells and by Kupffer macrophages in the liver (12, 13). Crypt cells are thought to sense body iron requirements, at least in part by transferrin-mediated iron uptake (25–27), and program iron absorption by mature enterocytes accordingly. Because iron absorption is increased in HH (28), and because some groups have found that HH enterocytes show features associated with iron deficiency, such as reduced ferritin (29) and enhanced levels of DMT-1 (17) and ferroportin1/IREG1/MTP1 (30), it seems that the crypt cells in HH behave as though they are iron-deficient, even though total body iron is increased. Kupffer macrophage cells have also been shown to be relatively iron-deficient compared with surrounding parenchymal hepatocytes in HH livers (14). Monocytes from HH patients have been found to have a higher iron response element-binding activity than controls, indicating that these cells are likewise relatively iron-deficient (16).
Thus in vivo, the cells that have been shown to express HFE protein behave as though they are iron-deficient in HH. These observations lead to the suggestion that the function of WT HFE in vivo, in these specialized cells, would be to raise iron. Consistent with this interpretation, Montosi et al. (20) found that expression of WT HFE in macrophages (achieved using recombinant Salmonella as a vector) increased iron accumulation by means of an unknown mechanism.
HFE could raise iron in cells of the macrophage (and crypt cell) lineage by enhancing iron uptake or by the inhibition of iron efflux. Moura et al. (31) found that monocytes from HH patients released more low molecular weight iron in vitro than control monocytes. Here we show that WT HFE inhibits the release of iron from ex vivo macrophages from HH patients and healthy individuals and monocytic THP-1 cells, but it does not enhance iron uptake.
Our analysis of HFE mutants shows that the ability to block iron release is destroyed by the substitution H41D, which is associated with iron overload in man, and for which no other functional effects are known. H41D HFE and two other mutants, R44E and W141A, which do not inhibit iron release to the same degree as WT HFE, bind to TfR1 as well as WT HFE (5). The residues H41, R44, and W141 do not contact TfR1 in the cocrystal of HFE:TfR1 (24). A fourth mutation, V78A, inhibits both TfR1 binding (5) and the ability to block iron release. A fifth mutation, S43C, which is found in a few HH patients (32) and binds TfR1 as well as WT HFE (5), also inhibits iron release as well as WT HFE. The three HFE mutants H41D, R44E, and W141A show that the ability to inhibit iron release does not precisely correlate with the ability to bind TfR1. Moreover, we show that, whereas the ability of HFE to inhibit iron uptake is blocked by increasing levels of iron loaded transferrin, transferrin does not block the ability of HFE to inhibit iron release.
These data indicate that HFE inhibits iron release independently of binding to TfR1, presumably by binding another ligand which is expressed by cells of the macrophage lineage, but not by other cell types such as HeLa and Chinese hamster ovary cells that have been used previously to investigate HFE function. We hypothesize that this second ligand is also expressed by crypt cells, and that HFE has the same function in macrophages and crypt cells. The putative second target for HFE is not known, but the ferroportin1 iron exporter is a possible candidate (19, 30, 33, 34). Like HFE, Ferroportin1 is strongly expressed by Kupffer cells in the liver (33, 34), and although most strongly expressed by mature enterocytes (which do not express HFE), ferroportin1 may also be present at lower levels in HFE expressing crypt cells from which mature enterocytes are derived (30). Recently, mutations in ferroportin1 have been found in HH patients expressing WT HFE (35, 36). It has been proposed that these mutations could have gain-of-function (35) or loss-of-function effects (36). Alternatively, if HFE does inhibit ferroportin1, the mutations could influence this interaction. Clearly, further experiments are required to elucidate how HFE inhibits iron release.
In contrast to our finding that HFE increases cellular iron by inhibiting iron release, Waheed et al. (37) showed that transferrin-bound iron uptake was increased by HFE expressed in hamster cells also expressing human TfR1. The authors hypothesized that these results with Chinese hamster ovary cells were representative of the function of HFE in crypt cells. They suggested that when HFE is dysfunctional, the enhancement of iron uptake is lost, so that the crypt cells are relatively iron-deficient, leading to the increased dietary iron absorption observed in HH. This idea and our scheme for HFE function are both consistent with the finding that the duodenum of Hfe knockout mice demonstrated reduced iron accumulation from serum (which could be due to reduced uptake or increased release of iron) (38). However, the enhanced uptake model for HFE function does not explain the iron status of mice homozygous for WT HFE but expressing only one copy of TfR1 (39). According to the hypothesis proposed by Waheed et al. (37), with only one copy of the TfR1 gene, iron uptake into crypt cells should be reduced, with the result that intestinal iron absorption by these mice should be increased. In fact Levy et al. (39) showed that TfR1+/− mice had significantly reduced body iron.
We have proposed that the inhibition of iron release by WT HFE expressed by Kupffer and duodenal crypt cells plays an important role in iron homeostasis, and that this function of HFE is lost in HH because of C260Y and H41D mutations (19). We suggest that the partitioning of HFE between inhibition of an iron exporter, or binding to TfR1, is a key homeostatic mechanism that will be influenced by the level of transferrin saturation in serum (19). In conditions of low transferrin saturation, HFE will be predominantly bound by TfR1, leading to release of iron from reticuloendothelial stores and up-regulation of iron absorption as a result of iron-deficient crypt cells. When transferrin is heavily saturated HFE would be prevented from binding to TfR1 and available to inhibit iron efflux, leading to enhanced iron storage and down-regulation of iron absorption. Loss of this regulatory feedback mechanism in HH leads to a reduced capacity for iron storage by reticuloendothelial cells and inappropriately low crypt iron for any degree of serum transferrin saturation. The crypt cells would thus program overabsorption of dietary iron by mature enterocytes. Moreover, with regard to the TfR1+/− mouse discussed above, our model argues that reduced TfR1 expression would free up HFE to inhibit iron release from crypt cells, with the result that iron-loaded crypt cells cause reduced iron absorption, in agreement with the findings of Levy et al. (39). The data shown here support our scheme for HFE function.
Acknowledgments
We thank Pamela Björkman, Jeremy Brock, Caroline Enns, Kathryn Robson, and David Weatherall for support, discussion, and advice, and the Wellcome Trust, the Howard Hughes Medical Institute, and the Medical Research Council for funding.
Abbreviations
HH, hereditary hemochromatosis
DFO, desferrioxamine
FAC, ferric ammonium citrate
TfR1, transferrin receptor-1
Numbering begins from the first amino acid of the mature protein, omitting the 22 amino acids of the signal sequence, so that C260 = C282 (and H41 = H63, etc) of the immature protein.
References
- 1.Sheldon J. H., (1935) Haemochromatosis (Oxford Univ. Press, Oxford).
- 2.Andrews N. C. (1999) N. Engl. J. Med. 341 1986-1995. [DOI] [PubMed] [Google Scholar]
- 3.Feder J. N., Gnirke, A., Thomas, W., Tsuchihashi, Z., Ruddy, D. A., Basava, A., Dormishian, F., Domingo, R., Jr., Ellis, M. C., Fullan, A., et al. (1996) Nat. Genet. 13 399-408. [DOI] [PubMed] [Google Scholar]
- 4.Lebrón J. A., Bennett, M. J., Vaughn, D. E., Chirino, A. J., Snow, P. M., Mintier, G. A., Feder, J. N. & Björkman, P. J. (1998) Cell 93 111-123. [DOI] [PubMed] [Google Scholar]
- 5.Lebron J. A. & Björkman, P. J. (1999) J. Mol. Biol. 289 1109-1118. [DOI] [PubMed] [Google Scholar]
- 6.Lebron J. A., West, A. J. & Björkman, P. J. (1999) J. Mol. Biol. 294 239-245. [DOI] [PubMed] [Google Scholar]
- 7.Gross C. N., Irrinki, A., Feder, J. N. & Enns, C. A. (1998) J. Biol. Chem. 273 22068-22074. [DOI] [PubMed] [Google Scholar]
- 8.Roy C. N., Penny, D. M., Feder, J. N. & Enns, C. A. (1999) J. Biol. Chem. 274 9022-9028. [DOI] [PubMed] [Google Scholar]
- 9.Salter C. L., Brunmark, A., Li, Y., Leturcq, D., Peterson, P. A., Jackson, M. R. & Yang, Y. (1999) Proc. Natl. Acad. Sci. USA 96 5434-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riedel H. D., Muckenthaler, M. U., Gehrke, S. G., Mohr, I., Brennan, K., Herrmann, T., Fitscher, B. A., Hentze, M. W. & Stremmel, W. (1999) Blood 94 3915-3921. [PubMed] [Google Scholar]
- 11.Corsi B., Levi, S., Cozzi, A., Corti, A., Altimare, D., Albertini, A. & Arosio, P. (1999) FEBS Lett. 460 149-152. [DOI] [PubMed] [Google Scholar]
- 12.Bastin J. M., Jones, M., O'Callaghan, C. A., Schimanski, L., Mason, D. Y. & Townsend, A. R. (1998) Br. J. Haematol. 103 931-941. [DOI] [PubMed] [Google Scholar]
- 13.Parkkila S., Waheed, A., Britton, R. S., Feder, J. N., Tsuchihashi, Z., Schatzman, R. C., Bacon, B. R. & Sly, W. S. (1997) Proc. Natl. Acad. Sci. USA 94 2534-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Block M. B., Moore, G., Wasi, P. & Haiby, G. (1965) Am. J. Pathol. 47 89-124. [PMC free article] [PubMed] [Google Scholar]
- 15.Brink B., Disler, P., Lynch, S., Jacobs, P., Charlton, R. & Bothwell, T. (1976) J. Lab. Clin. Med. 88 725-731. [PubMed] [Google Scholar]
- 16.Cairo G., Recalcati, S., Montosi, G., Castrusini, E., Conte, D. & Pietrangelo, A. (1997) Blood 89 2546-2553. [PubMed] [Google Scholar]
- 17.Zoller H., Pietrangelo, A., Vogel, W. & Weiss, G. (1999) Lancet 353 2120-2123. [DOI] [PubMed] [Google Scholar]
- 18.Drakesmith H. & Townsend, A. (2000) BioEssays 22 595-598. [DOI] [PubMed] [Google Scholar]
- 19.Townsend A. & Drakesmith, H. (2002) Lancet 359 786-790. [DOI] [PubMed] [Google Scholar]
- 20.Montosi G., Paglia, P., Garuti, C., Guzman, C. A., Bastin, J. M., Colombo, M. P. & Pietrangelo, A. (2000) Blood 96 1125-1129. [PubMed] [Google Scholar]
- 21.Ben-Arieh S. V., Zimerman, B., Smorodinsky, N. I., Yaacubovicz, M., Schechter, C., Bacik, I., Gibbs, J., Bennink, J. R., Yewdell, J. W., Coligan, J. E., et al. (2001) J. Virol. 75 10557-10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oria R., Alvarez-Hernandez, X., Liceaga, J. & Brock, J. H. (1988) Biochem. J. 252 221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuchiya S., Yamabe, M., Yamaguchi, Y., Kobayashi, Y., Konno, T. & Tada, K. (1980) Int. J. Cancer 26 171-176. [DOI] [PubMed] [Google Scholar]
- 24.Bennett M. J., Lebron, J. A. & Björkman, P. J. (2000) Nature 403 46-53. [DOI] [PubMed] [Google Scholar]
- 25.Taylor M. R. & Gatenby, P. B. (1966) Br. J. Haematol. 12 747-753. [DOI] [PubMed] [Google Scholar]
- 26.Cox T. M. & Peters, T. J. (1980) Br. J. Haematol. 44 75-86. [DOI] [PubMed] [Google Scholar]
- 27.Raja K. B., Pountney, D. J., Simpson, R. J. & Peters, T. J. (1999) Blood 94 3185-3192. [PubMed] [Google Scholar]
- 28.Cox T. M. & Peters, T. J. (1978) Lancet 1 123-124. [DOI] [PubMed] [Google Scholar]
- 29.Pietrangelo A., Rocchi, E., Casalgrandi, G., Rigo, G., Ferrari, A., Perini, M., Ventura, E. & Cairo, G. (1992) Gastroenterology 102 802-809. [DOI] [PubMed] [Google Scholar]
- 30.McKie A. T., Marciani, P., Rolfs, A., Brennan, K., Wehr, K., Barrow, D., Miret, S., Bomford, A., Peters, T. J., Farzaneh, F., et al. (2000) Mol. Cell 5 299-309. [DOI] [PubMed] [Google Scholar]
- 31.Moura E., Noordermeer, M. A., Verhoeven, N., Verheul, A. F. & Marx, J. J. (1998) Blood 92 2511-2519. [PubMed] [Google Scholar]
- 32.Mura C., Raguenes, O. & Ferec, C. (1999) Blood 93 2502-2505. [PubMed] [Google Scholar]
- 33.Donovan A., Brownlie, A., Zhou, Y., Shepard, J., Pratt, S. J., Moynihan, J., Paw, B. H., Drejer, A., Barut, B., Zapata, A., et al. (2000) Nature 403 776-781. [DOI] [PubMed] [Google Scholar]
- 34.Abboud S. & Haile, D. J. (2000) J. Biol. Chem. 275 19906-19912. [DOI] [PubMed] [Google Scholar]
- 35.Njajou O. T., Vaessen, N., Joosse, M., Berghuis, B., van Dongen, J. W., Breuning, M. H., Snijders, P. J., Rutten, W. P., Sandkuijl, L. A., Oostra, B. A., et al. (2001) Nat. Genet. 28 213-214. [DOI] [PubMed] [Google Scholar]
- 36.Montosi G., Donovan, A., Totaro, A., Garuti, C., Pignatti, E., Cassanelli, S., Trenor, C. C., Gasparini, P., Andrews, N. C. & Pietrangelo, A. (2001) J. Clin. Invest. 108 619-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waheed A., Grubb, J. H., Zhou, X. Y., Tomatsu, S., Fleming, R. E., Costaldi, M. E., Britton, R. S., Bacon, B. R. & Sly, W. S. (2002) Proc. Natl. Acad. Sci. USA 99 3117-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trinder D., Olynyk, J. K., Sly, W. S. & Morgan, E. H. (2002) Proc. Natl. Acad. Sci. USA 99 5622-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy J. E., Jin, O., Fujiwara, Y., Kuo, F. & Andrews, N. C. (1999) Nat. Genet. 21 396-399. [DOI] [PubMed] [Google Scholar]