Abstract
We have examined the progression of hepatitis C virus (HCV) infections by gene expression analysis of liver biopsies in acutely infected chimpanzees that developed persistent infection, transient viral clearance, or sustained clearance. Both common responses and outcome-specific changes in expression were observed. All chimpanzees showed gene expression patterns consistent with an IFN-α response that correlated with the magnitude and duration of infection. Transient and sustained viral clearance were uniquely associated with induction of IFN-γ-induced genes and other genes involved in antigen processing and presentation and the adaptive immune response. During the early stages of infection, host genes involved in lipid metabolism were also differentially regulated. We also show that drugs that affect these biosynthetic pathways can regulate HCV replication in HCV replicon systems. Our results reveal genome-wide transcriptional changes that reflect the establishment, spread, and control of infection, and they reveal potentially unique antiviral programs associated with clearance of HCV infection.
Hepatitis C virus (HCV) infection is a rapidly increasing public health problem, with an estimated 200 million chronically infected patients worldwide (1). No vaccines are currently available for HCV, and only a subset of HCV patients responds to IFN-α and Ribavirin treatment (1). Despite substantial research, the mechanisms of viral infection, persistence, and clearance are not well understood, primarily because of the absence of readily manipulatable animal models and cell culture systems. Although HCV replicons (2, 3) are currently being used as cell-based models to study HCV replication, they do not serve as models for HCV infection. Because chimpanzees can be infected by HCV, these studies provide a unique opportunity to track the virological, immunological, pathological, and genomic changes in vivo prospectively during infection.
Previous genomics studies have advanced our understanding of hepatitis C infections by characterizing the transcriptional changes associated with a clearance episode in an infected chimpanzee (4) and by characterizing the differences in gene expression between HCV and hepatitis B virus in chronically infected patients (5). In this study, we have examined the host response to HCV in three infected chimpanzees with different outcomes of infection, using gene expression analysis at multiple time points during the course of infection. Our goal was to identify and correlate the hepatic gene expression profiles in each of these animals to gain insight into the cellular and molecular mechanisms that determine the outcome of HCV infection.
Materials and Methods
HCV Infections.
Chimpanzee 1590 (Ch1590) was transfected intrahepatically with RNA transcribed from a plasmid DNA consisting of a full-length copy of the H77 clone (genotype 1a) lacking the hypervariable region 1 of HCV (6). Ch96A008 was inoculated intravenously with 90 ml of plasma from Ch1590 taken 4 wk after inoculation, containing a total of 900 genome equivalents of HCV (6). Ch1581 was inoculated intravenously with a serum pool containing one chimpanzee infectious dose of HCV (genotype 1a) from a chimpanzee that had been inoculated intravenously with serum from a patient with acute fulminant HCV infection (7). At baseline and each week after inoculation, needle liver biopsies and blood samples were obtained. HCV RNA was detected in chimpanzee serum as described (8) by using the RT-PCR with primers targeting the 5′-untranslated region of the HCV genome (HCV-Monitor Amplification KIT, Roche Diagnostics, Version 2.0). The details of these infections are described in a separate publication (8). Chimpanzees were handled according to humane use and care guidelines specified by the Animal Research Committees at the National Institutes of Health and the Scripps Research Institute.
Gene Expression Analysis.
For selected time points in each chimpanzee (Fig. 1, asterisks), liver-derived cRNA was prepared according to standard protocols and hybridized to high-density oligonucleotide arrays (Affymetrix U95A Human GeneChips), which interrogate the expression of ≈9,000 unique human genes. Each sample was hybridized in duplicate. Primary image analysis was performed by using genechip version 3.1 (Affymetrix, Santa Clara, CA) and each chip was scaled to an average difference value of 200. Genes whose average difference value did not exceed 150 in any sample, and genes that were not called “present” by both replicates in at least one time point were excluded from further analysis. Average difference values less than 1 were clipped, and all values were logarithm-transformed. Genes were identified whose expression was correlated (positively or negatively, by using Pearson's correlation coefficient) with the pattern of log-transformed HCV viremia. To address duplicate measurements at each time point, correlations were averaged over all combinations of replicates (if <300 total combinations) or sampled from the set of all possible combinations (300 iterations). For genes that correlated with HCV RNA levels over all time points across all chimpanzees, a background distribution of correlation coefficients was calculated by randomization of sample labels, from which significance values were empirically calculated; P < 0.05 was set as the threshold for significance. For shorter patterns, calculation of meaningful P-values by randomization was not possible, so correlation thresholds were set at 0.7 and −0.7 for positive correlation and negative correlation, respectively.
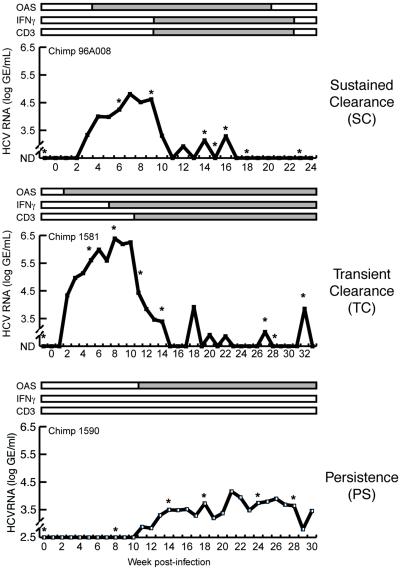
Fig 1.
The time course of infection for the three chimpanzees is shown. The x axis indicates the week postinfection at which a liver biopsy was taken. The y axis shows log HCV genome equivalents per milliliter of serum (GE/ml). Values below the limit of detection are shown as not detected (ND). Levels of oligoadenylate synthase, IFN-γ, and CD3 mRNA were also determined by RNase protection assays (8), and their presence in the liver is shown by the gray bars. In this study, gene expression profiling was performed on the biopsy samples at time points indicated by asterisks.
HCV Replicon Studies.
In vitro transcribed RNA was prepared from ScaI linearized pHCVrep1bBB7 (S1179I) plasmid (3) and electroporated into Huh-7 cells as described previously (9). Forty-eight hours after transfection, 500 μg/ml G418 was added to the culture medium. G418-resistant colonies were visible after 3 wk, and individual clones were isolated, expanded, and characterized. The presence of HCV RNA replication in Huh-7 S1179I clones was confirmed by Northern blot analysis for HCV RNA minus strands and 5-bromouridine 5′-triphosphate labeling. Huh-7 cells containing the HCV subgenomic replicon S1179I were grown in DMEM plus 500 μg/ml G418. After 3 days of cultivation, the G418 was removed, and the monolayers were treated with cerulenin (0, 4.5, 22.4, 44.8 μM), 25-hydroxycholesterol (0, 2.5, 6.25 μM), or nystatin (0, 16.2, 32.4 μM). Total RNA was extracted at several time points with Trizol reagent, and RNA samples (10 μg per lane) were separated by denaturing agarose gel electrophoresis. HCV plus strand RNA was detected by Northern blot analysis as described previously (3). In vitro transcribed replicons (109, 108, 107 copies) were serially diluted in total RNA from Huh-7 cells and analyzed in parallel. The effects of cerulenin, 25-hydroxycholesterol, and nystatin on Huh-7 cell cycling was analyzed by flow cytometry following standard procedures. Briefly, Huh-7 cells stably replicating the HCV subgenomic replicon S1179I were treated with cerulenin, 25-hydroxycholesterol, and nystatin at the indicated concentrations for 18 h. The cells were collected by trypsinization, fixed with ethanol, and stained with propidium iodide. The data were collected on a FACScan flow cytometer and analyzed by using the modfit lt software (Verity Software House, Topsham, ME).
A selectable replicon expressing firefly luciferase was generated from the pFK-I389/Neo/NS3–5B/NK5.1 plasmid (10), which was modified by restriction enzyme digestion/ligation at a unique site near the start site for translation of the neomycin gene. The corresponding sites were introduced, along with a unique sequence separating the luciferase and neomycin genes, to the luciferase gene at the 5′ and 3′ ends by PCR. The modification was confirmed by sequencing. The modified construct was then linearized with ScaI and in vitro transcribed (3, 10). Replicon RNA was transfected into Huh-7 cells by using a liposome-mediated transfection procedure (Lipofectin, GIBCO/BRL). Typically, 30 μl of Lipofectin was diluted into 400 μl of Optimem (GIBCO/BRL), incubated for 1 h, followed by addition of 5 μg of RNA that was incubated for an additional 15 min. Then 8.5 μl of the RNA–Lipofectin mixture was added to each well of a 96-well culture plate and incubated for 12 h, after which the transfection mixture was removed from the cells, and the cells were washed twice with PBS and grown in DMEM. Expression of HCV proteins was confirmed by Western blot analysis by using an NS3 antibody (gift from Chiron). After 4–8 h of recovery, the cells were treated with cerulenin, 25-hydroxycholesterol, or nystatin, with final concentrations of drug ranging from 0 to 50 μM. Luciferase assays were performed as described (10). Experiments were performed in triplicate. Typical measurements of luciferase activity were 3,000–5,000 light units/well for untreated cells.
Results and Discussion
Previously, three chimpanzees were infected with HCV, each of which exhibited a different outcome of infection (Fig. 1) (8). Ch96A008 successfully cleared the virus (SC) in the context of a vigorous intrahepatic immune response and IFNγ induction (8). Ch1581 became persistently infected but initially transiently cleared (TC) the virus also in the context of an intrahepatic immune response and IFN-γ induction, and subsequently controlled HCV at titers several orders of magnitude below initial peak levels (8). Ch1590 developed an unrestrained persistent HCV infection (PS) in the absence of both an intrahepatic antiviral immune response and IFN-γ induction (8). In this study, gene expression profiling was performed on selected samples from the three chimps during the time course of infection (Fig. 1).
To determine the set of genes whose expression most likely reflects the initial host response to HCV in the liver, we identified genes with expression patterns that strongly correlated with the amount of HCV RNA in the serum of all of the chimps over the entire time course profiled. We calculated the Pearson's correlation between the expression of each gene and the amount of HCV RNA at each time point in all chimpanzees. The P-values for correlation coefficients were estimated by randomizing the time-point labels.
We found 27 unique transcripts with highly significant correlation coefficients (P < 0.05) (Fig. 2A). These genes were induced with an average peak fold-change (FC) value of 8.8 (the primary data for all of the genes shown in Fig. 2 are published as Tables 1–6 in the supporting information on the PNAS web site, www.pnas.org). Consistent with the findings of Bigger et al. (4), expression of many of these genes is known to be stimulated by IFN-α. IFN-α-stimulated gene expression is an important component of the innate immune response to many viruses (11), including HCV (4, 8), and is mediated by binding of signal transducer and activator of transcription (STAT) heterodimers to IFN-stimulated response element (ISRE) sites (12). We therefore searched the upstream promoter regions of 20 of these genes, where promoter sequences could be retrieved from the human genome assembly for ISRE motifs (12). Nineteen genes, including STAT1, 2′-5′ oligoadenylate synthase, and Mx1, contained at least one probable ISRE motif in their promoter region (Tables 7 and 8, which are published as supporting information on the PNAS web site); all are well known to exert antiviral activity through inhibition of translation, activation and repression of transcriptional activity, and mRNA degradation (13). A complete list of gene names and raw data is available in Table 2. In some viral infections, for example encephalomyocarditis virus and vesicular stomatitis virus, the IFN-α-mediated response contributes to viral clearance (14). However, viruses such as poxviruses and papilloma viruses are able to evade the antiviral effects of IFN-α by blocking the induction and/or function of its targets (11, 15). Here, we show that HCV-infected cells successfully induce the transcription of many antiviral IFN-α-stimulated genes, but that this response has little or no effect on viral titer or outcome, possibly acting via HCV-induced mechanisms downstream of IFN-α-stimulated transcription (16).
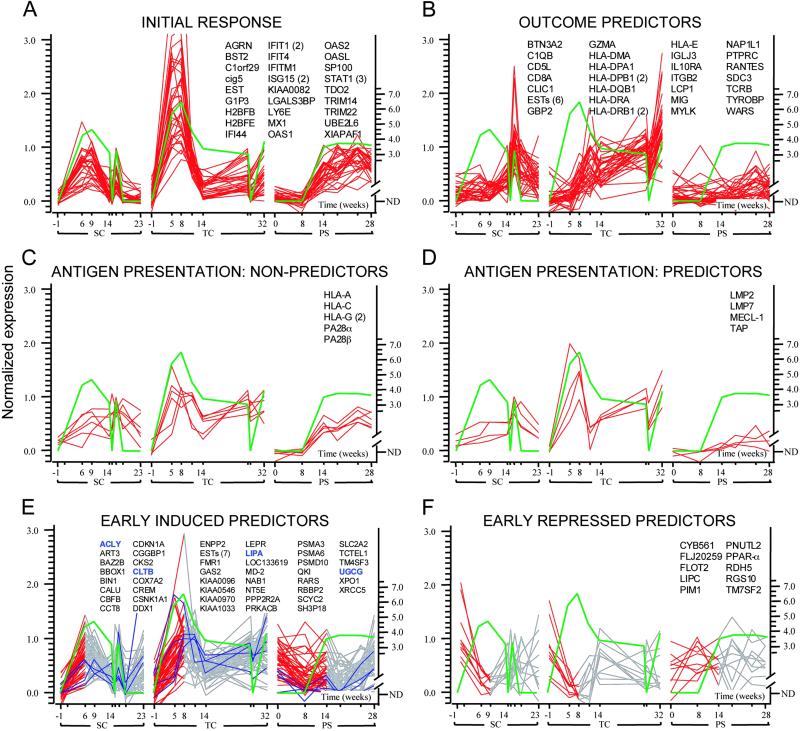
Fig 2.
Genes coordinately regulated during HCV infections in SC, TC, and PS. Green lines represent log (HCV RNA) patterns with values below the threshold for detection represented by zero. Data traces are shown for different clusters of genes: (A) genes correlating with HCV RNA in all chimpanzees; (B) genes that are negatively correlated with HCV RNA levels during clearance episodes and correlated with IFN-γ levels and T cell influx to the liver (Fig. 1); (C) MHC class I and proteasome nonpredictor genes; (D) antigen presentation-related genes that show outcome-specific expression; (E) early predictors correlated with increasing HCV RNA levels with representative hepatocellular early predictor genes associated with lipid metabolism highlighted in blue; and (F) early predictors negatively correlated with increasing HCV RNA levels. For the early predictor genes in E and F, the portion of the gene expression pattern depicted in red is the region that positively or negatively correlates with the initial rise of HCV RNA. If multiple probes interrogate the same gene, the probe count appears in parentheses. For optimal visualization, gene expression levels were normalized to the 10th and 90th percentiles for each gene. Values on the x axis represent days after inoculation with HCV. The primary data for all clusters can be found in Table 1, which is published as supporting information on the PNAS web site.
To identify genes whose expression patterns were predictive of the outcome of infection, we searched for genes that were induced during declining viremia in SC/TC and not in PS and were correlated with the pattern of IFN-γ and HCV-specific T cell infiltrates (Fig. 1) (8). We identified 27 unique genes and six EST clusters (Fig. 2B) that are highly induced in SC/TC (average peak FC = 12.7) and not in PS (average peak FC = 2.0), many of which are known to be induced by IFN-γ and to be expressed as a result of the homing and activation of immune cells to the liver (17). Although induction of IFN-γ itself is not detected in this microarray analysis because of its low expression levels, the IFN-γ-induced transcriptional effects are readily apparent. IFN-γ activates transcription by causing STAT1 homodimers to bind to γ activation sites (GAS) (17); probable GAS motifs were present in 17 of the 17 genes for which promoter sequences were available (Tables 9 and 10, which are published as supporting information on the PNAS web site). This cluster of genes is likely to be directly related to outcome of infection, because they are known to play a role in the activation of the immune system, recruitment of T cells, and antigen processing and presentation. Among the genes are CD8, several MHC Class II components, and the chemokines RANTES and MIG (Fig. 2B), as well as IP9 and IP10, which show slightly different kinetics (data not shown). The mRNA levels of granzyme A also increased with declining HCV RNA levels during clearance in an outcome-specific fashion. Granzyme A is serine protease that is delivered to the cytosol of infected cells by natural killer cells and cytotoxic T lymphocytes, where it induces antiviral and apoptotic pathways (18). That these genes are not induced in PS is consistent with the absence of a T cell response in that animal (8).
To be recognized by virus-specific T cells, infected hepatocytes must process, transport, and present HCV peptides in the context of MHC class I molecules. The process of antigen presentation involves proteasome subunits that cleave HCV proteins into antigenic peptides, transporter proteins that allow peptide entry into the endoplasmic reticulum, and MHC Class I molecules that bind the antigen for presentation (19). Given the importance of these genes in facilitating an adaptive immune response, we examined the expression of these genes in our profiling and found that most appear to be coordinately regulated. Some of these genes are induced in all chimpanzees regardless of outcome, including MHC class I subunits and proteasome activator subunits (PA28 α/β) (Fig. 2C), presumably reflecting their regulation by IFN-α/β. However, several other genes in this pathway are selectively induced in SC/TC, including the transporter associated with antigen processing (TAP) and the three unique subunits of the immunoproteasome (LMP2, LMP7, and MECL1), which are specifically involved in generating peptides for antigen presentation (Fig. 2D) (19). Although the kinetics of induction differ between the SC and TC chimpanzees, the level of induction in both of these animals (average FC = 4.6) far exceeds the flat pattern of expression in PS, presumably reflecting the differential induction of IFN-γ in these animals (8).
The differential regulation of immunological genes involved in antigen processing and T cell recruitment between SC/TC and PS suggests that transcriptional activation of the adaptive immune system is required for a successful outcome to HCV infection. These observations are consistent with previous studies, which show that the outcome of HCV infection is related to the magnitude and diversity of the cellular immune response and the ability of virus-specific T cells to enter the liver (8, 20). The induction of antigen processing and presentation genes within the liver ultimately controls how much HCV antigen is presented to the cells of the immune system and, together with the induction of T cell recruitment genes (e.g., MIG, RANTES, IP10), likely controls the strength and multiplicity of the intrahepatic T cell response. Clearly, IFN-γ secretion by intrahepatic lymphocytes contributes to and strengthens both the innate and adaptive immune responses to HCV and increases expression levels of genes critical to antigen processing by infected hepatocytes. Our results suggest that HCV avoids recognition by the immune response in PS either by keeping HCV replication below a threshold level required for gene induction or perhaps by disrupting the induction of genes that control antigen processing and T cell recruitment (11).
In contrast to studies of patients infected with HCV (5), we are able to examine gene expression changes at very early time points after infection. Although the genes described above are indicative of the immune response to HCV infection, the early gene expression changes in the initial weeks after infection may reflect virus-induced host cell responses that regulate HCV replication. Furthermore, genes that are differentially induced or repressed in SC/TC versus PS early in viral infection may represent prognostic markers of outcome. We found 45 genes and 7 EST clusters whose expression was positively correlated with the onset of viremia selectively in SC/TC (average FC = 3.9) (Fig. 2E) and 10 genes that were negatively correlated with the initial rise in HCV RNA levels selectively in the same animals (average FC = −3.9) (Fig. 2F). Many of the genes in both of these groups are associated with lipid metabolism. For example, peroxisome proliferator activated receptor α (PPAR-α) was down-regulated preferentially in SC/TC during the increase in viremia (Fig. 2F). PPAR-α modulates target genes encoding lipid metabolism enzymes, lipid transporters, or apolipoproteins, and is generally involved in lipid and glucose homeostasis (21). Flotillin 2 also shares this pattern of expression; this protein is an integral component of lipid rafts, membrane structures that are involved in vesicular trafficking and signal transduction. Transcription of hepatic lipase C is also repressed early in SC/TC; this enzyme is involved in the hydrolysis of lipoprotein triglycerides and phospholipids and may mediate interactions of lipoproteins with cell surface receptors.
We also identified many genes that are selectively up-regulated in SC/TC during the early onset of viremia, several of which are again related to lipid metabolism (Fig. 2E). The gene encoding UDP-glucose ceramide glucosyltransferase (UGCG) is induced early selectively in SC/TC. This gene is involved in membrane glycosphingolipid biosynthesis. Expression of UGCG can influence viral infectivity; for example, inhibitors of UGCG have previously been shown to reduce infectivity of HIV-1 (22). Other lipid metabolism genes related to the serum response element-binding protein (SREBP) signaling pathway were induced during the initial rise in viremia preferentially in SC/TC (Fig. 2E). Lipase A is involved in hydrolysis of cholesterol esters and triglycerides, which are known to be involved in cell membrane structure. ATP citrate lyase, showing similar induction, is responsible for generating the cytosolic acetyl CoA required for both cholesterol and fatty acid synthesis and is induced by SREBP (23). Interestingly, the fatty acid synthase (FAS) gene is expressed at higher levels in SC/TC, although the pattern is not well correlated with HCV RNA levels (data not shown). Several other genes associated with the SREBP pathway display similar expression patterns, including the SREBP-cleavage activated protein (23).
The roles in the HCV life cycle of nonimmunological genes that we have identified to be positively or negatively correlated with HCV RNA in SC/TC (Fig. 2 E and F) are not established. It is possible that these genes play a role in HCV replication or represent an early hepatic response that contributes to viral clearance. The functional cluster of lipid metabolism genes in our profiling studies is particularly interesting in the context of several previous studies. In particular, HCV core protein has been shown to negatively affect microsomal triglyceride transfer protein activity and the assembly and secretion of very low density lipoproteins (24), HCV RNA-containing capsids are contained within triglyceride-rich lipoprotein particles in the plasma of infected patients (25), HCV is known to cause the formation of hepatocellular lipid droplets (steatosis) onto which HCV structural proteins (26) and nonstructural proteins (27) have been shown to localize, and the induction of fatty acid and sterol biosynthesis via SREBP activation in liver and adipose tissues can also cause steatosis (28). In addition, expression of the entire HCV polyprotein in tissue culture cells induces morphological alterations of the cellular membranes leading to the formation of a membranous web. Because both structural and nonstructural proteins associate with the membranous web, it has been proposed to be the scaffolding on which viral replication occurs (29).
Because of the prominent association of lipid metabolism genes with the early onset of HCV viremia, we decided to examine the extent to which alterations in lipid metabolism might affect HCV replication. To test whether the SREBP pathway affects HCV replication, we examined the effects of several small molecules that are known to modulate this pathway on the replication of two subgenomic HCV replicons with different adaptive mutations (3, 10). Treatments were carried out in Huh-7 cells stably transfected with replicon RNA that had been transcribed from pHCVrep1bBB7 (S1179I) (3), and from Huh-7 cells transiently transfected with another replicon transcribed from a plasmid constructed from pFK-I389/Neo/NS3–5B/NK5.1 (10) containing a firefly luciferase-neomycin fusion protein controlled by the HCV IRES. Huh-7 cells harboring these replicons were treated with cerulenin, a FAS inhibitor; with 25-hydroxycholesterol, which inhibits cleavage and activation of the cytosolic portion of SREBP that in turn translocates to the nucleus to initiate transcription of genes associated with fatty acid and cholesterol biosynthesis (23); and with nystatin, a cholesterol sequestering compound that induces the SREBP pathway (as confirmed by Northern blot of SREBP-responsive genes HMG-CoA reductase and acetyl-CoA carboxylase after nystatin treatment; data not shown). The proteasome inhibitor MG132 was used as a positive control as it is known to inhibit HCV IRES-dependent translation (30).
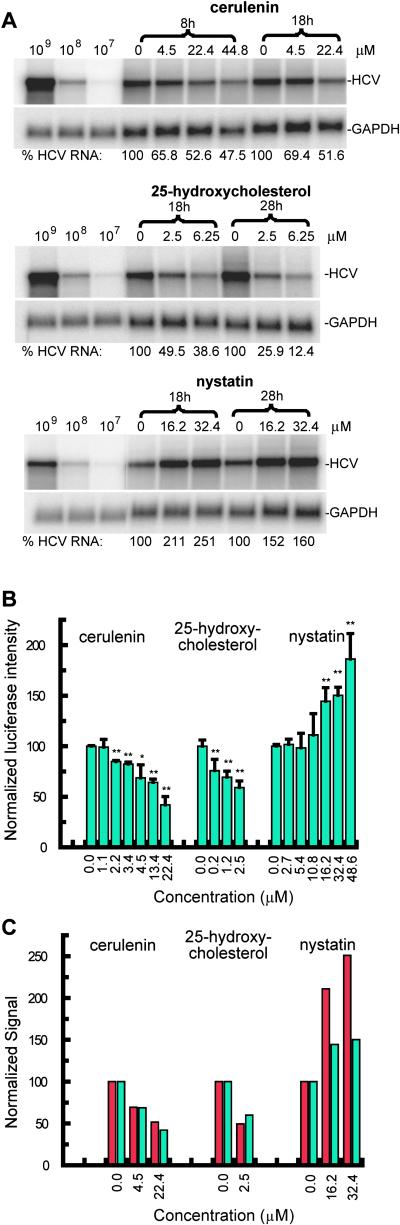
As shown in Fig. 3, cerulenin and 25-hydroxycholesterol, both of which block fatty acid biosynthesis, inhibited HCV replication in a dose-dependent manner at nontoxic concentrations [deduced indirectly from GAPDH levels and directly by MTS assays for cytotoxicity (Promega); data not shown] in both replicon systems as determined by Northern blot (Fig. 3A) and luciferase assays (Fig. 3B). Replication was inhibited by nearly 90% by 25-hydroxycholesterol after treatment at 6.25 μM for 28 h. In contrast, nystatin, which may activate the SREBP pathway through cholesterol sequestration, caused a dose-dependent increase in replication levels by nearly 100% compared with untreated cells (Fig. 3). Because the replicon systems contain IRES elements from both HCV and encephalomyocarditis virus (EMCV), we screened the regulatory activity of the same compounds in cells transfected with bicistronic plasmids that express reporter genes under the control of either HCV or EMCV IRES elements separately (30). These results confirmed that the effects of the drugs on the replicons are not specifically due to the presence of EMCV IRES (data not shown). In addition, to confirm that these effects do not simply reflect changes in cell replication that are known to influence the replication rate of the HCV replicon (31), cell counts and cell cycle analysis were performed on Huh-7 cells before and after treatment with cerulenin, 25-hydroxycholesterol, and nystatin. At all concentrations tested, none of the compounds altered the distribution of cells in the cell cycle compared with control cells (data not shown). Activation of SREBP increases both fatty acid and cholesterol biosynthesis (23). Both cerulenin, which directly inhibits FAS, and 25-hydroxycholesterol, which inhibits SREBP cleavage/translocation, cause a decrease in fatty acid biosynthesis. On the other hand, nystatin may cause an increase in fatty acid biosynthesis through cholesterol depletion, which may induce the cleavage/translocation of SREBP as a homeostatic response. These results demonstrate that up-regulation of genes specifically associated with fatty acid biosynthesis have a positive effect on the HCV replicons, suggesting that they may have a similar effect on HCV replication. The direct regulation of these genes, particularly those related to fatty acid biosynthesis, by HCV may also have implications to the development of steatosis in chronically infected patients (24–26, 28).
Fig 3.
Effects of drugs that affect cholesterol/lipid metabolism on HCV replicons. (A) Regulation of plus strand HCV replicon RNA levels after treatment of an Huh-7 cell line stably transfected with RNA derived from the pHCVrep1bBB7 (S1179I) plasmid. (B) Results of luciferase assays 18 h after treatment of transiently transfected Huh-7 cell with in vitro transcribed RNA from a modified version of the pFK-I389/Neo/NS3–5B/NK5.1 plasmid with cerulenin, 25-hydroxycholesterol, or nystatin at the indicated concentrations. Measurements were done in triplicate and error bars are shown with asterisks indicating statisitical significance (*, P < 0.05; **, P < 0.01). (C) Direct comparison of the effects of drugs on the different replicons. The data for the HCV subgenomic replicon S1179I are in red and those for the replicon based on pFK-I389/Neo/NS3–5B/NK5.1 are in blue.
Conclusion
Genome-wide transcriptional analyses of HCV infections in chimpanzees suggest that the induction of both immunological and hepatocellular genes can influence the course and outcome of the infection. Of the genes that are differentially regulated in SC/TC, specific genes that are related to the induction of IFN-γ, antigen processing and presentation, and T cell recruitment correlate with viral clearance and with the strength and multiplicity of the intrahepatic T cell response. Although these results are derived from a limited number of animals, the data suggest that higher viral titers may result in the induction of immunological genes that lead to a successful adaptive immune response. In contrast, virus-induced genes that reflect the induction of IFN-α do not appear to inhibit viral replication. Several genes that were differentially regulated early in the infection in animals that permanently or transiently cleared the virus may represent host genes that are essential for either the HCV life cycle or viral clearance. Many of these early predictors are known to be involved in lipid metabolism. We have shown that small molecules that perturb lipid metabolism, fatty acid biosynthesis, and SREBP signaling can specifically modulate HCV replication in replicon systems. These results illustrate a hitherto unappreciated critical role of cellular lipid metabolism in the HCV life cycle and demonstrate that genome-wide transcriptional analysis can yield insight into the host–virus interaction during HCV infection. The ability of small molecules that block FAS and SREBP activity to inhibit HCV replication represents new insight into the HCV life cycle that might be exploited for the development of novel antiviral drugs.
Supplementary Material
Acknowledgments
We thank R. Bartenschlager (Johannes Gutenberg University, Mainz, Germany) and C. Rice (The Rockefeller University) for providing materials for HCV replicon studies; M. Krüger (Medizinische Hochschule, Hannover, Germany) for providing dicistronic plasmids for IRES screening experiments; L. G. Guidotti for assistance with RNA preparation; and G. Hampton, D. Lockhart, K. Kuhen, N. Gekakis, and Z. Zhang for useful discussions during the course of this work. This work was supported by National Institutes of Health Grants AI20001 (to F.V.C.) and CA 76403 (to F.V.C.) and Novartis (P.G.S.). This is manuscript No. 15053-CH from The Scripps Research Institute.
Abbreviations
HCV, hepatitis C virus
Chn, chimpanzee n
SC, successfully cleared
TC, transiently cleared
PS, unrestrained persistent HCV infection
FC, fold change
ISRE, IFN-stimulated response elements
SREBP, serum response element-binding protein
IRES, internal ribosome entry site
STAT, signal transducer and activator of transcription
References
- 1.Hagedorn C. H. & Rice, C. M., (1999) Current Topics in Microbiology and Immunology (Springer, Berlin).
- 2.Bartenschlager R. & Lohmann, V. (2001) Antiviral Res. 52 1-17. [DOI] [PubMed] [Google Scholar]
- 3.Blight K. J., Kolykhalov, A. A. & Rice, C. M. (2000) Science 290 1972-1974. [DOI] [PubMed] [Google Scholar]
- 4.Bigger C. B., Brasky, K. M. & Lanford, R. E. (2001) J. Virol. 75 7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honda M., Kaneko, S., Kawai, H., Shirota, Y. & Kobayashi, K. (2001) Gastroenterology 120 955-966. [DOI] [PubMed] [Google Scholar]
- 6.Forns X., Thimme, R., Govindarajan, S., Emerson, S. U., Purcell, R. H., Chisari, F. V. & Bukh, J. (2000) Proc. Natl. Acad. Sci. USA 97 13318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farci P., Munoz, S. J., Shimoda, A., Govindarajan, S., Wong, D. C., Coiana, A., Peddis, G., Rubin, R. & Purcell, R. H. (1999) J. Infect. Dis. 179 1007-1011. [DOI] [PubMed] [Google Scholar]
- 8.Thimme R., Bukh, J., Spangenberg, H. C., Wieland, S., Pemberton, J., Steiger, C., Govindarajan, S., Purcell, R. H. & Chisari, F. V. (2002) Proc. Natl. Acad. Sci. USA 99 15661-15668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lohmann V., Korner, F., Koch, J. O., Herian, U., Theilmann, L. & Bartenschlager, R. (1999) Science 285 110-113. [DOI] [PubMed] [Google Scholar]
- 10.Krieger N., Lohmann, V. & Bartenschlager, R. (2001) J. Virol. 75 4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guidotti L. G. & Chisari, F. V. (2001) Annu. Rev. Immunol. 19 65-91. [DOI] [PubMed] [Google Scholar]
- 12.Levy D. E., Kessler, D. S., Pine, R., Reich, N. & Darnell, J. E., Jr. (1988) Genes Dev. 2 383-393. [DOI] [PubMed] [Google Scholar]
- 13.de Veer M. J., Holko, M., Frevel, M., Walker, E., Der, S., Paranjape, J. M., Silverman, R. H. & Williams, B. R. G. (2001) J. Leukocyte Biol. 69 912-920. [PubMed] [Google Scholar]
- 14.Shors S. T., Beattie, E., Paoletti, E., Tartaglia, J. & Jacobs, B. L. (1998) J. Interferon Cytokine Res. 18 721-729. [DOI] [PubMed] [Google Scholar]
- 15.Chang Y. E. & Laimins, L. A. (2000) J. Virol. 74 4174-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gale M., Blakely, C. M., Kwieciszewski, B., Tan, S. L., Dossett, M., Tang, N. M., Korth, M. J., Polyak, S. J., Gretch, D. R. & Katze, M. G. (1998) Mol. Cell. Biol. 18 5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boehm U., Klamp, T., Groot, M. & Howard, J. C. (1997) Annu. Rev. Immunol. 15 749-795. [DOI] [PubMed] [Google Scholar]
- 18.Fan Z., Beresford, P. J., Zhang, D. & Lieberman, J. (2002) Mol. Cell. Biol. 22 2810-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groettrup M., van den Broek, M., Schwarz, K., Macagno, A., Khan, S., de Giuli, R. & Schmidtke, G. (2001) Crit. Rev. Immunol. 21 339-358. [PubMed] [Google Scholar]
- 20.Cooper S., Erickson, A. L., Adams, E. J., Kansopon, J., Weiner, A. J., Chien, D. Y., Houghton, M., Parham, P. & Walker, C. M. (1999) Immunity 10 439-449. [DOI] [PubMed] [Google Scholar]
- 21.Xu H. E., Stanley, T. B., Montana, V. G., Lambert, M. H., Shearer, B. G., Cobb, J. E., McKee, D. D., Galardi, C. M., Plunket, K. D., Nolte, R. T., et al. (2002) Nature 415 813-817. [DOI] [PubMed] [Google Scholar]
- 22.Hug P., Lin, H. M. J., Korte, T., Xiao, X. D., Dimitrov, D. S., Wang, J. M., Puri, A. & Blumenthal, R. (2000) J. Virol. 74 6377-6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nohturfft A., Yabe, D., Goldstein, J. L., Brown, M. S. & Espenshade, P. J. (2000) Cell 102 315-323. [DOI] [PubMed] [Google Scholar]
- 24.Perlemuter G., Sabile, A., Letteron, P., Vona, G., Topilco, A., Chretien, Y., Koike, K., Pessayre, D., Chapman, J., Barba, G. & Brechot, C. (2002) FASEB J. 16 185-194. [DOI] [PubMed] [Google Scholar]
- 25.Andre P., Komurian-Pradel, F., Deforges, S., Perret, M., Berland, J. L., Sodoyer, M., Pol, S., Brechot, C., Paranhos-Baccala, G. & Lotteau, V. (2002) J. Virol. 76 6919-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mottola G., Cardinali, G., Ceccacci, A., Trozzi, C., Bartholomew, L., Torrisi, M. R., Pedrazzini, E., Bonatti, S. & Migliaccio, G. (2002) Virology 293 31-43. [DOI] [PubMed] [Google Scholar]
- 27.Shi S. T., Polyak, S. J., Tu, H., Taylor, D. R., Gretch, D. R. & Lai, M. M. (2002) Virology 292 198-210. [DOI] [PubMed] [Google Scholar]
- 28.Riddle T. M., Kuhel, D. G., Woollett, L. A., Fichtenbaum, C. J. & Hui, D. Y. (2001) J. Biol. Chem. 276 37514-37519. [DOI] [PubMed] [Google Scholar]
- 29.Egger D., Wolk, B., Gosert, R., Bianchi, L., Blum, H. E., Moradpour, D. & Bienz, K. (2002) J. Virol. 76 5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krüger M., Beger, C., Welch, P. J., Barber, J. R., Manns, M. P. & Wong-Staal, F. (2001) Mol. Cell. Biol. 21 8357-8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pietschmann T., Lohmann, V., Rutter, G., Kurpanek, K. & Bartenschlager, R. (2001) J. Virol. 75 1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.