Abstract
The recent application of molecular phylogeny to environmental samples has resulted in the discovery of an abundance of unique and previously unrecognized microorganisms. The vast majority of this microbial diversity has proved refractory to cultivation. Here, we describe a universal method that provides access to this immense reservoir of untapped microbial diversity. This technique combines encapsulation of cells in gel microdroplets for massively parallel microbial cultivation under low nutrient flux conditions, followed by flow cytometry to detect microdroplets containing microcolonies. The ability to grow and study previously uncultured organisms in pure culture will enhance our understanding of microbial physiology and metabolic adaptation and will provide new sources of microbial metabolites. We show that this technology can be applied to samples from several different environments, including seawater and soil.
Classification of microorganisms based on rRNA analysis has shown that the majority of microbes present in nature have no counterpart among previously cultured organisms (1–4). Establishing the metabolic properties and potential of these diverse organisms in the absence of pure culture presents an immense challenge for microbial ecologists (5). Although 16S rRNA studies combined with genomic analyses of naturally occurring marine bacterioplankton suggested the existence of metabolic functions (6, 7), a comprehensive understanding of the physiology of these organisms, and of the complex environmental processes in which they engage, will undoubtedly require their cultivation.
Conventional cultivation of microorganisms is laborious, time consuming, and, most important, selective and biased for the growth of specific microorganisms (8, 9). The majority of cells obtained from nature and visualized by microscopy are viable, but they do not generally form visible colonies on plates (9, 10). This phenomenon may reflect the artificial conditions inherent in most culture media (for example, extremely high substrate concentrations or the lack of specific nutrients required for growth). Thus, it was shown recently that previously uncultivable microorganisms could be grown in pure culture if provided with the chemical components of their natural environment (5, 11, 12). In addition, studies using modified media demonstrated the recovery of organisms not previously identified in culture by traditional cultivation methods (13, 14). Here, we describe a high throughput cultivation method based on the combination of a single cell encapsulation procedure with flow cytometry that enables cells to grow with nutrients that are present at environmental concentrations.
Materials and Methods
Sample Collection.
Water samples were collected in the Sargasso Sea (31°50′N 64°10′W and 32°05′ N 64°30′W) at a depth of 3 m. For each sample, a volume of 130 liters was concentrated by tangential flow filtration. Soil samples were collected from tropical forest (05°56′N 00°03′W) and (05°55′N 00°03′W) in Ghana and combined in equal amounts. Cells were separated from the soil matrix by repeated sheering cycles followed by density gradient centrifugation (15).
Cell Encapsulation and Growth Conditions.
Concentrated cell suspensions were used for encapsulation. Cells were diluted in sterile filtered sea water for marine samples and PBS buffer (pH 7.2; GIBCO) for soil samples to a final concentration of 107 cells per ml. Diluted cell suspensions (0.1 ml) were mixed with 0.5 ml of preheated agarose (at 40°C; OneCell System). Cell agarose mixtures were added into 15 ml of CellMix emulsion matrix (OneCell System) and emulsified at room temperature for 1 min by using the CellSys 100 microdrop maker at 2,200 rpm at room temperature followed by a 1-min emulsifying step (2,200 rpm) on ice. The oil-bacterial suspension was cooled with ice under stirring for 6 min at 1,100 rpm. This procedure results in ≈107 gel microdroplets (GMDs). Approximately 10% of formed GMDs are occupied by a single encapsulated cell. Encapsulation of single cells was monitored by microscopy. The GMDs were dispensed into sterile chromatography columns XK-16 (Amersham Pharmacia) containing 25 ml of media. Columns were equipped with two sets of filter membranes (0.1 μm at the inlet of the column and 8 μm at the outlet). The filters prevented free-living cells contaminating the media reservoir and retained GMDs in the column while allowing free-living cells to be washed out.
Media used for incubation of marine samples were: filter-sterilized Sargasso Sea water (SSW); SSW amended with NaNO3 (4.25 mg/liter), K2HPO4 (0.016 mg/liter), NH4Cl (0.27 mg/liter), trace metals, and vitamins (16); SSW amended with amino acids at concentrations between 6 and 30 nM (17) and marine medium (R2A, Difco) diluted in SSW 1:100 (vol/vol). Soil extracts were prepared as described (18) and added to the media at final concentrations of 25–40 ml/liter in 0.85% NaCl (vol/vol). Media were pumped through the column at a flow rate of 13 ml/h. GMDs were incubated in the columns for a period of at least 5 weeks. Microcolonies that were sorted individually into 96-well microtiter plates were grown with marine medium (R2A, Difco) in SSW or with soil extracts amended with glucose, peptone, yeast extract (1 g/liter), and humic acids extract 0.001% (vol/vol).
Flow Cytometry.
GMDs containing colonies were separated from free-living cells and empty GMDs by using a flow cytometer (MoFlo, Cytomation). Precise sorting was confirmed by microscopy. To determine the minimal number of cells required for encapsulation and detection by flow cytometry, a series of 1,000, 100, and 10 Escherichia coli cells obtained from liquid media (expressing a green fluorescent protein, ZsGreen, CLONTECH) were individually encapsulated as described above. Exact cell numbers of diluted cultures were determined by flow cytometry and direct cell counts. Encapsulated cells were incubated for 3 h to form microcolonies within the GMDs. GMDs were analyzed by flow cytometry and sorted.
Phylogenetic Analysis.
Ribosomal RNA genes from environmental samples, microcolonies, and cultures were amplified by PCR by using general oligonucleotide primers (27F and 1392R) for the domain Bacteria. To avoid nonspecific amplification, PCR reactions were irradiated with a UV Stratalinker (Stratagene) at maximum intensity before template addition. After cloning (TOPO-TA, Invitrogen), inserts were screened by their restriction pattern (restriction fragment length polymorphism) obtained with AvaI, BamHI, EcoRI, HindIII, KpnI, and XbaI. Nearly full-length 16S rRNA gene sequences were obtained and added to an aligned database of >12,000 homologous 16S rRNA primary structures maintained with the ARB software package (www.mikro.biologie.tu-muenchen.de). Phylogenetic relationships were evaluated by using evolutionary distance, parsimony, and maximum likelihood methods and were tested with a wide range of bacterial phyla as outgroups (19). Hypervariable regions were masked from the alignment. The phylogenetic trees (see Figs. 3–5) demonstrate the most robust relationships observed and were determined by using evolutionary distances calculated with the Kimura 2-parameter model for nucleotide change and neighbor-joining. Bootstrap proportions from 1,000 resamplings were determined by using both evolutionary distance and parsimony methods. Short reference sequences were added to the phylogenetic trees with the parsimony insertion tool of ARB and are indicated by dotted lines in the figures.
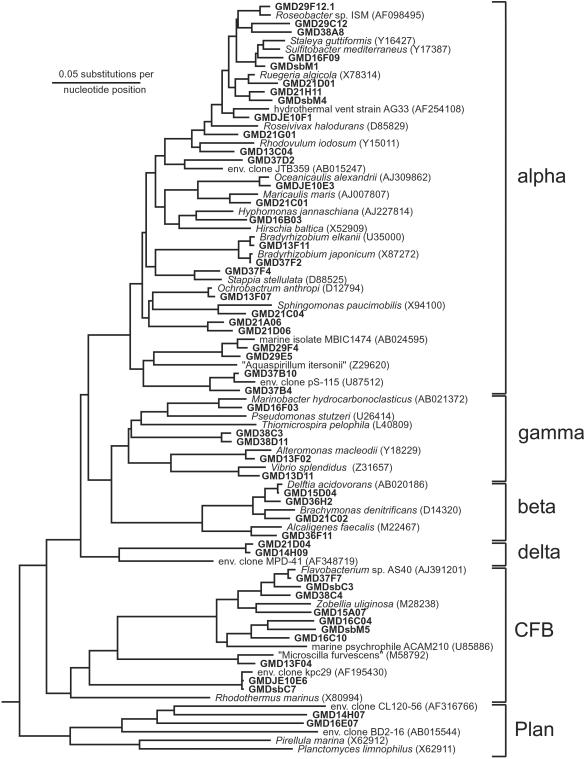
Fig 3.
Phylogenetic tree based on 16S rRNA sequences that were retrieved from GMDs obtained from the Sargasso Sea. Shown are groups of the alpha, beta, gamma, and delta subclasses of Proteobacteria, as well as of the Cytophaga–Flavobacterium–Bacteroides and relatives (CFB) and Planctomycetes and relatives (Plan).
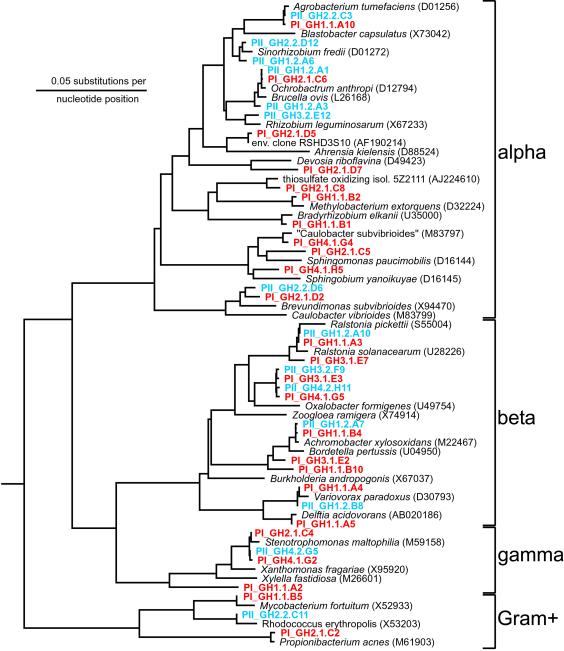
Fig 5.
Phylogenetic tree based on 16S rRNA sequences that were retrieved from GMDs obtained from a soil sample (Ghana). Shown are groups of the alpha, beta, and gamma subclasses of Proteobacteria, as well as of Gram-positive bacteria. Cultures obtained in phase I (PI) are marked in red; cultures obtained in phase II (PII) are marked in blue.
Results and Discussion
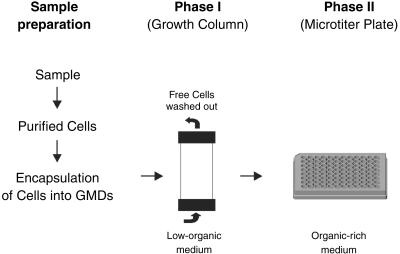
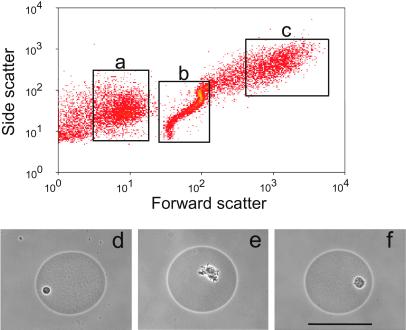
Seawater was collected from sites located in the Sargasso Sea. Individual cells were concentrated from this seawater by tangential flow filtration and encapsulated in GMDs. Similar GMDs have been used previously to grow traditionally isolated bacteria (e.g., members of the genera Bacillus, Escherichia, and Pseudomonas) for screening purposes (20–23). In the work reported here, we used GMDs and flow cytometry to isolate and culture microorganisms from environmental samples in a high throughput manner. Our approach differs from standard limiting dilution culturing (24) and modifications thereof (11–13) in several important respects. First, whereas each cell is encapsulated, all of the encapsulated cells from the environment are cultured together in a single vessel. This proximity simulates to some extent the natural environment; because the pore size of the GMD matrix is large, it allows the exchange of metabolites and other molecules such as signaling molecules. The importance of diffusing molecules produced by other microorganisms in enhancing culturability is gaining appreciation (5, 13). Second, GMD culturing is performed in an open, continuously fed system instead of a closed batch system which also helps simulate the open condition of most natural environments. Third, our culturing approach is capable of extremely high throughput and can be easily and inexpensively scaled to meet any downstream characterization needs, including industrial-scale screening and characterization platforms. Single encapsulated cells (see Materials and Methods) were transferred into chromatography columns (referred to henceforth as growth columns). Different culture media selective for aerobic, nonphototrophic organisms were pumped through the growth columns containing 10 million GMDs (Fig. 1). The pore size of the GMDs allows the free exchange of nutrients. The encapsulated microorganisms were able to divide and form microcolonies of ≈20–100 cells within the GMDs. Based on their distinctive light-scattering signature, these microcolonies were detected and separated by flow cytometry at a rate of 5,000 GMDs per second. The increase in forward and side scatter was shown by microscopy to be directly proportional to the size of the microcolony grown within the GMD. This property enabled discrimination between unencapsulated single cells, empty or singly occupied GMDs, and GMDs containing a microcolony (Fig. 2).
Fig 1.
Model of the experimental setup. Cells captured from environmental samples were encapsulated into GMDs and incubated in growth columns (phase I). GMDs containing microcolonies were detected and separated by flow cytometry into 96-well microtiter plates containing a rich organic medium (phase II).
Fig 2.
Discrimination among (a) free-living cells, (b) singly occupied or empty GMDs, and (c) GMDs containing microcolonies was accomplished by flow cytometry in forward and side light-scatter mode. (d–f) Phase contrast photomicrographs of separated GMDs containing microcolonies. (Bar = 50 μm.)
To determine the growth medium for a broad diversity of organisms, four media were tested in the growth columns. After 5 weeks of incubation, 1,200 GMDs, each containing a microcolony, were collected by flow cytometry from each of the four growth columns. A 16S rRNA gene clone library was generated from each group of 1,200 microcolonies. From each group, 140 randomly picked clones were analyzed by restriction fragment length polymorphism analysis, and 50 clones were then sequenced. In diluted marine medium, only four bacterial species were identified, belonging to the genera Vibrio, Marinobacter, or Cytophaga, all common sea water bacteria that have been cultivated previously (3, 9). The media containing amino acids or inorganic minerals revealed slightly more diversity (12 different bacterial sequences from the amino acid supplemented medium and 11 sequences from the inorganic medium). Filtered seawater alone (taken from the original sampling site) yielded the highest biodiversity (39 sequences of 50 clones analyzed), with many different phylogenetic groups represented. These results suggested that organisms capable of rapid growth outgrew their more fastidious neighbors in the presence of organic-rich medium.
Growth columns were next inoculated with GMDs again generated from samples obtained from the Sargasso Sea, but now using only filtered sea water as growth medium. From each of two growth columns, 500 GMDs containing microcolonies were sorted, and the 16S rRNA genes contained therein were amplified by PCR. A 16S rRNA gene library (167 clones analyzed) was also constructed from the original environmental sample from which the microorganisms were obtained for encapsulation. Almost all (166 of 167, or 99%) of the environmental 16S rRNA sequences derived from the latter sample fell within the nine common bacterioplankton groups (3). SAR11 was the most predominant group (28%), followed by sequences within the Cytophaga–Flavobacterium–Bacteroides group (13%) and the SAR116 clade (12%; ref. 3). In contrast, 16S rRNA gene sequences obtained from the microcolonies reflected a broader range of microorganisms (Fig. 3), with only 71 of 150 (47%) sequences representing the same nine common bacterioplankton groups. A number of GMD microcolony sequences fell into clades, which contain no previously cultivated representatives and could not be detected in the environmental gene library. Three of the most notable examples, described in more detail below, were clades affiliated with the Planctomycetes and relatives, the Cytophaga–Flavobacterium–Bacteroides and relatives, and the alpha subclass of Proteobacteria (Fig. 4). Sequences related to these groups were not detected within the environmental 16S rRNA gene clone library.
Fig 4.
Phylogenetic analysis of 16S rDNA sequences from GMDs obtained from Sargasso Sea water. Nodes supported by bootstrap proportions >90% in both distance and parsimony analyses are shown as filled circles; open circles indicate ≤70% support. Scale bars (a–c) correspond to 0.10 substitutions per nucleotide position. Dotted lines represent lineages added by the ARB parsimony insertion tool. (a) Planctomycetes and relatives. Verrucomicrobia and Chlamydiae sequences were used as outgroups. (b) Cytophaga–Flavobacterium–Bacteroides and relatives. Chlorobiaceae sequences were used as outgroups. (c) Alpha Proteobacteria. Gamma and beta Proteobacteria sequences were used as outgroups. (d) Phase contrast photomicrograph of strain GMDJE10E6. (Bar = 10 μm.)
Five microcolony 16S rRNA gene sequences were related to the Planctomycetales, one of the main phylogenetic branches of the domain Bacteria (ref. 3; Fig. 4a). Sequencing of cloned rRNA genes from marine environments had previously revealed several new, apparently uncultivated phylotypes within the Planctomycetales (25–27). Many of these new phylotypes fall within a single, highly diverse monophyletic clade that, before this study, contained no cultivated representatives. The five Planctomycetales-related microcolonies identified in this study form two separate lineages within this deep branching Planctomycetales clade. One lineage, represented by sequences GMD21C08, GMD14H10, and GMD14H07, was most closely related to 16S rRNA gene clone sequences recovered from bacteria associated with marine corals (84.9–89.2% similar; ref. 26). The second lineage, represented by GMD16E07 and GMD15D02, forms a unique line of descent within this clade and is <84% similar to all previously published 16S rRNA gene sequences.
Two microcolony 16S rRNA gene sequences fell within the Cytophaga–Flavobacterium–Bacteroides and their relatives (Fig. 4b). These two closely related sequences form a lineage within a cluster of gene clone sequences from predominantly marine and hypersaline environments (28–30). This cluster occupies one of the deepest phylogenetic branches of theCytophaga–Flavobacterium–Bacteroides and relatives group; only the Rhodothermus/Salinibacter lineage is deeper (29). Within this cluster, the two microcolony gene sequences were nearly identical (>99% similar) to environmental 16S rRNA gene clone sequences obtained from seawater collected off of the Atlantic coast of the United States (30). Analysis of phase II cultures (Fig. 1) obtained from these sorted microcolonies revealed a culture (strain GMDJE10E6) with an identical 16S rRNA gene sequence that reached an optical density (OD600) of 0.3 (Fig. 4d). This result indicates that some of the microcolonies can grow to high density when subsequently provided a high nutrient medium.
A cluster of six microcolonies was recovered that was phylogenetically affiliated with a previously uncultivated lineage of 16S rRNA gene clone sequences within the alpha subclass of the Proteobacteria (Fig. 4c). The microcolony sequences formed two subclusters; one was closely related to two 16S rRNA gene clone sequences recovered from marine samples taken from a coral reef (95.1–98.6% similar), and the second was moderately related to the same coral reef-associated environmental gene clones (87.9–95.7% similar).
Thus, the application of this high throughput cultivation method resulted in the growth and isolation of a broad range of bacteria, including previously uncultured phylotypes (Fig. 3). The GMDs likely permit the simultaneous and relatively noncompetitive growth of both slow- and fast-growing microorganisms in media with very low substrate concentrations, thereby preventing overgrowth by the fast-growing microorganisms (the “microbial weeds”; ref. 9). However, the most abundant microorganism in this sample based on environmental 16S rRNA gene library (SAR11) could not be detected within the analyzed GMDs. As demonstrated recently, a cultivated member of the SAR11 clade was not able to grow above 20°C (M.R., unpublished results). The slightly elevated incubation temperature (23°C) of the growth columns could have inhibited the growth of members of the SAR11 clade. However, the broad range of microorganisms cultivated within the GMDs might suggest that we were able to recover less abundant members of the microbial community more efficiently.
To test whether this high throughput cultivation method is applicable to different environments, we applied the technology to an alkaline lake sediment (Lake Bogoria, Kenya, data not shown) and to a soil sample (Ghana). Microorganisms from the soil sample were separated from the soil matrix, encapsulated, and incubated in the growth column under aerobic conditions in the dark. Diluted soil extract, obtained from the same sample, was used as growth medium. The microcolonies were analyzed by 16S rRNA gene sequencing. To cater for bacteria with disparate growth rates, microcolonies were separated from the growth column by flow cytometry at different time points. 16S rRNA gene sequence analysis revealed that many phylogenetically different microorganisms could be cultivated within the GMDs in phase I (Fig. 5). A number of microcolonies obtained in phase I had low blast similarities (≤96%) with 16S rRNA genes from isolates in the database.
Physiological studies, natural product screening, or studies of cell–cell interaction require the ability to grow microorganisms to a certain cell mass. Therefore, we designed experiments to determine whether these microcolonies are able to serve as inocula for larger scale microbial cultures (Fig. 1, phase II). Encouragingly, earlier microscopic analysis had revealed that encapsulated bacteria could indeed grow out of GMDs when provided with a rich supply of nutrients. A total of 960 microcolony-containing GMDs from the Ghana soil sample, each derived from a single organism, were individually sorted into 96-well microtiter plates filled with organic rich medium (Fig. 1, phase II). The 960 cultures were analyzed for growth by measuring optical densities (OD600). After 1 week of incubation, 67% of the cultures showed turbidity above OD 0.1, corresponding to at least 107 cells per ml. Cell densities were high enough to permit the detection of antifungal activity among some of the cultures (data not shown). To analyze the diversity within these cultures in more detail, 100 randomly picked cultures were analyzed by 16S rRNA gene sequencing, revealing many different species (Fig. 5). The remaining 33% of the cultures that did not grow to measurable densities (<106 cells per ml) showed bacterial growth when assessed microscopically. This result is consistent with recent reports indicating that certain bacteria do not grow to cell densities >106 cells per ml (12).
To maintain and access microcolonies for physiological studies, we evaluated the minimal number of cells required for passaging by reencapsulation and detection by flow cytometry. Flow cytometry analysis of 1,000 and 100 individually encapsulated cells resulted in the detection of 360 and 15 microcolonies, respectively. Even when using cultures comprising just 10 bacterial cells, this method allowed recovery of, on average, one viable bacterial culture. This experiment demonstrates that it is possible to transfer, and therefore maintain, a culture of 100 cells derived directly from a microcolony.
GMDs separate microorganisms from each other while still allowing the free flow of metabolites and signaling molecules between different microcolonies. Therefore, this method might be applicable for the analysis of interactions between different organisms under in situ conditions, for example by inserting the encapsulated cells back into the environment (e.g., the open ocean). The simultaneous encapsulation of more than one cell (prokaryotic as well as eukaryotic) into one GMD might also be used to mimic conditions found in nature, allowing analysis of cell–cell interactions. Another advantage of this technology is the very sensitive detection of growth. This high throughput cultivation method allows the detection of microcolonies containing as few as 20–100 cells. Nutrient-sparse media, such as sea water, were sufficient to support growth, and yet their carbon content was low enough to prevent “microbial weeds” from overgrowing slow-growing microorganisms. We have demonstrated that this technology can be used to culture thus far uncultivated microorganisms. The microcolonies obtained can be used as inocula for further cultivation. In addition, we believe that the new cultivation approach described can be extended to many other physiological and environmental conditions. For example, we have found that encapsulated cells of Methanococcus thermolithotrophicus can grow and form microcolonies within GMDs when incubated under strictly anaerobic conditions. In combination with rRNA analysis and mixed organism recombinant screening approaches (31, 32), this technology should permit a more complete understanding of unexplored microbial communities. It should find applications in environmental microbiology, whole cell engineering, and drug discovery. The combination of cultivation with direct DNA amplification from microcolonies will contribute to a broader understanding of microbial ecology by linking microbial diversity with metabolic potential.
Acknowledgments
We thank Melvin Simon and Mervyn Bibb for critical review of the manuscript, Debra Mertes and David Walsh for sequencing help, Trevin Holland for FACS support, and Greg Clark and Manuela Baumgartner for technical assistance. We are grateful to our partners at the Bermuda Biological Station for Research (Hank Trapido-Rosenthal and Sandra Zielke), at the University of Ghana (Yaa D. Osei and Sammy T. Sackey), at the International Center of Insect Physiology and Ecology, Kenya (James Kabii and Lucy Mackenzie), and at the Kenya Wildlife Service (Paul Mungai) for providing us with the samples.
Abbreviations
GMD, gel microdroplet
SSW, Sargasso Sea water
References
- 1.Pace N. R. (1997) Science 276 734-740. [DOI] [PubMed] [Google Scholar]
- 2.Amann R. I., Ludwig, W. & Schleifer, K.-H. (1995) Microbiol. Rev. 59 143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giovannoni S. J. & Rappe, M. (2000) in Microbial Ecology of the Ocean, ed. Kirchman, D. L. (Wiley, New York), pp. 47–84.
- 4.Fuhrman J. A., McCallum, K. & Davis, A. A. (1993) Appl. Environ. Microbiol. 59 1294-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaeberlein T., Lewis, K. & Epstein, S. S. (2002) Science 296 1127-1129. [DOI] [PubMed] [Google Scholar]
- 6.Beja O., Aravind, L., Koonin, E. V., Suzuki, M. T., Hadd, A., Nguyen, L. P., Jovanovich, S. B., Gates, C. M., Feldman, R. A., Spudich, J. L., et al. (2000) Science 289 1902-1906. [DOI] [PubMed] [Google Scholar]
- 7.Beja O., Suzuki, M. T., Heidelberg, J. F., Nelson, W. C., Preston, C. M., Hamada, T., Eisen, J. A., Fraser, C. M. & DeLong, E. F. (2002) Nature 415 630-633. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson R. L., Buckley, E. N. & Palumbo, A. V. (1984) Appl. Environ. Microbiol. 47 49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eilers H., Pernthaler, J., Glöckner, F. O. & Amann, R. (2000) Appl. Environ. Microbiol. 66 3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu H. S., Roberts, N., Singleton, F. L., Attwell, R. W., Grimes, D. J. & Colwell, R. R. (1982) Microb. Ecol. 8 313-323. [DOI] [PubMed] [Google Scholar]
- 11.Connon S. A. & Giovannoni, S. J. (2002) Appl. Environ. Microbiol. 68 3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rappé M. S., Connon, S. A., Vergin, K. L. & Giovannoni, S. J. (2002) Nature 418 630-633. [DOI] [PubMed] [Google Scholar]
- 13.Bruns A., Cypionka, H. & Overmann, J. (2002) Appl. Environ. Microbiol. 68 3978-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssen P. H., Yates, P. S., Grinton, B. E., Taylor, P. M. & Sait, M. (2002) Appl. Environ. Microbiol. 68 2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fægri A., Torsvik, V. L. & Goksöyr, J. (1977) Soil Biol. Biochem. 9 105-112. [Google Scholar]
- 16.Widdel F. & Bak, F. (1992) in The Prokaryotes, eds. Balows, A., Trüper, H. G., Dworkin, M., Harder, W. & Schleifer, K.-H. (Springer, New York), Vol. 4, pp. 3352–3392. [Google Scholar]
- 17.Ouverney C. C. & Fuhrman, J. A. (2000) Appl. Environ. Microbiol. 66 4829-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vobis G. (1992) in The Prokaryotes, eds. Balows, A., Trüper, H. G., Dworkin, M., Harder, W. & Schleifer, K.-H. (Springer, New York), Vol. 2, pp. 1029–1060. [Google Scholar]
- 19.Ludwig W., Bauer, S. H., Bauer, M., Held, I., Kirchhof, G., Schulze, R., Huber, I., Spring, S., Hartmann, A. & Schleifer, K.-H. (1997) FEMS Microbiol. Lett. 153 181-190. [DOI] [PubMed] [Google Scholar]
- 20.Short, J. M. & Keller, M. (2001) U.S. Patent US 6,174,673 B1.
- 21.Manome A., Zhang, H., Tani, Y., Katsuragi, T., Kurane, R. & Tsuchida, T. (2001) FEMS Microbiol. Lett. 197 29-33. [DOI] [PubMed] [Google Scholar]
- 22.Ryan C., Nguyen, B. T. & Sullivan, S. J. (1995) J. Clin. Microbiol. 33 1720-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powell K. T. & Weaver, J. C. (1990) Bio/Technology 8 333-337. [DOI] [PubMed] [Google Scholar]
- 24.Button D. K., Schut, F., Quang, P., Martin, R. & Robertson, B. R. (1993) Appl. Environ. Micobiol. 59 881-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowman J. P., Rea, S. M., McCammon, S. A. & McMeekin, T. A. (2000) Environ. Microbiol. 2 227-237. [DOI] [PubMed] [Google Scholar]
- 26.Frias-Lopez J., Zerkle, A. L., Bonheyo, G. T. & Fouke, B. W. (2002) Appl. Environ. Microbiol. 68 2214-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravenschlag K., Sahm, K., Pernthaler, J. & Amann, R. (1999) Appl. Environ. Microbiol. 65 3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanner M. A., Everett, C. L., Coleman, W. J., Yang, M. M. & Youvan, D. C. (2000) Bio/Technology 8 1-16. [Google Scholar]
- 29.de Souza M. P., Amini, A., Dojka, M. A., Pickering, I. J., Dawson, S. C., Pace, N. R. & Terry, N. (2001) Appl. Environ. Microbiol. 67 3785-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly K. M. & Chistoserdov, A. Y. (2001) FEMS Microbiol. Ecol. 35 85-95. [DOI] [PubMed] [Google Scholar]
- 31.Short J. M. (1997) Nat. Biotechnol. 15 1322-1323. [DOI] [PubMed] [Google Scholar]
- 32.Robertson D. E., Mathur, E. J., Swanson, R. V., Marrs, B. L. & Short, J. M. (1996) SIM News 46 3-8. [Google Scholar]