Abstract
MinD is a widely conserved ATPase that has been demonstrated to play a pivotal role in selection of the division site in eubacteria and chloroplasts. It is a member of the large ParA superfamily of ATPases that are characterized by a deviant Walker-type ATP-binding motif. MinD localizes to the cytoplasmic face of the inner membrane in Escherichia coli, and its association with the inner membrane is a prerequisite for membrane recruitment of the septation inhibitor MinC. However, the mechanism by which MinD associates with the membrane has proved enigmatic; it seems to lack a transmembrane domain and the amino acid sequence is devoid of hydrophobic tracts that might predispose the protein to interaction with lipids. In this study, we show that the extreme C-terminal region of MinD contains a highly conserved 8- to 12-residue sequence motif that is essential for membrane localization of the protein. We provide evidence that this motif forms an amphipathic helix that most likely mediates a direct interaction between MinD and membrane phospholipids. A model is proposed whereby the membrane-targeting motif mediates the rapid cycles of membrane attachment–release–reattachment that are presumed to occur during pole-to-pole oscillation of MinD in E. coli.
During vegetative growth, most bacteria form a division septum at the center of the cell by coordinated ingrowth of the cytoplasmic membrane, the rigid murein (peptidoglycan) layer, and, in Gram-negative bacteria, the outer membrane of the cell envelope (1). In the rod-shaped bacterium Escherichia coli, placement of the division septum is negatively regulated by the three proteins encoded by the minB operon: MinC, MinD, and MinE (2, 3). MinC and MinD act in concert to form a global division inhibitor whose activity is restricted to polar sites by MinE (2). Studies of GFP-labeled Min proteins have revealed that they undergo a complex bipolar oscillation that causes the time-averaged concentration of the MinC-MinD division inhibitor to be lowest at midcell (4–8). This dynamic distribution of MinC-MinD makes midcell the preferred site for construction of a circumferential ring of polymerized FtsZ, which is the initiating event in bacterial cytokinesis (9).
MinD is the best conserved and most widely distributed of the Min proteins, being found in all domains of life (eubacteria, archaea, and eukaryotes). It is a member of the ParA superfamily of ATPases, most of which (apart from the MinD subgroup) are involved in plasmid or chromosome partitioning (10–12). The ATPase activity of MinD is presumed to provide the driving force for pole-to-pole oscillation of the MinC-MinD division inhibitor (13, 14). This activity is stimulated by MinE but only in the presence of phospholipids (13, 14). MinD is a peripheral membrane protein (10), and its association with the inner membrane is a prerequisite for subsequent recruitment of both MinC and MinE to the membrane. However, the mechanism by which MinD associates with the inner membrane and subsequently recruits MinC and MinE remains enigmatic; it seems to lack a transmembrane domain, and the amino acid sequence is devoid of hydrophobic stretches that might predispose the protein to interaction with lipids. It has been suggested that MinD might be localized to the inner membrane by means of interactions with a membrane protein (15). However, recent studies have demonstrated conclusively that, at least in vitro, MinD is capable of binding directly to lipid bilayers (13, 14). Furthermore, this interaction induces extensive MinD polymerization (14).
Crystal structures have been determined for presumptive MinD homologs from the hyperthermophilic archaeons Archaeoglobus fulgidus (15), Pyrococcus furiosus (16), and Pyrococcus horikoshii (17). Unfortunately, these structures provide little insight into the mechanism by which MinD associates with the membrane. Intriguingly, however, all three structures are incomplete in the C-terminal region. No electron density was observed for the C-terminal 30 and 8 residues of A. fulgidus and P. furiosus MinD, respectively, presumably because of the high intrinsic flexibility of these residues. Although electron density was obtained for all but the final two residues in the crystal of P. horikoshii MinD, the C-terminal residues 235–243 had very high B factors (17), indicative of significant motion in the crystal. On the basis of these crystal structures, we speculated that the C-terminal region of MinD might be involved in membrane attachment in vivo, and that this region might be structured only when associated with lipid. In this study, we show that the C-terminal region of MinD, but not that of other members of the ParA superfamily, contains a highly conserved motif that is essential for membrane localization. We propose that this motif forms an amphipathic helix that mediates a direct interaction between MinD and membrane phospholipids.
Experimental Procedures
Construction of GFP Fusions.
All gfp:minD genes used in this study were placed downstream of the Para promoter and were therefore inducible with arabinose. GFP fusions to E. coli MinD (EcMinD) were derived from pSLR22 (7), a pBAD33-derived plasmid encoding Para-GFP:EcMinD. Various C-terminal truncation mutants (EcMinDΔ19, EcMinDΔ5, EcMinDΔ3, and EcMinDΔ2) were generated by PCR using pSLR22 as the template and primers that incorporated flanking XbaI/HindIII restriction sites. An EcMinD(L267E) point mutant and several insertion mutants were generated in a similar manner by using 3′ primers incorporating the desired mutations.
Bacillus subtilis MinD (BsMinD) was obtained by PCR using the full-length minD gene as template and 5′ and 3′ primers incorporating XbaI and PstI restriction sites, respectively. This construct was then ligated, along with a PCR-generated GFPmut2 gene incorporating 5′ EcoRI and 3′ XbaI sites, into plasmid pSJ4 (18) that had been digested with EcoRI/PstI. This ligation yielded plasmid pTS26 in which BsMinD is encoded as in-frame fusion to the C terminus of GFP. Various C-terminal truncation mutants (BsMinDΔ24, BsMinDΔ5, BsMinDΔ4, BsMinDΔ3, and BsMinDΔ2) were then generated by PCR using pTS26 as the template and primers that incorporated flanking XbaI/PstI restriction sites.
Fluorescence Microscopy.
For each GFP-fusion construct, cells from a single colony were grown in LB supplemented with 20 μg/ml chloramphenicol and 1% glucose at 37°C for 4 h (OD600 ≈ 0.5–0.7). Harvested cells (1 ml) were washed in medium lacking glucose before inoculating fresh LB medium containing 20 μg/ml chloramphenicol and 0.005% arabinose to a starting OD600 of 0.05. Cultures were grown at 30°C for 4–5 h. Cells were then viewed live and after fixation for 45 min in a final concentration of 1.7% (vol/vol) formaldehyde and 0.17% (vol/vol) glutaraldehyde. Cells were viewed by using an Olympus BX40 microscope (Olympus, New Hyde Park, NY), and images were captured by using a MagnaFire digital camera (Optronics International, Chelmsford, MA).
Western Blot Analysis.
Cells collected from fluorescence experiments were pelleted, resuspended in 0.5% SDS, boiled for 5 min, then stored at −80°C until use. The total protein content of each strain was determined by bicinchoninic acid assay (Pierce). SDS/polyacrylamide gels were loaded with 5 μg of total cell protein for each GFP-fusion construct. The gel was electroblotted onto Hybond ECL nitrocellulose membrane (Amersham Pharmacia), and GFP-fusion proteins were probed by using rabbit anti-GFP primary antibodies (Molecular Probes) and anti-rabbit horseradish peroxidase-conjugated secondary antibodies (Amersham Pharmacia). Immunoblots were developed on Hyperfilm ECL (Amersham Pharmacia) by using a chemiluminescent substrate.
Overexpression and Purification of MinD.
An NdeI–BamHI PCR fragment encompassing the entire E. coli minD gene was cloned into pET-15b (Novagen) to produce plasmid pTS21, in which EcMinD is encoded as an N-terminal His6-tag fusion protein. His6-EcMinD was overproduced by expression from pTS21 in E. coli BL21ΔminCDE:kan(λDE3) cells (a kind gift from D. RayChaudhuri, Tufts University, Boston). An NdeI–EcoRI PCR fragment encompassing the first 251 codons of the E. coli minD gene followed by a stop codon was cloned, in-frame, into pET28a to produce the plasmid pSLR90, in which EcMinDΔ19 is encoded as an N-terminal His6-tag fusion protein. Expression of His6-EcMinDΔ19 and His6-EcMinD in either BL21(λDE3) or its ΔminCDE derivative, respectively, was induced with 1 mM isopropyl β-d-thiogalactoside after growth at 34°C to an OD600 of 0.6. Cells were grown at 30–34°C for a further 3 h before being harvested by centrifugation, resuspended in binding buffer (5 mM imidazole/500 mM NaCl/20 mM Tris⋅HCl, pH 7.9), and lysed by using a French press. MinD fusion protein was purified from the soluble fraction by nickel affinity chromatography with His-bind resin (Novagen). Washes with 60–120 mM imidazole were used to elute contaminating proteins. His6-EcMinD (or His6-EcMinΔD19) was then eluted by using elution buffer (190 mM imidazole/500 mM NaCl/20 mM Tris⋅HCl, pH 7.9) and dialyzed against appropriate buffer in preparation for CD spectroscopy. Final protein preparations were judged from Coomassie blue-stained SDS/polyacrylamide gels to be >98% pure.
CD Spectropolarimetry.
CD spectra were recorded at 4°C by using a Jasco J-715 spectropolarimeter (Jasco, Easton, MD). Protein samples were 10 μM in 1 mM NaPi/20 mM NaF, pH 7.9. Spectra were the average of 16 scans acquired by using a scan rate of 20 nm⋅min−1 and a response time of 8 s.
Results
The C-Terminal Region of MinD Is Essential for Membrane Attachment.
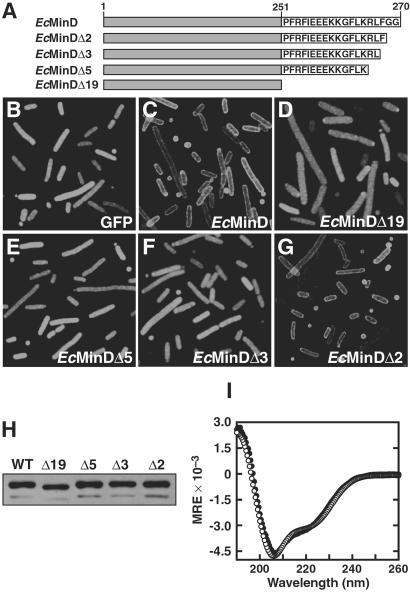
MinD localizes to the inner membrane (10), even in the absence of MinC and MinE (4, 7). To investigate whether the C-terminal region of MinD plays a role in membrane attachment of the protein, we used fluorescence microscopy to examine the cellular distribution of several C-terminal truncation mutants (Fig. 1A) fused to GFP. Because our focus was elucidation of determinants responsible for membrane attachment of MinD, all localization experiments were performed in the host strain PB114, which contains a deletion of the minB operon (2). This strategy allowed us to avoid potential complications that might result from MinE-induced oscillation of MinD and from truncation mutants that had aberrant interactions with MinC and/or MinE. Note that deletion of the minB operon causes cultures of PB114 to exhibit a classical “minicelling” phenotype (a mixture of normal-length cells, short filaments, and minicells), because the cells divide randomly at either midcell or polar sites (2).
Fig 1.
Localization of EcMinD truncation mutants. (A) Illustration of the EcMinD constructs used in this study. EcMinDΔX indicates a C-terminal deletion of X residues. (B–G) Fluorescence micrographs showing localization in E. coli of unfused GFP, GFP-EcMinD, and GFP-tagged C-terminal truncation mutants of EcMinD (see text for details). (H) Western blot comparing the cellular levels of EcMinD and various C-terminal truncation mutants. The intense upper band is MinD and the weak lower band is presumed to be a MinD degradation product. (I) Far-UV CD spectra of His6-EcMinD (○) and His6-EcMinDΔ19 (•). MRE is the mean residue ellipticity in degrees⋅cm2⋅dmol−1.
It has been shown that fusion of EcMinD to the C terminus of GFP has no deleterious effects on MinD function (4, 7). Thus, as expected, we found that GFP-EcMinD exhibited a distinct peripheral localization pattern extending around the entire circumference of the cell (Fig. 1C), whereas GFP alone was uniformly distributed throughout the cytoplasm (Fig. 1B). The peripheral distribution of GFP-EcMinD was observed in all cell types (WT, minicells, and filaments) and is consistent with previous immunoelectron studies showing that MinD localizes to the cytoplasmic face of the inner membrane (10). To examine the importance of the C-terminal region in membrane localization of MinD, we aimed to delete as much of the C terminus as possible without perturbing the 3D fold of the protein. In all three MinD structures solved to date, the final structural element preceding the disordered C-terminal region is an α-helix (designated α11 in the two Pyrococcus structures) (16, 17). Sequence alignments show that the C terminus of α11 corresponds to Leu-247 of E. coli MinD. Thus, we initially examined the localization of a mutant EcMinD truncated by 19 residues at Arg-251; this conservative truncation allows for the possibility that α11 might be slightly longer in the E. coli protein.
Removal of the C-terminal 19 residues yielded a mutant (GFP-EcMinDΔ19) that failed to exhibit a peripheral localization pattern (Fig. 1D). Instead, the cells appeared uniformly green, suggesting that at least some portion of the C-terminal 19 residues is essential for membrane localization of EcMinD. To delineate better the portion of the C terminus contributing to membrane localization, shorter MinD truncation mutants were generated. A GFP-EcMinDΔ5 truncation mutant failed to localize properly (Fig. 1E), indicating that key determinants for membrane localization exist in the C-terminal five residues of EcMinD. Significantly, a two-residue C-terminal truncation (GFP-EcMinDΔ2) localized normally to the periphery of the cell (Fig. 1G), whereas a three-residue C-terminal truncation (GFP-EcMinDΔ3) was diffusely distributed throughout the cytoplasm (Fig. 1F). This finding indicates that the third residue from the C terminus (Phe-268) is critical for attachment of EcMinD to the inner membrane, whereas the two C-terminal glycine residues are dispensable for this function.
Western blot analysis of cells expressing each of the GFP-EcMinD truncation mutants indicated that the MinD protein levels were similar to those observed in cells expressing WT GFP-EcMinD (Fig. 1H). Furthermore, the CD spectrum of purified His6-EcMinDΔ19 was very similar to that of His6-EcMinD (Fig. 1I), indicating that deletion of the C-terminal 19 residues does not perturb the global fold of the protein. The high thermal stability of the protein was also unaffected by deletion of the C-terminal 19 residues; the thermal unfolding temperature (Tm) of both EcMinDΔ19 and EcMinD was >90°C (CD data not shown). Because Δ19 was the largest deletion studied, it is safe to assume that the smaller truncations (Δ2, Δ3, and Δ5) also have no effect on the global fold or thermodynamic stability of MinD. We conclude that the aberrant localization observed for the Δ3, Δ5, and Δ19 truncation mutants was not due to an effect on the stability or global fold of the protein but rather was due to the loss of key determinants for membrane localization that are present in the C-terminal region of EcMinD.
A Conserved Membrane-Targeting Sequence in the C-Terminal Region of MinD.
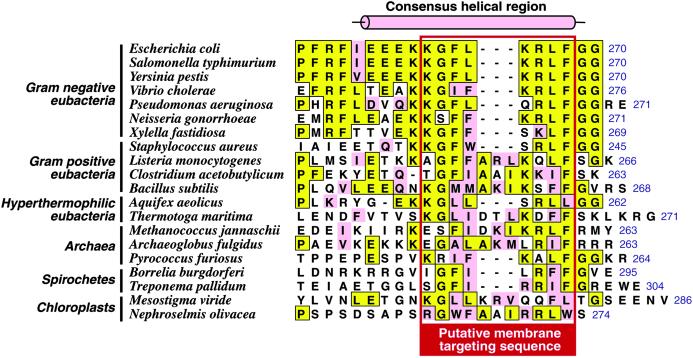
Although MinD is the best conserved of the Min proteins, the highest levels of sequence conservation are restricted to the Walker-type ATPase motifs located in the N-terminal region of the protein. However, close inspection of the C-terminal region reveals a short sequence motif that is phylogenetically well conserved (Fig. 2). Although the 10-residue sequence spanning residues Lys-261–Gly-270 of the E. coli sequence is remarkably well conserved among Gram-negative eubacteria, we showed that the terminal diglycine sequence is dispensable for membrane localization of EcMinD, and the sequence alignment in Fig. 2 shows that these residues are not highly conserved outside of Gram-negative bacteria. Thus, we propose that a slightly more restricted sequence motif corresponding to Lys-261–Phe-268 of EcMinD is responsible for membrane localization of MinD in eubacteria, archaea, and plastids. We refer to this as the MinD membrane-targeting sequence (MTS).
Fig 2.
The C terminus of MinD contains a widely conserved sequence motif. An alignment of the extreme C-terminal region of EcMinD with the corresponding region of MinD from various organisms is shown. Residues identical to the E. coli sequence are boxed in yellow and conservative substitutions are pink. The C-terminal residue is numbered (blue text to the right of each sequence). The consensus helical region (see text) is indicated by the cylinder above the sequences. The putative MTS, which is boxed in red, is conserved across eubacteria, archaea, and chloroplasts.
An obvious prediction of our hypothesis is that any MinD with the MTS should be capable of localizing correctly in E. coli. MinD from B. subtilis contains a good match with the proposed E. coli MTS (50% identity; see Fig. 2), and hence we examined the localization of BsMinD in the E. coli Δmin strain PB114. As shown in Fig. 3A, GFP-BsMinD exhibited a distinct peripheral localization pattern extending around the entire circumference of the cell; the pattern of localization in E. coli was indistinguishable from that of GFP-EcMinD (Fig. 1C). To confirm that this localization was indeed due to the proposed MTS, we examined the localization of several truncation mutants (Fig. 3E). First, we examined the localization of GFP-BsMinDΔ24 in which the C-terminal 24 residues of BsMinD were removed. BsMinD contains a small insertion in the C-terminal region compared with EcMinD, as well as a longer extension beyond the proposed MTS (see Fig. 2), so BsMinDΔ24 effectively corresponds to the EcMinDΔ19 truncation mutant. GFP-BsMinDΔ24 failed to exhibit a peripheral localization pattern (Fig. 3B), indicating that key determinants for membrane localization are located, as predicted, in the terminal 24 residues of the protein.
Fig 3.
B. subtilis MinD localizes to the membrane in E. coli. Fluorescence micrographs show localization in E. coli of BsMinD (A), BsMinDΔ24 (B), BsMinDΔ5 (C), and BsMinDΔ3 (D). (E) Illustration of two of the BsMinD constructs used in this study.
Phe-264 of BsMinD corresponds to the functionally important Phe-268 residue of EcMinD. Although the E. coli sequence extends only two residues beyond the phenylalanine, BsMinD contains an additional four residues (GVRS), which provided us with an opportunity to demarcate the C-terminal boundary of the MTS. If the phenylalanine residue corresponds to the C-terminal end of the MTS, then a five-residue truncation of BsMinD that eliminates the phenylalanine should not localize to the E. coli membrane, whereas shorter truncations should exhibit a peripheral localization pattern. As predicted, we found that GFP-BsMinDΔ5 was diffusely distributed throughout the cytoplasm (Fig. 3C), whereas GFP-BsMinDΔ3 (Fig. 3D) and GFP-BsMinDΔ2 (not shown) exhibited a peripheral localization pattern that was indistinguishable from that of GFP-BsMinD (Fig. 3A) and GFP-EcMinD (Fig. 1C). Cells expressing GFP-BsMinDΔ4 exhibited an intermediate phenotype in which only 10–20% of cells showed distinct peripheral localization of the fusion protein (not shown). This effect was not due to poor expression of the fusion protein because Western blot analyses (not shown) indicated that the MinD protein levels were very similar in cells expressing WT GFP-BsMinD and each of the truncation mutants. Thus, it seems that the MTS highlighted in Fig. 2 corresponds to the absolute minimal region required for membrane localization of MinD, but that optimal localization of some MinD proteins may require additional C-terminal residues.
We conclude that the C-terminal region of MinD contains a membrane-targeting sequence that is conserved at least in Gram-negative and Gram-positive bacteria, and most likely also in archaea and plastids. It is salient to note that MinD from the Gram-negative coccus Neisseria gonorrhoeae was recently shown to be functional in E. coli (19), which indicates that N. gonorrhoeae MinD must be able to localize to the E. coli inner membrane, consistent with the fact that it contains a canonical MTS (Fig. 2).
The MinD MTS Is an Amphipathic Helix.
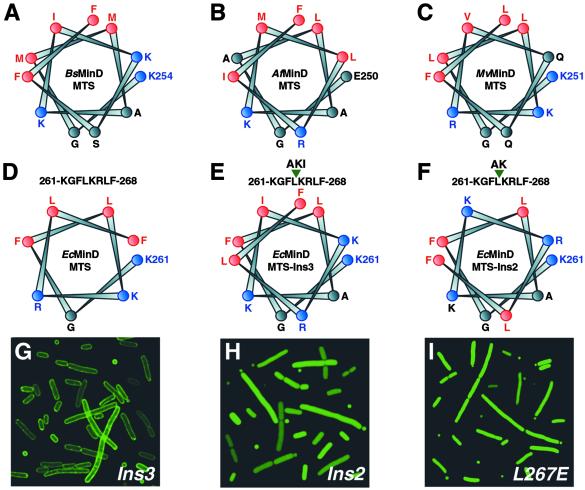
The MTS regions of MinD from archaea, eubacteria, and chloroplasts are predicted to be helical by numerous secondary structure prediction programs. When representative MinD MTSs from each domain of life are arranged on a helical wheel (Fig. 4 A–C) they form highly amphipathic helices; one face of the helix is highly hydrophobic and consists primarily of Phe, Leu, and Met residues, whereas the opposing face is polar and is generally dominated by positively charged Lys and Arg residues. Protein interactions with the lipid bilayer are commonly mediated by amphipathic helices, which are often structured only in the presence of phospholipid membranes (20, 21). We therefore wondered whether the predicted helical structure of the MTS was important for membrane localization of MinD.
Fig 4.
The helicity and amphipathicity of the MTS is important for its membrane-targeting function. (A–F) Helical-wheel representations of the putative MinD MTS from the Gram-positive eubacterium B. subtilis (BsMinD) (A), the hyperthermophilic archaeon A. fulgidus (AfMinD) (B), the chloroplast of the unicellular green flagellate Mesostigma viride (MvMinD) (C), the Gram-negative eubacterium E. coli (EcMinD) (D); a mutant of EcMinD containing a three-residue insertion (Ala-Lys-Ile) between residues Leu-264 and Lys-265 of the MTS (EcMinDIns3) (E); and a mutant of EcMinD containing a two-residue insertion (Ala-Lys) between residues Leu-264 and Lys-265 of the MTS (EcMinDIns2) (F). Strongly hydrophobic residues are shown in red and positively charged residues are shown in blue. The N-terminal residue of each helix is numbered. Note the pronounced amphipathic nature of the WT sequences; one face of the helix is highly hydrophobic, whereas the other face is strongly polar and usually comprises several cationic residues. (G–I) Fluorescence micrographs showing localization in E. coli of EcMinDIns3 (G), EcMinDIns2 (H), and an L267E mutant of EcMinD (I).
We were intrigued that many of the MTSs from Gram-positive bacteria, archaea, and chloroplasts contain a three- or four-residue insertion relative to the MTSs from Gram-negative bacteria (Fig. 2). The insertion always occurs between residues corresponding to Leu-264 and Lys-265 of the E. coli sequence (Fig. 2). We postulated that, if the MTS is helical with 3.6 residues per turn, then a three- to four-residue insertion would maintain the helical phase of the MTS. Smaller or larger insertions would alter the helical phase of the MTS and reduce or eliminate the amphipathicity of the helix, which might diminish the ability of the MTS to target MinD to the membrane. This postulate suggested an experimental strategy for testing whether the helical nature of the MTS was important for MinD localization.
First, we constructed a mutant of EcMinD in which three helix-compatible residues (Ala-Lys-Ile) were inserted between Leu-264 and Lys-265 of the MTS; these residues are equivalent to the three extra residues in the B. subtilis MTS. This mutant (EcMinDIns3) is predicted to be helical and, furthermore, the size of the insertion ensures that the amphipathicity of the MTS is maintained (see helical wheel in Fig. 4E). GFP-EcMinDIns3 exhibited a distinct peripheral localization pattern that was indistinguishable from that of GFP-EcMinD (Fig. 4G). Next, we examined the localization of a mutant in which two helix-compatible residues (Ala-Lys) were inserted between Leu-264 and Lys-265 of the MTS; this insertion should maintain the helicity of the MTS, but it destroys the polar-apolar residue phasing so that a Lys residue is positioned on the otherwise hydrophobic face of the helix and a hydrophobic Leu residue is positioned on the polar face (see helical wheel in Fig. 4F). We found that this mutant (EcMinDIns2) was diffusely distributed throughout the cytoplasm (Fig. 4H). Insertion of two helix-breaking residues (Gly-Asn) between Leu-264 and Lys-265 of the MTS also caused aberrant cytoplasmic localization of MinD (data not shown).
We further explored whether the amphipathic nature of the MTS helix is critical for membrane targeting by examining the localization of an EcMinD mutant in which Leu-267, a highly conserved residue located on the hydrophobic face of the putative MTS helix (see Fig. 4D), was mutated to a negatively charged Glu residue. Glu was chosen because it is compatible with helix formation, and a Leu → Glu mutation on the hydrophobic face of the N-terminal amphipathic helix of sterol carrier protein-2 has been shown to abolish its membrane-targeting function (20). As shown in Fig. 4I, the GFP-EcMinD(L267E) mutant was found to be localized diffusely throughout the E. coli cytoplasm. We conclude that both the helicity and the amphipathicity of the MTS is critical for its membrane-targeting function.
Discussion
MinD is a ubiquitous ATPase that has been demonstrated to play a pivotal role in selection of the division site in eubacteria (2, 19, 22) and chloroplasts (23–25). MinD associates with the inner membrane of E. coli (10), and this interaction is a prerequisite for subsequent recruitment of MinC to the membrane (5, 6). This interaction is a critical step in the topological regulation of bacterial cytokinesis because, at normal cellular concentrations, MinC can function as an inhibitor of cell division only when recruited to the membrane by MinD (5).
Until now, the mechanism by which MinD is recruited to the membrane has proved elusive. In this study we have shown that localization of MinD to the bacterial membrane is mediated by a highly conserved sequence in the C-terminal region of the protein that we refer to as the membrane-targeting sequence or MTS (Fig. 2). The consensus MTS is KG[FLI][LFI]X3–4KR[LFI][FL], where X3–4 is a three- or four-residue insertion found in some MinD proteins. Significant conservation occurs outside this region within Gram-negative eubacteria, but the MTS is the only portion of the C-terminal region that is conserved across phyla. The MinD MTS spans only 8–12 residues, but MTSs in several other proteins are similarly small; for example, the membrane-anchoring sequence of the E. coli signal-transducing enzyme IIAGlucose comprises only nine residues at the extreme N terminus of the protein (26), whereas a 12-residue amphipathic helix mediates membrane attachment of ADP-ribosylation factor 1 (ARF1) (27).
The observation that the extreme C-terminal region of MinD contains a MTS explains why large C-terminal fusions, such as GFP, abolish MinD function (22); we presume that these fusions sterically impede the interaction of the MinD MTS with the cytoplasmic membrane, thus abrogating the membrane association–dissociation cycle necessary for the biological function of MinD (13, 14).
The MinD MTS Is Not Present in Other Members of the ParA SuperFamily.
MinD is a member of the large ParA superfamily of ATPases that are characterized by a deviant Walker-type ATP-binding motif (11, 12, 28). Examination of previously compiled ParA alignments (e.g., see supporting information for ref. 11, www.pnas.org/cgi/content/full/97/26/14656/DC1/3), as well as extensive blast searches using the MinD MTS, revealed that this motif is confined specifically to MinD and is absent from other members of the ParA superfamily. Consistent with this observation, we are not aware of any ParA superfamily proteins, apart from MinD, that specifically localize to the membrane. Several ParA proteins involved in chromosome and plasmid partitioning undergo a bipolar oscillation like MinD, but they associate with the nucleoid rather than the bacterial membrane (29–31).
The MinD MTS might be a useful criterion for distinguishing between true MinD orthologs and other ParA superfamily members in organisms that are recalcitrant to genetic manipulation. For example, the annotated genome sequence of the spirochete Borrelia burgdorferi includes three chromosomally encoded “MinD-related proteins” (YLXH-1, YLXH-2, and YLXH-3), but only one of these (YLXH-1) contains the MinD MTS signature. Similarly, P. furiosus contains three apparent MinD homologs, but only one contains the MinD MTS signature. The MinD MTS does not seem to be present in either of the Pyrococcus proteins chosen for structure determination (16, 17), and hence, these proteins may not be functional homologs of MinD. However, the MTS is present in the A. fulgidus MinD homolog used for structure determination (15), and hence, this protein is likely to be an authentic MinD ortholog.
A Model for Membrane Association of MinD.
The MinD MTSs are predicted to be helical and, when displayed on a helical wheel, the putative helices are highly amphipathic. One face of the helix is composed entirely of hydrophobic residues (most often Phe and Leu), whereas the opposing polar face usually contains several positively charged Arg and Lys residues and is generally devoid of anionic residues (Fig. 4 A–D). Experiments with several insertion mutants (Fig. 4 E–H) and an EcMinD(L267E) point mutant (Fig. 4I) suggested that both the helicity and amphipathicity of the MinD MTS are important for its membrane-targeting function.
Amphipathic helices mediate the association of numerous proteins with biological membranes (32, 33). These helices are generally unstructured in the absence of membrane (20, 21, 26, 34) and, in most cases that have been examined experimentally, they preferentially associate with anionic phospholipids because of the large number of positively charged residues on the polar face of the helix; examples include sterol carrier protein-2 (20), CTP:phosphocholine cytidyltransferase (34), and the mammalian GTPase RGS4 (21). The marked preponderance of cationic residues on the polar face of the MinD MTS suggests that this motif most likely mediates the interaction of MinD with biological membranes by preferentially interacting with anionic phospholipids. Consistent with this hypothesis, it was previously shown that mutation of Gly-262 on the polar (cationic) surface of the EcMinD MTS to a negatively charged Asp residue results in a minicelling phenotype (35). The cellular localization of the G262D mutant has not been examined, but we predict that the minicelling phenotype is a direct result of impaired membrane association of EcMinD.
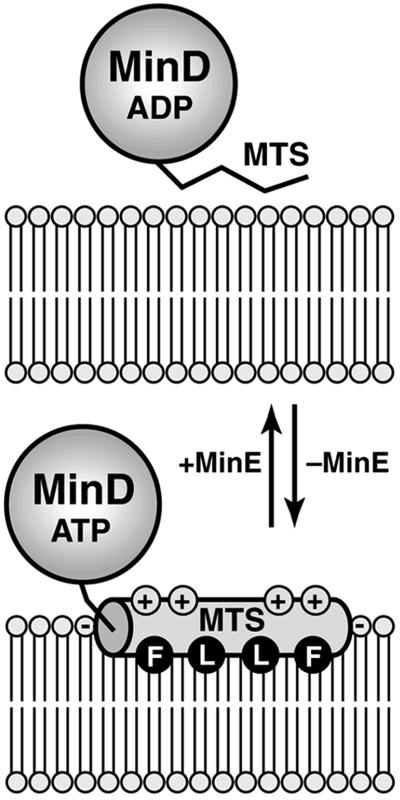
Amphipathic helices that contain a large number of positively charged residues on their polar face usually align parallel to the membrane surface, because it is thermodynamically highly unfavorable to insert Lys and Arg residues into the hydrophobic interior of the bilayer (33). We anticipate that this is likely to be the case for the MinD MTS. As shown in Fig. 5, the helix orientation is likely to be such that the hydrophobic residues interact directly with lipid acyl chains, whereas the cationic residues on the opposite face of the helix interact with the headgroups of anionic phospholipids (21, 33). This type of superficial protein–lipid interaction is often used by proteins whose activity is regulated by reversible membrane association; for example, the amphipathic membrane-targeting helix of CTP:phosphocholine cytidyltransferase mediates interconversion between the inactive cytoplasmic and active membrane-bound forms of the enzyme (34).
Fig 5.
Model of the MinD membrane attachment–detachment cycle. In the absence of MinE (Lower), the C-terminal MTS of MinD forms an amphipathic helix (shaded cylinder) that interacts with the lipid bilayer. The helix most likely orients parallel to the membrane surface. Partial insertion of the helix into the cytoplasmic monolayer would allow residues such as Phe (F) and Leu (L) on the hydrophobic face of the amphipathic helix to interact with lipid acyl chains, whereas residues on the opposing polar face of the helix could interact with lipid headgroups. The numerous cationic residues (indicated by +) on the polar face of the MinD MTS probably make specific contacts with the headgroups of anionic phospholipids (indicated by −). We presume that MinE causes detachment of the MTS, thus releasing MinD from the membrane (Upper). It is unclear at present whether this release involves a direct interaction between MinE and the MTS, a conformational change in MinD provoked by MinE-induced ATP hydrolysis, or some other mechanism.
MinD reversibly associates with the membrane during its catalytic cycle: ATP binding promotes association with phospholipid bilayers, whereas MinE-stimulated ATP hydrolysis leads to release of MinD from lipid bilayers (13, 14). This cycle of attachment–release–reattachment occurs throughout the cell cycle and must be very rapid because bipolar oscillation of the protein (i.e., from one cell pole to the other and back again) can occur in a time frame of less than a minute (4, 13). Superficial attachment of MinD to the bilayer by an amphipathic helix is more compatible with such rapid membrane trafficking than stable attachment to the membrane by some form of transmembrane anchor.
What Regulates MTS Association–Dissociation Cycles?
We presume that MinE-stimulated ATP hydrolysis leads to detachment of the MinD MTS from the membrane, which raises two critical questions: What is the molecular basis of MTS detachment from the membrane, and what prevents it from immediately reassociating with the bilayer? We suggest two possibilities. First, it is possible that MinE interacts directly with the MTS, thereby causing it to detach from the membrane and remain cytosolic so long as it continues to associate with MinE. Second, it is possible that ATP hydrolysis provokes a change in the conformation and/or polymerization state of MinD that makes it less favorable for the MTS to interact with the bilayer. One possible scenario is that, after detachment from the membrane, the MTS associates with the core of the MinD protein such that the hydrophobic face of the helix becomes buried. This mechanism is used by several GTPases to modulate their interaction with biological membranes. For example, the Ras-related GTPase ARF1 associates reversibly with membranes in a GTP-dependent manner and this interaction is mediated by an N-terminal amphipathic helix (27). In the crystal structure of the GDP-bound form of ARF1 (36), the N-terminal helix is tightly associated with the rest of the protein, with residues on the hydrophobic face directed toward the protein core. However, GTP binding provokes a conformational change in ARF1 that exposes these hydrophobic residues and allows them to interact with membrane lipids (27, 37).
Only minor differences exist between the structures of a presumptive P. furiosus MinD homolog (16) complexed with ADP and the nonhydrolyzable ATP analog adenosine 5′-[β,γ-methylene]triphosphate (AMPPCP), which suggests that membrane association of the MinD MTS is not controlled by an ATP-induced conformational switch. However, AMPPCP may not be a good ATP analog in the case of MinD because ATP and adenosine 5′-O-(3-thiotriphosphate) (ATPγS) promote interaction of MinD with phospholipid vesicles, whereas AMPPCP does not (14). Furthermore, the P. furiosus protein may not be a true MinD ortholog because it lacks a canonical MinD MTS, and therefore its crystal structure may not be indicative of the conformational changes that occur when MinD binds ATP. Consequently, it remains possible that ATP binding induces a conformational change in MinD that promotes membrane attachment of the MTS. Structures of an authentic MinD complexed with ADP and ATPγS should help to resolve this issue.
Acknowledgments
This manuscript is dedicated to the memory of our good friend and colleague, Dr. Gregory P. Mullen. We thank Dr. Debabrata RayChaudhuri for the BL21ΔminCDE strain. This work was supported by National Institutes of Health Grants GM-48583 (to G.F.K.) and GM-60632 (to L.I.R.).
Abbreviations
ARF1, ADP-ribosylation factor-1
BsMinD, Bacillus subtilis MinD
EcMinD, Escherichia coli MinD
MTS, membrane-targeting sequence
References
- 1.Rothfield L. I. & Justice, S. S. (1997) Cell 88 581-584. [DOI] [PubMed] [Google Scholar]
- 2.de Boer P. A. J., Crossley, R. E. & Rothfield, L. I. (1989) Cell 56 641-649. [DOI] [PubMed] [Google Scholar]
- 3.Rothfield L. I., Shih, Y.-L. & King, G. F. (2001) Cell 103 13-16. [DOI] [PubMed] [Google Scholar]
- 4.Raskin D. M. & de Boer, P. A. J. (1999) Proc. Natl. Acad. Sci. USA 96 4971-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raskin D. M. & de Boer, P. A. J. (1999) J. Bacteriol. 181 6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Z. & Lutkenhaus, J. (1999) Mol. Microbiol. 34 82-90. [DOI] [PubMed] [Google Scholar]
- 7.Rowland S. L., Fu, X., Sayed, M. A., Zhang, Y., Cook, W. R. & Rothfield, L. I. (2000) J. Bacteriol. 182 613-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu X., Shih, Y.-L., Zhang, Y. & Rothfield, L. I. (2001) Proc. Natl. Acad. Sci. USA 98 981-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothfield L. I., Justice, S. & Garcia-Lara, J. (1999) Annu. Rev. Genet. 33 423-448. [DOI] [PubMed] [Google Scholar]
- 10.de Boer P. A. J., Crossley, R. E., Hand, A. R. & Rothfield, L. I. (1991) EMBO J. 10 4371-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamaichi Y. & Niki, H. (2000) Proc. Natl. Acad. Sci. USA 97 14656-14661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerdes K., Møller-Jensen, J. & Jensen, R. B. (2001) Mol. Microbiol. 37 455-466. [DOI] [PubMed] [Google Scholar]
- 13.Hu Z. & Lutkenhaus, J. (2001) Mol. Cell 7 1337-1343. [DOI] [PubMed] [Google Scholar]
- 14.Hu Z., Gogol, E. P. & Lutkenhaus, J. (2002) Proc. Natl. Acad. Sci. USA 99 6761-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordell S. C. & Lowe, J. (2001) FEBS Lett. 492 160-165. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi I., Oyama, T. & Morikawa, K. (2001) EMBO J. 20 1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakai N., Yao, M., Itou, H., Watanabe, N., Yumoto, F., Tanokura, M. & Tanaka, I. (2001) Structure (London) 9 817-826. [DOI] [PubMed] [Google Scholar]
- 18.Justice S. S., García-Lara, J. & Rothfield, L. I. (2000) Mol. Microbiol. 37 410-423. [DOI] [PubMed] [Google Scholar]
- 19.Szeto J., Ramirez-Arcos, S., Raymond, C., Hicks, L. D., Kay, C. M. & Dillon, J. A. (2001) J. Bacteriol. 183 6253-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang H., Ball, J. M., Billheiner, J. T. & Schroeder, F. (1999) Biochemistry 38 13231-13243. [DOI] [PubMed] [Google Scholar]
- 21.Bernstein L. S., Grillo, A. A., Loranger, S. S. & Linder, M. E. (2000) J. Biol. Chem. 275 18520-18526. [DOI] [PubMed] [Google Scholar]
- 22.Marston A. L., Thomaides, H. B., Edwards, D. H., Sharpe, M. E. & Errington, J. (1998) Genes Dev. 12 3419-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colletti K. S., Tattersall, E. A., Pyke, K. A., Froelich, J. E., Stokes, K. D. & Osteryoung, K. W. (2000) Curr. Biol. 4 507-516. [DOI] [PubMed] [Google Scholar]
- 24.Kanamaru K., Fujiwara, M., Kim, M., Nagashima, A., Nakazato, E., Tanaka, K. & Takahashi, H. (2000) Plant Cell Physiol. 41 1119-1128. [DOI] [PubMed] [Google Scholar]
- 25.Dinkins R., Reddy, M. S., Leng, M. & Collins, G. B. (2001) Planta 214 180-188. [DOI] [PubMed] [Google Scholar]
- 26.Wang G., Peterkofsky, A. & Clore, G. M. (2000) J. Biol. Chem. 275 39811-39814. [DOI] [PubMed] [Google Scholar]
- 27.Antonny B., Beraud-Dufour, S., Chardin, P. & Chabre, M. (1997) Biochemistry 36 4675-4684. [DOI] [PubMed] [Google Scholar]
- 28.Koonin E. V. (1993) J. Mol. Biol. 229 1165-1174. [DOI] [PubMed] [Google Scholar]
- 29.Quisel J. D., Lin, D. C.-H. & Grossman, A. D. (1999) Mol. Cell 4 665-672. [DOI] [PubMed] [Google Scholar]
- 30.Marsten A. L. & Errington, J. (1999) Mol. Cell 4 673-682. [DOI] [PubMed] [Google Scholar]
- 31.Ebersbach G. & Gerdes, K. (2001) Proc. Natl. Acad. Sci. USA 98 15078-15083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson J. E. & Cornell, R. B. (1999) Mol. Membr. Biol. 16 217-235. [DOI] [PubMed] [Google Scholar]
- 33.Bechinger B. (2000) Curr. Opin. Chem. Biol. 4 639-644. [DOI] [PubMed] [Google Scholar]
- 34.Dunne S. J., Cornell, R. B., Johnson, J. E., Glover, N. R. & Tracey, A. S. (1996) Biochemistry 35 11975-11984. [DOI] [PubMed] [Google Scholar]
- 35.Labie C., Bouché, F. & Bouché, J.-P. (1990) J. Bacteriol. 172 5852-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amor J. C., Harrison, D. H., Kahn, R. A. & Ringe, D. (1994) Nature 372 704-708. [DOI] [PubMed] [Google Scholar]
- 37.Goldberg J. (1998) Cell 95 237-248. [DOI] [PubMed] [Google Scholar]