Abstract
In this study we have developed bioluminescence-imaging strategies to noninvasively and quantitatively image protein—protein interactions in living mice by using a cooled charge-coupled device camera and split reporter technology. We validate both complementation and intein-mediated reconstitution of split firefly luciferase proteins driven by the interaction of two strongly interacting proteins, MyoD and Id. We use transient transfection of cells and image MyoD–Id interaction after induction of gene expression in cell culture and in cells implanted into living mice. Techniques to study protein–protein interactions in living subjects will allow the study of cellular networks, including signal transduction pathways, as well as development and optimization of pharmaceuticals for modulating protein–protein interactions.
Protein–protein interactions are important determining factors in the control of many cellular processes. To understand these interactions, several techniques have been developed and studied by using intact cells and cell extracts. The yeast two-hybrid system is extremely useful in screening for protein–protein interactions (1–3). The limitation of this system is that it requires the protein interaction to occur in the nucleus (4). To circumvent this limitation, other techniques have been developed, including the split ubiquitin system (5, 6), Sos recruitment system (7–9), dihydrofolate reductase complementation (10, 11), β-galactosidase complementation (4), β-lactamase complementation (12), and the G protein fusion system (13, 14).
A split reporter protein approach can be used for studying protein–protein interactions through either complementation or reconstitution strategies (Fig. 1). Complementation strategies do not require the formation of a mature protein from split proteins (15). Intracistronic complementation of β-galactosidase by using interacting proteins has been used to measure the rate of interaction between two proteins (4). Reconstitution strategies attempt to reconstitute the mature reporter protein. Protein splicing is a posttranslational process that releases matured protein after proper ligation without altering protein activity (16). Inteins are protein domains that perform a cis-splicing reaction to excise themselves posttranslationally from nascent polypeptide chains, thus forming new peptide bonds between the exteins (17). Inteins also can be split into two parts and expressed as inactive forms that can regain their activity when brought together again (18–21). Intein-mediated protein splicing of reporter proteins has been reported (14, 22) using green fluorescent protein (GFP) and firefly luciferase (Fluc) in vitro. Note that Fluc refers to the protein/enzyme and fluc to the gene. A complementation approach using Fluc has not been previously reported.
Fig 1.
Schematic diagram of two strategies for using split reporters to monitor protein–protein interactions. (A) Complementation-mediated restoration of firefly luciferase activity. N-terminal half of firefly luciferase is attached to protein X through a short peptide FFAGYC, and the C-terminal half of firefly luciferase is connected to protein Y through the peptide CLKS. Interaction of protein X and Y recovers Fluc activity through protein complementation. (B) Split Intein (DnaE)-mediated protein splicing leads to firefly luciferase reconstitution. The N-terminal half of firefly luciferase is connected to the N-terminal half of DnaE (DnaE-n) with peptide FFAGYC. The N-terminal half of DnaE in turn is connected to protein X. Similarly, the C-terminal half of firefly luciferase is connected to the C-terminal half of DnaE (DnaE-c) with peptide CLKS, and the C-terminal half of intein is in turn connected to protein Y. The interaction of proteins X and Y mediates reconstitution through splicing of the N and C halves of DnaE.
We and others have been developing methods to image reporter gene expression in living subjects by using technologies such as positron-emission tomography (PET), single photon emission-computed tomography, magnetic resonance imaging, as well as optical approaches (23–29). These approaches allow the possibility of imaging cellular and molecular events within cells that are in their natural in vivo environment. We have been using optical bioluminescence approaches to validate approaches related to signal amplification, gene delivery, and expression (27, 30). These approaches require the use of bioluminescence reporters and injection of the appropriate substrate into the subject (27). We recently reported on a method to extend the yeast two-hybrid approach to an inducible system for optical imaging of protein–protein interactions in living subjects by using a bioluminescence reporter (31). A similar approach also has been studied recently for use with PET imaging (32).
In the current study, we validate an approach whereby split firefly luciferase reporter proteins consisting of the N-terminal (NFluc, 1–437 amino acids; note that NFluc refers to the protein/enzyme, and Nfluc refers to the gene) and the C-terminal (CFluc, 438–554 amino acids; note that CFluc refers to the protein/enzyme, and Cfluc refers to the gene) are inactive until closely approximated (complementation strategy) or spliced together (reconstitution strategy), through the interaction of two test proteins that are known to interact strongly (MyoD and Id). MyoD and Id are members of the helix-loop-helix (HLH) family of nuclear proteins. MyoD is expressed in skeletal muscle and is a myogenic regulatory protein (33, 34). The Id protein acts as a negative regulator of myogenic differentiation and can associate with three HLH proteins: MyoD, E12, and E47 (35, 36). We validate the complementation and reconstitution strategies in cell culture and then demonstrate the ability to noninvasively image protein–protein interactions with cell implants in living mice.
Materials and Methods
Plasmid Constructs and Reagents.
The N-part of firefly luciferase gene (with C-terminal linker peptide FFAGYC) was released from the vector pIRES DSL (Y/S; ref. 14) by NheI and HindIII restriction enzymes and ligated to pcDNA 3.1(+) vector backbone to construct vector PA. The cDNA of genes Id released from pBIND-Id of Promega's mammalian two-hybrid system kit containing vector by BamHI and XhoI and cloned in the C- terminal of vector PA to construct vector PC. The N-half of DnaE was PCR-amplified by using the template pIRES DSL (Y/S) and cloned in the HindIII site of vector PC to construct vector PE. The cytomegalovirus (CMV) promoter of the vectors PC and PE were replaced by cloning the NFκB promoter/enhancer elements subcloned from the vector pNFκB-Luc of Stratagene in pET15b at BglII/HindIII restriction enzyme sites to construct vectors PG and PH. The amino acids between 72–390 of murine p53 gene were released from the vector supplied in the Mammalian Two-Hybrid Assay Kit (Stratagene) and cloned to vectors PC and PE by replacing the fragment Id with restriction enzymes HindIII and XhoI and constructed vectors PI and PJ. The vector PK was constructed by ligating the fluc gene released from vector pNFkB-Luc by NheI and XhoI to pcDNA 3.1 (+). The PCR-amplified fragment of Cfluc containing start codon was cloned in the NheI and XhoI site of pcDNA 3.1 to generate vector PB. The PCR-amplified fragment of MyoD with start codon was ligated to pcDNA 3.1 (+) in NheI/BamHI site and further inserted with the PCR product of Cfluc with linker peptide CLKS in the BamHI and XhoI site to construct vector PD. The PCR-amplified C-half of DnaE was cloned at the BamHI site of vector PD to construct vector PF (Fig. 2). Superfect transfection reagent, plasmid extraction kit, and DNA gel extraction kit were purchased from Qiagen (Chatsworth, CA). TNF-α, HRP substrates, and antibiotics for bacterial culture were purchased from Sigma. Luciferase assay kit, monoclonal antibody against firefly luciferase, and anti-mouse IgG-horseradish peroxidase (HRP) conjugate, and CheckMate mammalian two-hybrid kit were purchased from Promega. Mammalian two-hybrid kit was purchased form Stratagene. d-luciferin was purchased from Xenogen (Alameda, CA). Bacterial culture media were purchased from Difco. Enhanced chemiluminescence (ECL) kit was purchased from Amersham Pharmacia.
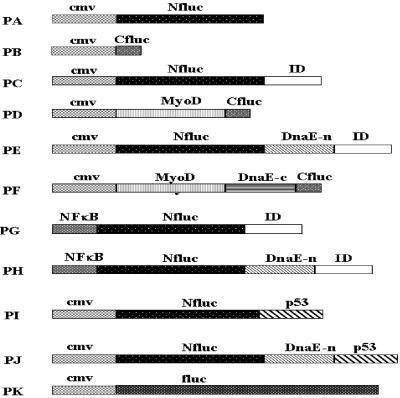
Fig 2.
Schematic representation of the plasmid constructs made and used in this study. Shown on top of each bar are the parts of genes (Nfluc, N-terminal half of firefly luciferase; Cfluc, C-terminal half of firefly luciferase; fluc, firefly luciferase; DnaE-n, N-terminal half of intein DnaE; DnaE-c, C-terminal half of intein DnaE; MyoD, cDNA sequence of amino acids 1–318 of myogenic regulatory protein; Id, cDNA sequence of amino acids 29–148 of negative regulatory protein of myogenic differentiation; p53, amino acids 72–390 of murine p53 gene) and promoter sequences (cmv, cytomegalovirus promoter; NFκB promoter).
Cell Culture.
Human embryonic kidney cancer cells 293T (American Type Culture Collection), were grown in MEM supplemented with 10% (vol/vol) FBS and 1% (vol/vol) penicillin/streptomycin solution. The N2a cells (mouse neuroblastoma cells) were obtained from V. P. Mauro (The Scripps Research Institute, La Jolla, CA) and COS-1 (monkey kidney cells) cells were grown in DMEM (high glucose) supplemented with 10% (vol/vol) FBS and 1% (vol/vol) penicillin/streptomycin.
Cell Transfection and Luciferase Assay.
Transfections were performed in 80% confluent 24-h cultures of 293T, COS-1, and N2a cells. In 12-well plates, 200 ng per well of Nfluc and 300 ng per well of Cfluc were used for transfection. For the transfection in 100-mm Petri dishes, 2 and 3 μg of Nfluc and Cfluc, respectively, were used. Volumes of superfect used were as recommended by the manufacturer. For cell induction, 0.05 μg/ml TNF-α were added immediately after transfection and assayed 24 h later.
Western Blot Analysis.
Transfection (PC, PD, PE, PF, and PK) and cotransfection (PC plus PD and PE plus PF) of plasmid constructs were made in 293T cells. After a 24-h incubation at 37°C and 5% CO2, cells were washed twice in PBS and lysed mechanically in a buffer containing 10 mM Tris⋅HCl (pH 8.0), 1 mM EDTA, 1 mM DTT with 20% glycerol, and 0.1 mM PMSF. The samples were centrifuged at 4°C and 9,300 × g for 5 min. Protein was estimated, and 10 mg of protein from each sample was mixed with two volumes of sample buffer and boiled for 5 min. Denatured samples were electrophoresed in 12% acrylamide gel and transferred to poly(vinylidene difluoride) membrane by using Hoefer semiblot apparatus. Membrane was immediately transferred to PBS containing 3% milk powder and blocked for 3 h with proper mixing. Membrane was incubated with primary antibody (monoclonal anti-Fluc antibody) overnight at room temperature with proper shaking. Washed membrane was incubated for 1 h with donkey anti-mouse IgG-HRP conjugate for 1 h. Immunochemical detection was carried out by using the substrates from the Amersham Pharmacia ECL kit for 30 sec and 5 min.
Imaging Split fluc Expression in Living Mice by Using a Cooled Charge-Coupled Device (CCD) Camera.
The 293T cells were transiently transfected with plasmids PC and PD separately and cotransfected with PC plus PD. The cells were harvested after incubating in the medium with serum for 2 h after transfection. Cells were suspended in phosphate buffered saline. An aliquot of 1 × 106 cells from each combination (PC, PD, PC plus PD, and mock transfected cells) were implanted s.c. in four different sites in the ventral side of anesthetized (ketamine-xylazine, 4:1) nude mice. Immediately after cell implantation, 100 μl of d-luciferin (30 mg/ml) was injected i.p., and the mice were imaged at one-minute intervals until reaching the maximum photon counts. For modulating in vivo imaging signals, 293T cells were transfected with plasmids PD, PG, and PD plus PG for evaluating the complementation strategy and with plasmids PF, PH, and PF plus PH for evaluating the reconstitution strategy. After transfection, cells were harvested and implanted s.c. in mice, as described above. After the first scan, the mice were injected i.p. with 0.5 μg of TNF-α and imaged 18 h later. The animals then were reinduced with equivalent concentrations of TNF-α and scanned 18 h later (i.e., at 36 h after implantation). A total of six mice were used for each strategy with equal numbers of controls.
All mice were imaged by using a cooled CCD camera (Xenogen IVIS, Xenogen, Alameda, CA). The animals were placed supine in a light-tight chamber, and a gray scale reference image was obtained under low-level illumination. Photons emitted from cells implanted in the mice were collected and integrated for a period of 1 min. Images were obtained by using LIVING IMAGE software (Xenogen, Alameda, CA) and IGOR image analysis software (WaveMetrics, Lake Oswego, OR). To quantify the measured light, regions of interest were drawn over the tumor region showing light signal, and maximum photons/sec/cm2/steridian (sr) were obtained, as previously validated (27).
Results
Cells Transiently Expressing Nfluc Give Greater Activity than Cells Expressing Cfluc, but both Are Markedly Less than Cells Expressing the Full fluc Reporter Gene.
To achieve a low-background signal, it would be ideal for cells expressing each split half of fluc to produce minimal activity when exposed to the substrate d-luciferin. Transient transfection studies in COS-1, N2a, and 293T cells with either plasmid PA or PB alone show that NFluc produces 15 ± 5-fold higher background activity than CFluc alone (P < 0.01). Furthermore, the NFluc and CFluc activities were 25-fold and >1,000-fold, respectively, less than in cells transfected with plasmid PK (Fluc), indicating relatively low-background activity of each split reporter as compared with the intact full reporter. To minimize the background activity of NFluc, we determined that 200 ng of DNA per well for any plasmid containing Nfluc is optimal when using 12-well culture plates (data not shown). This amount was used for all subsequent transfection studies.
Complementation and Reconstitution of Fluc Activity in Transient Transfection Cell Culture Studies Can Be Achieved Through the Interaction of Proteins Id and MyoD.
Cotransfection of plasmid constructs PC plus PD was studied in COS-1, 293T, and N2a cells to test the complementation strategy. Cotransfection of plasmid constructs PC plus PD shows a 15 ± 5-fold or a 500 ± 50-fold, respectively, higher Fluc activity than in cells transfected with either PC or PD alone (Fig. 3A, 293T cells; the S.E.M. is across all three cell lines). The complementation activity achieved for PC plus PD is ≈40–60% of that for cells transfected with the plasmid encoding the full reporter (PK). Cotransfection of constructs PD plus PI shows Fluc activity which is ≈10-fold less than PC alone and ≈100-fold less than the cotransfection of PC plus PD, which is consistent with a lack of any significant complementation when using two noninteracting proteins (p53 and MyoD).
Fig 3.
(A) Complementation-based split luciferase activity in transiently transfected 293T cells. 293T cells were transiently transfected with plasmid constructs PC, PD, PI (only parts of firefly luciferase), PC plus PD (complementation), PD plus PI (no interaction), and PK (full firefly luciferase). The cells were harvested after 24 h and assayed for Fluc activity. The relative light units (RLU) per microgram of protein are represented. Error bars represent SEM for triplicate measurements. (B) Reconstitution of split luciferase in transiently transfected 293T cells. 293T cells were transiently transfected with plasmid constructs PE, PF, PJ (only parts of firefly luciferase), PE plus PF (reconstitution), PF plus PJ (no interaction), and PK (full firefly luciferase). The cells were harvested after 24 h and assayed for Fluc activity. The RLU per microgram of protein is represented. Error bars represent SEM for triplicate measurements. (C) Western blot of protein extracts from transient transfection studies in 293T cells. The protein band of ≈80 kDa was detected only in the cells transfected with both PE and PF (reconstitution) or PK and not from other studies in which the full Fluc protein was not recovered.
The Fluc activity measured when cotransfecting with the plasmid constructs PE plus PF (reconstitution strategy) is not significantly higher than that from the constructs without intein (complementation strategy) in all three cell lines tested (Fig. 3B, 293T cells). Again, the activity seen when using plasmids PE plus PF is significantly higher (P < 0.01) than when using PE or PF alone or PF plus PJ (noninteracting protein control) and is ≈45–60% of that for cells transfected with the plasmid encoding the full reporter (PK). Similar results were obtained across all cell lines tested, except that the absolute level of Fluc activity is highest with 293T cells; consequently, 293T cells were used for all subsequent studies. The variation in the Fluc activity observed for different cell lines is likely caused by differences in transfection and/or transcriptional efficiencies. These results demonstrate that both the complementation and reconstitution strategies are capable of producing significant specific signal after the interaction of MyoD and Id proteins in cell culture.
Western Blot Analysis from Cell Transfection Studies Shows the Full Fluc Protein Is Recovered When Using the Reconstitution Strategy.
To verify the difference between the complementation and reconstitution strategies at the protein level, proteins isolated from 293T cells transfected with a combination of vector constructs were separated by SDS/PAGE. Membrane-transferred proteins were detected by using a monoclonal antibody against firefly luciferase. These results show a band position of ≈80 kDa from firefly luciferase, and the protein reconstituted from the cells cotransfected with vector constructs PE plus PF (Fig. 3C). The cells transfected with vector constructs PC, PE, and PC plus PD (complementation strategy) synthesized fusion proteins carrying Id, MyoD, DnaE, and parts of NFluc and CFluc show no visible bands at low exposure times (Fig. 3C, 30 sec), but very weak bands are seen with longer exposure (5 min; data not shown) because of low specificity against the monoclonal antibody used for detection. The cells transfected with vector constructs PD and PF show no detectable bands. These data support the finding that the monoclonal antibody specifically detected the complete Fluc synthesized by cells transfected with plasmid PK as well as luciferase protein reconstituted from the vectors carrying Nfluc and Cfluc with DnaE.
Fluc Activity Can Be Modulated by TNF-α in Cell Culture for Both the Complementation and Reconstitution Strategies.
To modulate the interaction of the split proteins, the CMV promoter in plasmid constructs PC and PE was replaced with NF-κB promoter/enhancer elements (κB4-PTAL) to create plasmids PG and PH, respectively (Fig. 2). To test the ability to modulate the system, plasmids PD plus PG (complementation strategy) or PF plus PH (reconstitution strategy) were transfected into 293T cells and induced with TNF-α for a 24-h period. Fluc activity obtained with Nfluc under the NF-κB promoter/enhancer element is 50 ± 10% less than with the CMV promoter. The activity is significantly (P < 0.01) higher (13 ± 2-fold) than that of preinduction levels in both strategies (Fig. 4). There is no significant difference when transiently transfecting cells with the plasmids PD plus PG vs. PF plus PH. Cotransfection of plasmid constructs with CMV promoter PC plus PD or PE plus PF show smaller but significant induction (P = 0.009 and 0.02, respectively) with TNF-α, which is much less than the constructs with NF-κB promoter.
Fig 4.
Effect of TNF-α on activation of fluc expression. (A) Complementation strategy. 293T cells were transiently transfected with plasmids PC, PD, PG, PD plus PG, and PC plus PD. The cells were harvested after 24 h in the presence or absence of TNF-α and assayed for Fluc activity. The relative light units (RLU) per microgram of protein were estimated and compared for the induction. The cells transfected with plasmid constructs PG and PD plus PG carrying NFκB promoter/enhancer elements showed a significant increase upon TNF-α induction. The cells transfected with plasmid constructs PC plus PD carrying CMV promoter showed a significant increase upon TNF-α induction, but were significantly less than the constructs carrying NFκB promoter/enhancer elements. (B) Reconstitution strategy. 293T cells transiently transfected with plasmids PE, PF, PH, PE plus PF, and PF plus PH were harvested after 24 h in the presence and absence of TNF-α and assayed for Fluc activity. The relative light units (RLU) per microgram of protein were estimated and compared for the induction. The cells transfected with plasmid constructs PH and PF plus PH carrying NFκB promoter/enhancer elements showed significant increase in fluc expression upon TNF-α induction. The cells transfected with plasmid constructs PE plus PF carrying CMV promoter also showed significant increase upon induction but were significantly less than the cells transfected with plasmid constructs carrying NFκB promoter/enhancer elements.
Fluc Activity Recovered Through Protein–Protein Interaction-Mediated Complementation/Reconstitution Can Be Imaged in Living Mice.
Cooled CCD imaging of mice implanted with one million 293T cells mock transfected or transiently transfected with constructs PC, PD, or PC plus PD (complementation) show low background signal at time 0 [<2.9 ± 0.7 × 103 p/s/cm2/steridian (sr)] and significant (P < 0.05) signal only from the cells cotransfected with constructs PC plus PD (2.5 ± 0.87 × 105 p/s/cm2/sr) at time 16–24 h. The signal from cells transfected with either PC or PD alone at 16–24 h are 1.22 ± 0.3 × 104 p/s/cm2/sr and 6.08 ± 1.2 × 103 p/s/cm2/sr, respectively. The animals implanted with cells transfected with PE plus PF (reconstitution) show the signal that is 10 ± 2% greater with PC plus PD (complementation), but this is not statistically significant (images not shown).
Fluc Activity Recovered Through Protein–Protein Interaction-Mediated Complementation/Reconstitution and Modulated by TNF-α Can Be Imaged in Living Mice.
To test the effect of in vivo modulation on the split firefly luciferase system, we s.c. implanted nude mice in each of four separate body locations with one million transiently transfected 293T cells containing the constructs PD, PG, PD plus PG (complementation strategy), and mock-transfected cells. Similarly, another set of mice was implanted with 293T cells transfected with plasmid constructs PF, PH, PF plus PH (reconstitution strategy), and mock-transfected cells. Mice implanted with cells that did not receive TNF-α show relatively low signal over the course of 36 h in both the complementation strategy (Fig. 5) and reconstitution strategy (see Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). Mice i.p. injected with TNF-α show significant increase in signal over the study period (P < 0.05). The split luciferase system-mediated fluc expression shows a more significant gain (P < 0.05) in the induced group than the uninduced group did at 18 and 36 h. These results demonstrate that it is possible to image protein–protein interactions in living subjects by both the inducible complementation and reconstitution strategies.
Fig 5.
In vivo optical CCD imaging of mice carrying transiently transfected 293T cells for induction of the complementation-based split luciferase system. All images shown are the visible light image superimposed on the optical CCD bioluminescence image with a scale in photons/sec/cm2/steridian (sr). Mice were imaged in a supine position after i.p. injection of d-luciferin. (A) Set of nude mice were repetitively imaged after s.c. implantation of 293T cells transiently transfected with plasmids PD (site B), PG (site C), PD plus PG (site D), and mock transfected cells (site A). One group of mice was induced with TNF-α and the other group was not induced. Images are from one representative mouse from each group immediately after implanting cells (0 h) and 18 and 36 h after TNF-α induction. The induced mouse showed higher Fluc signal at site D when compared with the mouse not receiving TNF-α. The Fluc signal significantly increases after receiving TNF-α. (B) Graphs showing the uninduced (Upper) and induced (Lower) group with mean values across six mice from each group. The error bars represent SEM. The induced group showed a significantly higher signal at 18 and 36 h compared with the uninduced group from the site containing 293T cells transfected with plasmids PD plus PG.
Discussion
This study validates the ability to use split firefly luciferase to monitor the interaction of two proteins in cell culture and in living mice by using both complementation and reconstitution strategies. Although previous studies have validated the use of various split reporters in cell culture (4, 14, 22, 37), this study does so in living subjects. This study also demonstrates that complementation using split firefly luciferase is feasible. This complementation was demonstrated both in cell culture and in cells implanted in living subjects. We chose two proteins, MyoD and Id, known to be strongly interacting as our two test proteins, but it is likely that this system would be sensitive enough for detection of other protein partners with weaker interactions, based on the robust levels of signal obtained. Future studies will need to characterize the ability to image the interaction of different protein partners with varying degrees of affinity. Importantly, the background level of split reporter signal is relatively low compared with the signal after protein interaction, both in cell culture and in living subjects. We used transient transfection studies to readily validate our approach, but stable clones, viral gene delivery, as well as transgenic models could also be used. The optical bioluminescence approaches have been shown to be relatively sensitive at all depths and locations within a living mouse (28), so that the current approach could potentially be applied to study protein–protein interactions anywhere within a mouse model. The choice of the NF-κB promoter in the current work was because of the ability to induce this promoter in vivo, as characterized in our previous study (31). Future applications could potentially link expression of each split reporter to different endogenous promoters so as to better mimic endogenous protein levels.
The reconstitution strategy results in the formation of a new complete reporter protein that maintains its activity even in the absence of continuing interaction between the protein partners. A portion of the optical signal obtained from the intein-mediated split reporter protein strategy may include activity obtained from complementation as opposed to that obtained solely from reconstitution. The Western blot analysis supports the idea that significant reconstitution is occurring in the reconstitution strategy (PE+PF), but quantitation of the exact amount will require further investigation. The reduction in the optical signal observed as compared with using the fully intact reporter protein may in part be caused by the use of split intein with split exteins and also because of the efficiency of the two interacting proteins in bringing the inteins together. It has been reported that the intact intein-mediated transsplicing of exteins occurs rapidly even at a wide range of temperatures, and the kinetics of the intermediate steps have been investigated (38). Further work will be needed to characterize better the generalizability of the reconstitution strategy for different protein partners based on the intrinsic rate of protein association as compared with the intrinsic rate of complementation of the DnaE fragments.
In the complementation strategy, fusion proteins need protein interaction to be maintained to retain reporter activity. The reconstitution and complementation strategies yield comparable signal, thus allowing for the use of either approach without a compromise in reporter sensitivity. One might expect that reconstitution might be more sensitive because it produces intact reporter protein, but we found that complementation is a comparable approach for Fluc with the current choice of protein partners (MyoD and Id). The ability of the complementation approach to work in cell culture and in vivo should allow this robust strategy to be used in various future applications. The complementation strategy is easier to implement and, if it is proven to be as robust with many different protein partners, may be the preferred strategy over reconstitution. Moreover, the split reporter strategies (complementation and reconstitution) can be used to study cellular events that occur in any part of the cell, solving a key limitation of the yeast two-hybrid approach. Further studies will be needed to contrast the relative merits of the reconstitution, complementation, and yeast two-hybrid approaches.
We are actively investigating approaches where other split reporters can be used with other noninvasive imaging modalities (e.g., split herpes simplex virus type-1 thymidine kinase reporter proteins for use with PET), as well as approaches to link split reporters to small antisense oligodeoxynucleotides for potential imaging of endogenous mRNA levels. Systems-imaging approaches to study cells in their normal environment within an animal subject should be facilitated with the approaches developed in this work. The approaches developed should provide an efficient system for the continuous observation of protein–protein interactions in a particular network pathway under different conditions in vivo. Direct extensions of the current approach should also lead to the ability to study pharmaceuticals that modulate protein–protein interactions in living subjects to accelerate and expand the scope of drug development and testing in an in vivo setting.
Supplementary Material
Acknowledgments
We thank Dr. Frank Berger, Dr. David Stout, Jim Strommer, and Xiaoman Lewis for technical assistance. We also thank Drs. Herschman, Massoud, Kaufman, and Carey for helping us to improve the manuscript. This work was supported in part by National Institutes of Health Grants P50 CA86306, R0-1 CA82214, SAIRP R24 CA92865, and Department of Energy Contract DE-FC03-87ER60615 (all to S.S.G.).
Abbreviations
PET, positron-emission tomography
CMV, cytomegalovirus
CCD, charge-coupled device
TNF-α, tumor necrosis factor α
References
- 1.Chien C. T., Bartel, P. L., Sternglanz, R. & Fields, S. (1991) Proc. Natl. Acad. Sci. USA 88 9578-9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai C. & Elledge, S. J. (1996) Methods Enzymol. 273 331-347. [DOI] [PubMed] [Google Scholar]
- 3.Fields S. & Song, O. (1989) Nature 340 245-246. [DOI] [PubMed] [Google Scholar]
- 4.Rossi F., Charlton, C. A. & Blau, H. M. (1997) Proc. Natl. Acad. Sci. USA 94 8405-8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunnwald M., Varshavsky, A. & Johnsson, N. (1999) Mol. Biol. Cell 10 329-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnsson N. & Varshavsky, A. (1994) Proc. Natl. Acad. Sci. USA 91 10340-10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aronheim A. (1997) Nucleic Acids Res. 25 3373-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aronheim A., Zandi, E., Hennemann, H., Elledge, S. J. & Karin, M. (1997) Mol. Cell. Biol. 17 3094-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broder Y. C., Katz, S. & Aronheim, A. (1998) Curr. Biol. 8 1121-1124. [DOI] [PubMed] [Google Scholar]
- 10.Pelletier J. N., Arndt, K. M., Pluckthun, A. & Michnick, S. W. (1999) Nat. Biotechnol. 17 683-690. [DOI] [PubMed] [Google Scholar]
- 11.Remy I. & Michnick, S. W. (1999) Proc. Natl. Acad. Sci. USA 96 5394-5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wehrman T., Kleaveland, B., Her, J. H., Balint, R. F. & Blau, H. M. (2002) Proc. Natl. Acad. Sci. USA 99 3469-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrhard K. N., Jacoby, J. J., Fu, X. Y., Jahn, R. & Dohlman, H. G. (2000) Nat. Biotechnol. 18 1075-1079. [DOI] [PubMed] [Google Scholar]
- 14.Ozawa T., Kaihara, A., Sato, M., Tachihara, K. & Umezawa, Y. (2001) Anal. Chem. 73 2516-2521. [DOI] [PubMed] [Google Scholar]
- 15.Mohler W. A. & Blau, H. M. (1996) Proc. Natl. Acad. Sci. USA 93 12423-12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gimble F. S. (1998) Chem. Biol. 5 R251-R256. [DOI] [PubMed] [Google Scholar]
- 17.Paulus H. (2000) Annu. Rev. Biochem. 69 447-496. [DOI] [PubMed] [Google Scholar]
- 18.Mills K. V. & Paulus, H. (2001) J. Biol. Chem. 276 10832-10838. [DOI] [PubMed] [Google Scholar]
- 19.Lew B. M., Mills, K. V. & Paulus, H. (1998) J. Biol. Chem. 273 15887-15890. [DOI] [PubMed] [Google Scholar]
- 20.Lew B. M., Mills, K. V. & Paulus, H. (1999) Biopolymers 51 355-362. [DOI] [PubMed] [Google Scholar]
- 21.Shingledecker K., Jiang, S. & Paulus, H. (2000) Arch. Biochem. Biophys. 375 138-144. [DOI] [PubMed] [Google Scholar]
- 22.Ozawa T., Takeuchi, M., Kaihara, A., Sato, M. & Umezawa, Y. (2001) Anal. Chem. 73 5866-5874. [DOI] [PubMed] [Google Scholar]
- 23.Elias S. & Ciechanover, A. (1990) J. Biol. Chem. 265 15511-15517. [PubMed] [Google Scholar]
- 24.MacLaren D. C., Gambhir, S. S., Satyamurthy, N., Barrio, J. R., Sharfstein, S., Toyokuni, T., Wu, L., Berk, A. J., Cherry, S. R., Phelps, M. E. & Herschman, H. R. (1999) Gene Ther. 6 785-791. [DOI] [PubMed] [Google Scholar]
- 25.Gambhir S. S., Barrio, J. R., Phelps, M. E., Iyer, M., Namavari, M., Satyamurthy, N., Wu, L., Green, L. A., Bauer, E., MacLaren, D. C., et al. (1999) Proc. Natl. Acad. Sci. USA 96 2333-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gambhir S. S., Barrio, J. R., Wu, L., Iyer, M., Namavari, M., Satyamurthy, N., Bauer, E., Parrish, C., MacLaren, D. C., Borghei, A. R., et al. (1998) J. Nucl. Med. 39 2003-2011. [PubMed] [Google Scholar]
- 27.Bhaumik S. & Gambhir, S. S. (2002) Proc. Natl. Acad. Sci. USA 99 377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu J. C., Sundaresan, G., Iyer, M. & Gambhir, S. S. (2001) Mol. Ther. 4 297-306. [DOI] [PubMed] [Google Scholar]
- 29.Tjuvajev J. G., Avril, N., Oku, T., Sasajima, T., Miyagawa, T., Joshi, R., Safer, M., Beattie, B., DiResta, G., Daghighian, F., et al. (1998) Cancer Res. 58 4333-4341. [PubMed] [Google Scholar]
- 30.Iyer M., Wu, L., Carey, M., Wang, Y., Smallwood, A. & Gambhir, S. S. (2001) Proc. Natl. Acad. Sci. USA 98 14595-14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray P., Pimenta, H., Paulmurugan, R., Berger, F., Phelps, M. E., Iyer, M. & Gambhir, S. S. (2002) Proc. Natl. Acad. Sci. USA 99 3105-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luker G. D., Sharma, V., Pica, C. M., Dahlheimer, J. L., Li, W., Ochesky, J., Ryan, C. E., Piwnica-Worms, H. & Piwnica-Worms, D. (2002) Proc. Natl. Acad. Sci. USA 99 6961-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis R. L., Weintraub, H. & Lassar, A. B. (1987) Cell 51 987-1000. [DOI] [PubMed] [Google Scholar]
- 34.Weintraub H., Davis, R., Tapscott, S., Thayer, M., Krause, M., Benezra, R., Blackwell, T. K., Turner, D., Rupp, R., Hollenberg, S., et al. (1991) Science 251 761-766. [DOI] [PubMed] [Google Scholar]
- 35.Benezra R., Davis, R. L., Lassar, A., Tapscott, S., Thayer, M., Lockshon, D. & Weintraub, H. (1990) Ann. N.Y. Acad. Sci. 599 1-11. [DOI] [PubMed] [Google Scholar]
- 36.Benezra R., Davis, R. L., Lockshon, D., Turner, D. L. & Weintraub, H. (1990) Cell 61 49-59. [DOI] [PubMed] [Google Scholar]
- 37.Rossi F. M., Blakely, B. T., Charlton, C. A. & Blau, H. M. (2000) Methods Enzymol. 328 231-251. [DOI] [PubMed] [Google Scholar]
- 38.Martin D. D., Xu, M. Q. & Evans, T. C., Jr. (2001) Biochemistry 40 1393-1402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.