Abstract
We have investigated the effect of sequence-specific antisense phosphorothioate-modified oligodeoxyribonucleotides (PS-ODNs) targeting different regions of each of the 30/32-kDa protein complex (antigen 85 complex) encoding genes on the multiplication of Mycobacterium tuberculosis. Single PS-ODNs to one of the three mycolyl transferase transcripts, added either once or weekly over the 6-wk observation period, inhibited bacterial growth by up to 1 log unit. A combination of three PS-ODNs specifically targeting all three transcripts inhibited bacterial growth by ≈2 logs; the addition of these PS-ODNs weekly for 6 wk was somewhat more effective than a one-time addition. Targeting the 5′ end of the transcripts was more inhibitory than targeting internal sites; the most effective PS-ODNs and target sites had minimal or no secondary structure. The effect of the PS-ODNs was specific, as mismatched PS-ODNs had little or no inhibitory activity. The antisense PS-ODNs, which were highly stable in M. tuberculosis cultures, specifically blocked protein expression by their gene target. PS-ODNs targeting the transcript of a related 24-kDa protein (mpt51) had little inhibitory effect by themselves and did not increase the effect of PS-ODNs against the three members of the 30/32-kDa protein complex. The addition of PS-ODNs against the transcripts of glutamine synthetase I (glnA1) and alanine racemase (alr) modestly increased the inhibitory efficacy of the 30/32-kDa protein complex-specific PS-ODNs to ≈2.5 logs. This study shows that the three mycolyl transferases are highly promising targets for antituberculous therapy by using antisense or other antimicrobial technologies.
Keywords: phosphorothioate oligodeoxyribonucleotides, mRNA secondary structure, inhibition of translation initiation, drug design
Tuberculosis, caused primarily by the facultative intracellular bacterium Mycobacterium tuberculosis, is the leading cause of death from a single infectious agent. The emergence of multidrug-resistant strains of M. tuberculosis has given urgency to the need for novel antimicrobial agents to combat this pathogen (1).
M. tuberculosis expresses and secretes three closely related mycolyl transferases of 30–32 kDa mass, also known as the antigen 85 protein complex (antigen 85A, 85B, and 85C are referred to herein as the 32A, 30, and 32B proteins, respectively). All three enzymes catalyze the transfer of mycolic acid from one trehalose 6-monomycolate to another, resulting in trehalose 6,6′-dimycolate and free trehalose (2). The 3D structure of two of the enzymes, the 30 and 32A proteins, their catalytic sites, and their reaction mechanisms have been described in detail (3, 4). All three proteins are abundantly secreted. The 30-kDa protein is the most abundant extracellular protein of M. tuberculosis, responsible for nearly one quarter of the total extracellular protein in broth culture, and the 32A-kDa protein is the second most abundant extracellular protein (5). The three mycolyl transferases are also abundantly expressed in human macrophages infected with M. tuberculosis (6). In M. tuberculosis phagosomes within human macrophages, the presence of the proteins in the phagosomal space and on the bacterial cell wall was demonstrated by the cryosection immunogold technique (5). Disruption of each individual gene of the 30/32-kDa protein complex has resulted in an altered bacterial cell wall or impairment of growth in minimal media or inside host cells but not in cell death (7, 8), suggesting that it may be necessary to block more than one enzyme to achieve growth inhibition.
The 30/32-kDA complex proteins are leading tuberculosis vaccine candidates. Vaccination of guinea pigs with purified 30-kDa protein induces substantial protective immunity against aerosol challenge with M. tuberculosis (9). Vaccination of guinea pigs with a recombinant Mycobacterium bovis bacillus Calmette–Guérin vaccine expressing and secreting the M. tuberculosis 30-kDa protein (rBCG30) induces stronger protective immunity against aerosol challenge with M. tuberculosis than conventional M. bovis bacillus Calmette–Guérin vaccine (10); this recombinant vaccine is the only vaccine thus far demonstrated superior to bacillus Calmette–Guérin.
In a previous study, we demonstrated that antisense technology is a feasible approach to combating M. tuberculosis. Incubation of M. tuberculosis with antisense phosphorothioate-modified oligodeoxyribonucleotides (PS-ODNs) directed against the M. tuberculosis enzyme glutamine synthetase I (glnA1 gene product), an enzyme central to the bacterium's nitrogen metabolism, decreased glutamine synthetase activity in the culture, reduced the amount of the poly-l-glutamate/glutamine heteropolymer in the M. tuberculosis cell wall, and inhibited bacterial growth (11). A combination of three different antisense PS-ODNs against three different segments of the glutamine synthetase I transcript was more effective than single antisense PS-ODNs, but even the combination of PS-ODNs resulted in only a modest reduction in bacterial growth. This result prompted us to search for more promising targets for antisense technology.
In this report, we demonstrate that a combination of three different PS-ODNs targeting the first 24 aa-encoding bases of each of the three mycolyl transferase gene transcripts markedly inhibits M. tuberculosis growth in broth culture. The biological effect is specific because mismatched PS-ODNs have no inhibitory effect at all. This study not only identifies an exceptionally effective combination of PS-ODNs for use against M. tuberculosis, but provides strong evidence for the concept that targeting the 30/32-kDa complex proteins is a feasible strategy for developing new antimicrobial agents against tuberculosis.
Materials and Methods
PS-ODN Selection and Preparation.
Several target sites along the gene transcripts of the 30/32-kDa proteins were chosen, based on the sequence similarity between all three transcripts and their propensity to remain in a linear conformation as determined by the oligo-5 secondary structure analysis program (Sigma–Genosys, The Woodlands, TX). The PS-ODNs' DNA sequences and their target sites are described in Table 1. Antisense PS-ODNs were synthesized on a 394 DNA/RNA synthesizer (Applied Biosystems) by using standard phosphoroamidite chemistry. Phosphorothioate bonds were introduced by oxidation with the Beaucage thiolating reagent (12), and assembled PS-ODNs were purified by HPLC and lyophilized. Mismatched PS-ODNs were synthesized in the same way except that the middle base of each base triplet was mismatched with regard to its complementary base in the mRNA strand. PS-ODN stock solutions were prepared just before their use and added to mycobacterial cultures after sterilization through 0.45-μm HT Tuffryn membrane filters (Gelman). Poly(G) and 2′-O-methyl modifications of PS-ODNs were synthesized as described (13–16).
Table 1.
Target sites of M. tuberculosis gene transcript-specific PS-ODNs
| mRNA target 5′ > 3′ | PS-ODN 5′ > 3′ | Propensity for linearity |
|---|---|---|
| 30:157–183 | GTCGCGGCCCATCGACGGCGACGGCAC (27) | Strong |
| 30:718–744 | CCATAGCCGGGTGTTGTTTGCGACCAG (27) | Weak |
| 30:871–897 | GTTGGGCGGGAAGTTGAACACGGCGTT (27) | Very weak |
| 30:157–183 (sense) | GTGCCGTCGCCGTCGATGGGCCGCGAC (27) | Strong |
| 30:718–744 (sense) | CTGGTCGCAAACAACACCCGGCTATGG (27) | Weak |
| 30:871–897 (sense) | AACGCCGTGTTCAACTTCCCGCCCAAC (27) | Very weak |
| 30:267–290 | GCCGGGGTGTTGATATCCCAGCCG (24) | Moderate |
| 32A:276–299 | GCCGGGGTGTTGATGTCCCAGCCG (24) | Moderate |
| 32B:279–302 | GCCGGGGTGTTGATGTCCCAGCCG (24) | Moderate |
| 24:225–248 | GCGTTACCCGCGGTGACCCAGTTA (24) | Moderate |
| 32A:276–303 | GAACGCCGGGGTGTTGATGTCCCAGCCG (28) | Moderate |
| 30:1–24 | AATCTTTCGGCTCACGTCTGTCAT (24) | None |
| 32A:1–24 | ACGAACCCTGTCAACAAGCTGCAT (24) | None |
| 32B:1–24 | TCGCACCTGTTCGAAGAACGTCAT (24) | Moderate |
| 24:1–24 | CAGCAGCGCCGACCGACCCTTCAT (24) | None |
| 30:1–24 | AATCTTTCGGCTCACGTCTGTCAT (24) | None |
| 32A:1–24 | ACGAACCCTGTCAACAAGCTGCAT (24) | None |
| 32A:1–24 | ACGAACCCTGTCAACAAGCTGCAT-5G (29) | Moderate |
| GSI:805–825 | CATGCCGGACCCGTTGTCGCC (21) | Weak |
| ALR:1–24 | GACATTCTCCCAGAACCGTTTCAC (24) | Weak |
Number of nucleotides appears in parentheses.
Based on OLIGO-5 SECONDARY STRUCTURE ANALYSIS program.
mRNA nucleotide numbers always given in ascending order and denote first and last hybridizing nucleotide.
The six 5′- and 3′-terminal nucleotides are 2′-O-methyl-modified nucleotides.
5G, PS-ODN-modified at the 3′ terminus by the addition of 5 Gs.
GSI, glutamine synthetase I (glnA1-Rv2220 gene product).
ALR, alanine racemase (alr-Rv3423c gene product).
Bacterial Inhibition Assay.
M. tuberculosis strain Erdman (American Type Culture Collection 35801) was maintained in 7H9 medium (Difco) supplemented with 2% glucose at 37°C in a 5% CO2–95% air atmosphere as unshaken cultures. For the analyses of the inhibitory effects of various PS-ODNs on bacterial growth, M. tuberculosis cultures were set up in duplicates as 2-ml broth cultures in polystyrene tubes (Fisher) and maintained for 6 wk. PS-ODNs were added to the medium just before the inoculation with mycobacteria at final concentrations of 1, 5, or 10 μM. Growth of the bacteria was monitored by gently sonicating the culture to break up bacterial clumps, removing small aliquots, washing the bacteria by centrifugation, plating serial dilutions of washed bacteria on 7H11 agar (Difco), and enumerating viable bacteria [colony-forming units (cfu)] after incubation for 2 wk.
Expression of 30/32-kDa Complex Proteins.
Bacterial cultures of M. tuberculosis were set up as described above, and individual PS-ODNs or combinations thereof were added to the culture medium at a final concentration of 10 μM for each PS-ODN. After 4 wk, 1 ml of each culture was removed and centrifuged to pellet bacteria, and the supernatant was concentrated to 125 μl. Half of the supernatant sample was subjected to SDS/PAGE, after which the proteins were transferred to a nylon membrane and probed with 30/32-kDa protein-specific immunoglobulins at a dilution of 1:100,000. Immunocomplexes were detected with Sigma's enhanced chemiluminescent kit, visualized by exposure to Kodak BioMax Light Film for 10–30 s, and digitized as a photoshop 5.5 (Adobe Systems, Mountain View, CA) file. Protein-specific bands were quantitated densitometrically using nih image 1.62 software.
PS-ODN Stability Assay.
Bacterial cultures of M. tuberculosis were set up, and individual PS-ODNs or combinations thereof were added to the medium at a final concentration of 10 μM for each PS-ODN as described above. Immediately and 14, 28, and 42 days later, a 20-μl aliquot (1% of the total culture volume) was removed from each culture and directly loaded onto a 1% agarose gel. Samples were electrophoresed at 50 volts in Tris/borate/EDTA (pH 8.3) until the bromophenol blue dye marker had migrated ≈70% of the distance between loading well and bottom of the gel. Gels were stained with ethidium bromide, photographed, and digitized as a photoshop 5.5 file, and PS-ODN bands were quantitated densitometrically.
Results
Selection of PS-ODN Target Sites and Stoichiometric Considerations.
In a previous study (11), using antisense PS-ODNs specifically targeting the transcript of the M. tuberculosis glnA1 gene, we demonstrated the importance of several parameters for a successful application of this technology to mycobacteria. First, PS-ODNs were most effective at ≥10 μM. Second, PS-ODNs should be at least 18 bases but preferably 24 bases in length to minimize random hybridization to the M. tuberculosis genome, which is 4.4 million base pairs in length (17). Third, in general, PS-ODNs and the corresponding mRNA targets should have a low propensity to form a stable secondary structure at 37°C. The latter consideration is only a general guideline because it does not take into account the fact that the in vivo structure of mRNA is not wholly predictable from sequence data and, for example, may be influenced by the binding of proteins.
The M. tuberculosis genome contains four genetic loci that encode proteins of the mycolyl transferase complex (17); three of these genes and their encoded proteins are very closely related to each other, whereas the fourth gene and protein are distantly related to the other three members of the complex. The three 30/32-kDa proteins are encoded by different, unlinked, single operon genes. The 30-kDa mycolyl transferase is encoded by the fbpB gene at map location Rv1886c; the 32A-kDa protein is encoded by fbpA at map location Rv3804c; and the 32B-kDa protein is encoded by fbpC at map location Rv0129c. All three of these proteins have been demonstrated to have mycolyl transferase-specific activity in pure form in vitro (2). It is unclear whether the fourth member of the complex, a 24-kDa protein (mpt51) encoded by fbpD at map location Rv3803c, exhibits mycolyl transferase activity. We refer in the text to the mycolyl transferase complex as the enzyme complex of the three 30/32-kDa proteins. The three proteins are constitutively expressed by M. tuberculosis throughout its growth in broth culture and in macrophages (5, 6); in broth culture, the proteins are expressed in a molar ratio of 3:2:1 for 30:32A:32B (5). The relative ratios of expression at the protein level are mirrored at the mRNA level; RNA analyses also have shown that each of the three genes is transcribed as a single transcription unit of ≈1 kb, although there is the theoretical possibility that the transcript starting from fbpA could continue into the related downstream gene Rv3803c (fbpD; ref. 5). Based on densitometric analyses of Northern blot hybridizations with RNA isolated from broth culture bacteria, we calculated that there are ≈100 transcripts specific for the 30-kDa protein, 50 transcripts specific for the 32A-kDa protein, and 30 transcripts specific for the 32B-kDa protein per cell in a mid-log phase culture.
Effect on M. tuberculosis Proliferation of Antisense PS-ODNs Against Internal Sites of the 30-kDa Protein Gene Transcript.
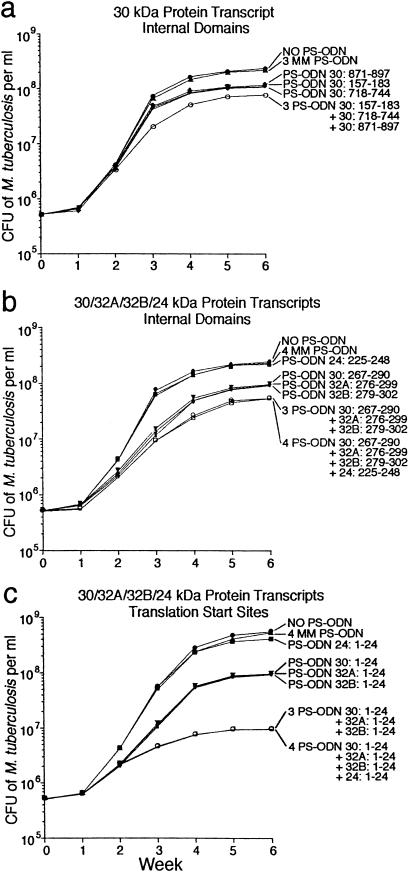
We first investigated the effect on M. tuberculosis growth of antisense PS-ODNs directed against three different internal sites of the 30-kDa protein gene (30:157–183, 718–744, 871–897). All three target sites were mismatched against the target sites of the other members of the 30/32/24-kDa protein complex by ≥3 nucleotides with the exception of PS-ODN 30:157–183, which was mismatched in only one position along the 32A-kDa protein gene transcript and in two positions along the 24-kDa protein gene transcript. We added the PS-ODNs at the beginning of the growth phase and evaluated cfu weekly for 6 wk. Each individual PS-ODN exerted a small but significant and reproducible inhibitory effect on M. tuberculosis growth; by the end of the 6-wk observation period, the growth of treated cultures was ≈0.25 log units below that of untreated cultures or cultures treated with a combination of three mismatched PS-ODNs to the 30-kDa protein gene transcript (Fig. 1a). The combination of all three PS-ODNs was slightly more effective (≈0.5 log units inhibition) than each individual PS-ODN, but still modest.
Fig 1.
Inhibition of M. tuberculosis growth by antisense PS-ODNs specific for the 30/32-kDa protein gene complex. Duplicate bacterial cultures were grown for 6 wk in 2 ml of 7H9 medium in the presence of antisense (PS-ODN) or mismatched PS-ODN (MM PS-ODN) at concentrations of 10 μM. For cultures with multiple PS-ODNs, each of the PS-ODNs or MM PS-ODNs was added at a concentration of 10 μM. All PS-ODNs were added once at the start of the observation period. At each time point, culture aliquots were harvested, washed, and plated on 7H11 agar medium. The cfu were enumerated after incubating plates for 2 wk. The maximal standard deviation for all of the data points was ≤6.7%. (a) PS-ODNs specific for various internal regions of the transcript of the 30-kDa protein gene. (b) PS-ODNs specific for an internal domain of each gene transcript of the mycolic acid transferase complex (30/32A/32B/24-kDa proteins). (c) PS-ODNs specific for the 5′ end of each gene transcript of the mycolic acid transferase complex (30/32A/32B/24-kDa proteins).
We considered the possibility that sense PS-ODNs against the 30-kDa protein gene may also exert a growth inhibitory effect on M. tuberculosis by promoting D-loop formation and hybridization to the antisense strand of a transcription bubble. The latter possibility was considered long ago (18) and was later shown to induce effective inhibition in an in vitro T7 transcription system (19). To investigate this possibility, we synthesized sense PS-ODNs to the same three regions of the 30-kDa protein gene studied above (30:157–183, 718–744, 871–897). Although the other genes of the 30/32-kDa protein gene complex show a high degree of sequence homology in the three selected regions, the presence of one to nine mismatches excluded stable hybridization of the PS-ODNs to the other genes. Surprisingly, none of the sense PS-ODNs had any effect on the growth of M. tuberculosis; nor did they interfere with the expression of the full-length 30-kDa protein molecule, because the amount of mature 30-kDa protein secreted into the culture supernatant fluid was not diminished compared with untreated cultures (data not shown).
Effect on M. tuberculosis Proliferation of Antisense PS-ODNs Directed Against Internal Sites of All Four 30/32/24-kDa Protein Complex Gene Transcripts.
We suspected that PS-ODNs against the 30-kDa protein gene transcript had only a modest inhibitory effect on M. tuberculosis growth because of the presence of uninhibited mycolyl transferase gene transcripts derived from the 32A- and 32B-kDa protein genes and possibly the 24-kDa protein gene. We therefore explored the inhibitory capacity of antisense PS-ODNs directed against all four members of the 30/32/24-kDa protein complex family. Each PS-ODN targeted a homologous internal site of one of these gene transcripts (30:267–290, 32A:276–299, 32B:279–302, and 24:225–248) and was 100% complementary to the mRNA of its target. We added the PS-ODNs once at the beginning of the growth phase and evaluated cfu weekly for 6 wk (Fig. 1b). Individually, the PS-ODNs targeting the 30/32-kDa complex exerted a small but significant and reproducible inhibitory effect of ≈0.35 log units on M. tuberculosis growth compared with untreated cultures or cultures treated with four mismatched PS-ODNs against the 30/32/24-kDa protein gene transcripts; however, the antisense PS-ODN specific for the 24-kDa protein gene transcript did not show any inhibitory activity. Combined, the three PS-ODNs against the 30/32-kDa complex exerted a growth inhibitory effect of ≈0.7 log units; the addition to this combination of the PS-ODN against the 24-kDa protein transcript did not further augment the combination's growth inhibitory capacity (Fig. 1b).
We attempted to replicate the effect of the three PS-ODNs directed against the 30/32-kDa protein complex gene transcripts with a single PS-ODN directed against an internal region of all three transcripts that had a high degree of sequence identity, such that the application of one PS-ODN would potentially lead to the inhibition of translation of all three mRNAs. The PS-ODN (32A:276–303) was 100% complementary only to the mRNA encoding the 32A-kDa protein, whereas it was mismatched in one position of the mRNAs encoding both the 30- and 32B-kDa proteins. The PS-ODN was mismatched in more than five positions to the mRNA encoding the 24-kDa protein, excluding any possibility of the PS-ODN hybridizing to this mRNA. Added once at the beginning of the growth phase, this PS-ODN exerted an inhibitory effect on M. tuberculosis growth of ≈0.5 log units, only slightly less than that of the combination of three perfectly matched PS-ODNs against each of the 30/32-kDa protein complex gene transcripts (data not shown).
Effect on M. tuberculosis Proliferation of PS-ODNs Targeting the 5′ Termini of the Mycolyl Transferase Complex Gene Transcripts.
We next explored the inhibitory capacity of PS-ODNs directed against the first 24 aa encoding nucleotides of each of the four mRNA transcripts of the mycolyl transferase complex genes. We hypothesized that PS-ODNs directed against this region may block the translation initiation complex and therefore be particularly effective. In addition, this region had a very low propensity to form a secondary structure. Individual PS-ODNs specific for the 30/32-kDa complex (30:1–24, 32A:1–24, and 32B:1–24) exerted a very significant inhibitory effect on M. tuberculosis growth of ≈0.6 log units (Fig. 1c). Combinations of two antisense PS-ODNs specific for any two of the three transcripts of the 30/32-kDa complex genes were at most only slightly more inhibitory than the single PS-ODNs (data not shown). However, when all three antisense PS-ODNs were administered in combination, the three PS-ODNs exerted a growth inhibitory effect of ≈1.7 log units compared with untreated cultures or cultures treated with four mismatched PS-ODNs against the 30/32A/32B/24-kDa protein complex gene transcripts. As in the previously described experiments (Fig. 1b), the antisense PS-ODN against the transcript coding for the 24-kDa protein (24:1–24) had little or no influence on bacterial proliferation either alone or in combination with the three PS-ODNs against the 30/32-kDa complex gene transcripts.
This experiment was remarkable because it showed the high potency of individual antisense PS-ODNs directed against the 5′ end of the 30/32-kDa protein gene transcripts and the remarkably high potency of a combination of antisense PS-ODNs against all three 30/32-kDa protein gene transcripts.
Effect of PS-ODNs Directed Against the 30/32-kDa Complex Genes on Protein Synthesis.
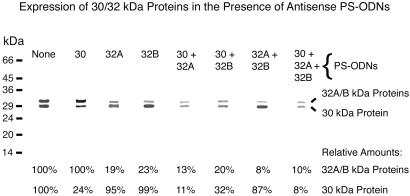
To determine whether the PS-ODNs directed against the mycolyl transferase genes inhibit expression of the encoded proteins, we assayed the amount of 30/32-kDa protein present in cultures treated with these PS-ODNs (Fig. 2). The PS-ODNs specifically inhibited protein synthesis by their target genes, i.e., a significant reduction in expression of the 30-kDa protein occurred only when a PS-ODN against the transcript of the 30-kDa protein gene was present, and a significant reduction in 32A/32B protein expression occurred only in the presence of a PS-ODN against one of the transcripts of these protein genes. The further reduction in protein expression observed in the presence of combinations of two or three PS-ODNs compared with single PS-ODNs may reflect a slightly greater inhibitory impact of the PS-ODN combinations on M. tuberculosis growth.
Fig 2.
Specific inhibition of protein synthesis by antisense PS-ODNs. Single or multiple PS-ODNs specific for the 5′ end of the gene transcripts of the mycolic acid transferase complex (30/32A/32B-kDa proteins), as indicated, were added to M. tuberculosis cultures at a concentration of 10 μM. Four weeks later, aliquots of the culture supernates were electrophoresed and immunoblotted, and the amount of 30/32-kDa complex protein present was quantitated densitometrically. Data below the immunoblot are the percentage of 30/32-kDa proteins present in each culture treated with one or more PS-ODNs compared with the amount of these proteins in an untreated culture.
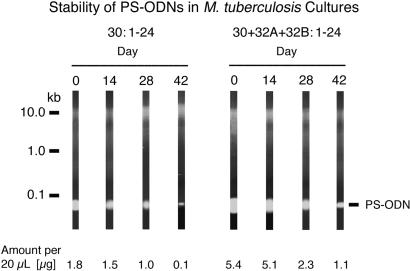
Stability of PS-ODNs in M. tuberculosis Cultures.
That PS-ODNs added once to M. tuberculosis cultures maintained their inhibitory capacity for 6 wk thereafter suggested that the PS-ODNs were fairly stable in the cultures. To explore the stability of PS-ODNs further, we added PS-ODNs to M. tuberculosis cultures and assayed the amount present initially and at 2-wk intervals thereafter up to 6 wk (Fig. 3). A two-phase decay curve was observed. During the first 4 wk (weeks 0, 2, and 4), the half-life was ≈23–25 days for either a single PS-ODN (30:1–24) or a combination of three PS-ODNs directed against the 5′ end of the transcripts of the 30/32-kDa protein genes. During the last 4 wk (weeks 2, 4, and 6), the half-life was ≈6 days for a single PS-ODN and ≈13 days for the combination of three PS-ODNs. The more rapid decay of the single PS-ODN during the final 4 wk of culture likely reflected the greater density of the M. tuberculosis culture treated with only one PS-ODN during this period, which might result in greater adsorption and uptake of the PS-ODN by the larger number of bacteria and greater degradation as a result of cell lysis and release of DNA degrading enzymes.
Fig 3.
Stability of PS-ODNs in M. tuberculosis cultures. A single mycolyl transferase-specific PS-ODN (Left) or three such PS-ODNs (Right) were added to M. tuberculosis cultures once at the start of a 6-wk incubation. Immediately and 14, 28, and 42 days later, 20-μl samples of the culture filtrates were electrophoresed on agarose gels, the gels were stained with ethidium bromide, and the amount of PS-ODN was quantitated densitometrically.
Effect on M. tuberculosis Proliferation of Weekly Administration of Mycolyl Transferase-Specific Antisense PS-ODNs.
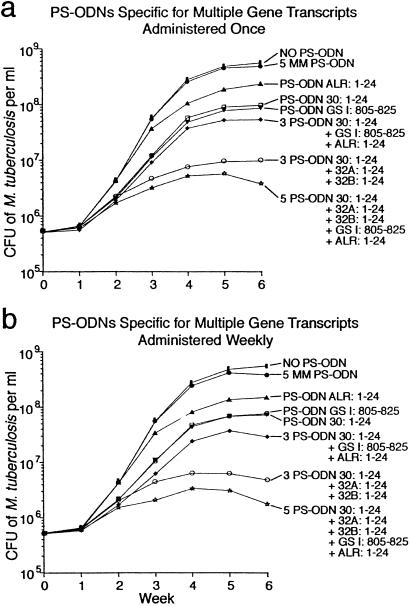
In studies described thus far, PS-ODNs were added only once at the start of the 6-wk observation period. To determine whether more frequent administration of PS-ODNs to bacterial cultures would result in greater growth inhibition, we compared the inhibitory capacity of PS-ODNs added once vs. weekly for 6 wk. For these studies, we used the highly effective PS-ODNs targeting the 5′ end of the 30/32-kDa protein gene transcripts (Fig. 4). When a single PS-ODN directed against the 30-kDa protein gene transcript was added to cultures at a concentration of 10 μM, weekly addition was more effective (≈1 log inhibition) than a one-time addition at the start of the 6-wk observation period (0.5–0.6 logs inhibition). Interestingly, weekly addition of a lesser concentration, either 1 or 5 μM, had minimal inhibitory capacity even though the cumulative dose (30 μM in the case of the 5 μM dose) exceeded 10 μM, the effective dose when the PS-ODN was added once at the start of the observation period. Similarly, when a combination of 3 PS-ODNs directed against the 30/32-kDa protein complex gene transcripts was added to cultures at a concentration of 10 μM, weekly addition was more effective than a one-time administration (2.3 logs inhibition vs. 1.7 logs inhibition). Weekly addition of a lower concentration of the 3 PS-ODNs (5 μM) was also more effective than a single administration (0.7 logs inhibition vs. 0.2 logs inhibition), but this dose was not nearly as effective as the 10-μM dose. Weekly addition of an even lower dose (1 μM) had no inhibitory effect (data not shown for 1- and 5-μM doses).
Fig 4.
Inhibition of cell proliferation of M. tuberculosis growth by antisense PS-ODNs specific for the gene transcripts of the mycolic acid transferase complex (30/32A/32B-kDa proteins), glutamine synthetase, and alanine racemase. PS-ODNs at a concentration of 10 μM were added once at the start of the observation period (a) or weekly for 6 wk (b). Growth inhibition was assayed in duplicate bacterial cultures as in the previous figures. The maximal standard deviation for all of the data points was 11.9%.
Effect on M. tuberculosis Proliferation of Modified PS-ODNs Specific for the Mycolyl Transferase Complex.
Two modifications were introduced in some of the PS-ODNs specific for the 30/32-kDa protein complex transcripts. (i) The PS-ODNs complementary to the 5′ terminal 24 aa-encoding bases of both the 30-kDa and 32A-kDa gene transcripts were modified such that the six 5′ and 3′ terminal nucleotides contained 2′-O-methyl ribose residues whereas the internal 12 bases remained unmodified with deoxyribose residues. (ii) The PS-ODN complementary to the 5′ terminal 24 aa-encoding bases of the 32A-kDa gene transcript was extended at the 3′ end beyond its standard length of 24 bases by 5 additional nonhybridizing G nucleotides. The first modification promotes PS-ODN stability and diminishes animal toxicity (14, 15). The second modification was postulated to enhance uptake of the PS-ODN based on evidence from eukaryotic systems, showing increased uptake of PS-ODNs modified by the terminal G residues (16). In general, both modifications only slightly changed the pattern of M. tuberculosis growth inhibition observed with the unmodified PS-ODN counterparts. Surprisingly, the 2′-O-methyl-modified PS-ODNs were slightly less effective (0.1–0.2 log units) than their unmodified counterparts, regardless of how the PS-ODNs were administered: individually or in combination, once or weekly. Interestingly, the G-tailed PS-ODN specific for the 32A-kDa gene transcript effected a slight increase in growth inhibition by ≈0.2 log units, possibly as a result of increased uptake into the bacteria. In the case of both types of modifications, mismatched PS-ODNs did not inhibit bacterial growth, just as was the case for their unmodified mismatched counterparts.
Effect on M. tuberculosis Proliferation of Additional, Non-Mycolyl Transferase Complex-Specific Antisense PS-ODNs.
To determine whether the addition of other PS-ODNs to M. tuberculosis cultures in combination with mycolyl transferase-specific PS-ODNs could further reduce bacterial growth, we studied the effect of adding a PS-ODN against the M. tuberculosis glnA1 transcript (GSI:805–825), previously shown to be inhibitory (11), and a PS-ODN against the alr transcript (alanine racemase, Rv3423c) (ALR:1–24) to the 30/32-kDa protein specific PS-ODN combination. As previously noted, glutamine synthetase is central to nitrogen metabolism and is evidently also involved in the synthesis of a poly-l-glutamate/glutamine cell wall structure. Alanine racemase is needed in the synthesis of the mycobacterial peptidoglycan chain, which contains l- and d-alanine. That these targets, like the mycolyl transferases, are involved in cell wall homeostasis might offer an additional advantage, because targeting them might theoretically “soften” the cell wall and promote the uptake of the other PS-ODNs. Used individually, the two PS-ODNs (GSI:805–825 and ALR:1–24) inhibited M. tuberculosis growth by 0.2–0.7 logs when added once (Fig. 4a) or weekly (Fig. 4b). When the two PS-ODNs were added in combination with a single PS-ODN directed against the 30-kDa protein gene transcript or in combination with three 30/32A/32B protein complex-specific PS-ODNs, growth inhibition was enhanced by 0.3–0.5 logs over the level obtained without them (Fig. 4). As previously observed, weekly addition of the various combinations of PS-ODNs tested was slightly more effective than a one-time addition at the start of the observation period.
Discussion
Our study demonstrates that the 30/32A/32B mycolyl transferase protein complex is a highly promising target for antimicrobial drug development by using antisense or more conventional technologies. We have studied >25 M. tuberculosis targets to date, and PS-ODNs against the mycolyl transferases have exerted by far the greatest inhibitory effect on M. tuberculosis growth, nearly achieving bacteriostasis of the cultures. As a drug target, the mycolyl transferase complex has several advantages. First, the enzymes are essential for mycobacterial cell wall synthesis, and the bacteria have no bypass mechanism. Second, the enzymes are unique to mycobacteria; hence, in contrast to more conserved molecules, drugs targeting them, including antisense PS-ODNs are less likely to crossreact with host molecules. Third, from the standpoint of antisense technology, each of the three genes encoding the enzymes is in a single gene unit. We have acquired considerable evidence that it is more difficult for a PS-ODN to bind to its cognate target if it is located in the middle of a multigene operon rather than at the beginning of it or in a single gene unit.
In the case of all three mycolyl transferase gene transcripts, PS-ODNs directed against the 5′ end of the transcript were clearly superior to PS-ODNs complementary to internal domains of the transcripts. Whereas more than one factor likely underlies the enhanced effectiveness of the PS-ODNs to the 5′ region, including the previously noted low propensity of the region and PS-ODNs targeting it to form a secondary structure, we postulate that a major factor is the PS-ODNs' capacity to block the translation initiation complex.
From the standpoint of antisense technology, it is encouraging that the effects of PS-ODNs against disparate targets were cumulative in our study. In the case of the mycolyl transferases, it seems likely that the greatly enhanced effect of targeting all three enzymes derives from preventing any one of them from compensating for the absence of another. However, the addition of PS-ODNs targeting the unrelated enzymes glutamine synthetase and alanine racemase to the PS-ODNs targeting the mycolyl transferases yielded still greater inhibition of M. tuberculosis growth. This finding suggests that large cocktails of antisense PS-ODNs targeting many different gene transcripts are likely to be even more effective. The use of multiple PS-ODNs should also minimize the likelihood of the mycobacteria developing resistance to this therapeutic strategy. Tuberculosis is typically treated with multiple antibiotics to prevent the emergence of resistant organisms. Because mycobacteria could potentially develop resistance to PS-ODN technology as a whole rather than to specific PS-ODNs (e.g., resistance to uptake of the PS-ODNs), it likely will be necessary to administer PS-ODNs in conjunction with conventional antibiotics. Along this line, in a previous study (11), we found that PS-ODNs could add to the inhibitory effect of conventional antibiotics.
A major challenge for antisense technology is to enhance their uptake into the target organism, in this case a Mycobacterium. In a previous study (11), we attempted to enhance uptake by “softening” up the mycobacterial cell wall with antibiotics. However, whereas the effects of the antibiotics and PS-ODNs were additive, no synergistic effect was observed. In the present study, we attempted to enhance uptake by modifications of the PS-ODNs. Adding 2′-O-methyl groups was unsuccessful, but adding a poly(G) tail slightly enhanced efficacy. In unpublished studies, we have tested a number of additional modifications of PS-ODNs. Adding a diaminooctane group to the 3′ terminus of a PS-ODN against glnA1 (805–825) slightly enhanced efficacy of the PS-ODN against M. tuberculosis. However, using tethers to add 1 to 5 amino groups to the 5′, 3′, or both terminal positions of the same PS-ODN, adding two to four modified deoxythymidines with an aminohexane linker to the 5′ position of this PS-ODN, or covalently attaching a lipophilic group to the 5′ end of this PS-ODN did not enhance its inhibitory capacity. Clearly, effective new approaches are needed for enhancing uptake of PS-ODNs into mycobacteria. Moreover, to be fully effective in vivo, PS-ODNs must also access the M. tuberculosis phagosome in host mononuclear phagocytes. In vivo studies in mice indicate that PS-ODNs are preferentially taken up by mononuclear phagocytes (20); however, the extent of their delivery to the M. tuberculosis phagosome is not known. Whereas these and other technical obstacles to the utilization of antisense technology remain to be surmounted, the present results hold promise for the therapeutic use of antisense technology in the treatment of tuberculosis in the future.
Acknowledgments
This work was supported by National Institutes of Health Grant AI 42925 and a grant from the G. Harold and Leila V. Mathers Foundation.
Abbreviations
PS-ODN, phosphorothioate-modified oligodeoxyribonucleotide
cfu, colony-forming units
glnA1, glutamine synthetase I
References
- 1.Pablo-Mendez A., Raviglione, M. C., Laszlo, A., Binkin, N., Rieder, H. L., Bustreo, F., Cohn, D. L., Lambregts-van Weezenbeek, C. S. B., Kim, S. J., Chaulet, P. & Nunn, P. (1998) N. Engl. J. Med. 338 1641-1649. [DOI] [PubMed] [Google Scholar]
- 2.Belisle J. T., Vissa, V. D., Sievert, T., Takayama, K., Brennan, P. J. & Besra, G. S. (1997) Science 276 1420-1422. [DOI] [PubMed] [Google Scholar]
- 3.Ronning D. R., Klabunde, T., Besra, G. S., Vissa, V. D., Belisle, J. T. & Sacchettini, J. C. (2000) Nat. Struct. Biol. 7 141-146. [DOI] [PubMed] [Google Scholar]
- 4.Anderson D. H., Harth, G., Horwitz, M. A. & Eisenberg, D. (2001) J. Mol. Biol. 307 671-681. [DOI] [PubMed] [Google Scholar]
- 5.Harth G., Lee, B.-Y., Wang, J., Clemens, D. L. & Horwitz, M. A. (1996) Infect. Immun. 64 3038-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee B.-Y. & Horwitz, M. A. (1995) J. Clin. Invest. 96 245-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armitige L. Y., Jagannath, C., Wanger, A. R. & Norris, S. J. (2000) Infect. Immun. 68 767-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson M., Raynaud, C., Lanéelle, M. A., Guilhot, C., Laurent-Winter, C., Ensergueix, D., Gicquel, B. & Daffé, M. (1999) Mol. Microbiol. 31 1573-1587. [DOI] [PubMed] [Google Scholar]
- 9.Horwitz M. A., Lee, B.-W. E., Dillon, B. J. & Harth, G. (1995) Proc. Natl. Acad. Sci. USA 92 1530-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horwitz M. A., Harth, G., Dillon, B. J. & Maslesa-Galic, S. (2000) Proc. Natl. Acad. Sci. USA 97 13853-13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harth G., Zamecnik, P. C., Tang, J.-Y., Tabatadze, D. & Horwitz, M. A. (2000) Proc. Natl. Acad. Sci. USA 97 418-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Padmapriya A. A., Tang, J. & Agrawal, S. (1994) Antisense Res. Dev. 4 185-199. [DOI] [PubMed] [Google Scholar]
- 13.Metelev V., Lisziewicz, J. & Agrawal, S. (1994) Bioorg. Med. Chem. Lett. 4 2929-2934. [Google Scholar]
- 14.Agrawal S., Tan, W., Cai, Q., Xie, X. & Zhang, R. (1997) Antisense Nucleic Acid Drug Dev. 7 245-249. [DOI] [PubMed] [Google Scholar]
- 15.Agrawal S., Iadarola, P. L., Temsamani, J., Zhao, Q. & Shaw, D. R. (1996) Bioorg. Med. Chem. Lett. 6 2219-2224. [Google Scholar]
- 16.Agrawal S. & Zhao, Q. (1998) Antisense Nucleic Acid Drug Dev. 8 135-139. [DOI] [PubMed] [Google Scholar]
- 17.Cole S. T., Brosch, R., Parkhill, L., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S., Barry, C., III, et al. (1998) Nature 393 537-544. [DOI] [PubMed] [Google Scholar]
- 18.Zamecnik P. C. & Stephenson, M. L. (1978) Proc. Natl. Acad. Sci. USA 75 280-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Temsamani J., Metelev, V., Levina, A., Agrawal, S. & Zamecnik, P. C. (1994) Antisense Res. Dev. 4 279-284. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Q., Zhou, R., Temsamani, J., Zhang, Z., Rosky, A. & Agrawal, S. (1998) Antisense Nucleic Acid Drug Dev. 8 451-458. [DOI] [PubMed] [Google Scholar]