Abstract
Nramp1 (Natural resistance-associated macrophage protein-1; also known as Slc11a1) is a host resistance gene that provides protection against several intracellular pathogens, including Salmonella enterica serovar Typhimurium. Little is known about the dynamic interplay that occurs between mammalian host resistance determinants such as Nramp1 and pathogens during infection. To explore these interactions, we examined the effect of Nramp1 on expression of Salmonella typhimurium (STM) virulence factors. We demonstrate that Salmonella pathogenicity island 2 (SPI2) is essential for replication of STM in spleens of infected Nramp1+/+ mice. Furthermore, the presence of Nramp1 in transfected cell lines and congenic knockout mice resulted in the up-regulation of STM SPI2-associated virulence genes critical for intramacrophage survival. This Nramp1-dependent up-regulation of SPI2 was mimicked in vitro by chelation of iron, demonstrating the iron-responsive nature of expression of STM SPI2-associated virulence genes. We propose that acquisition of SPI2 by S. enterica not only enabled this bacterium to become an effective intracellular pathogen but also allowed the bacterium to withstand the effects of macrophage defense mechanisms such as Nramp1 early in the evolution of its pathogenic character. These dynamic Nramp1–pathogen interactions may be essential for regulating the course of an infection. This study demonstrates the presence of a previously undescribed direct influence of a mammalian innate host resistance locus on a pathogen at the genetic level.
It has become apparent in recent years that the study of the dynamic interplay between host and pathogen is crucial to our understanding of the disease process. In particular, the means by which innate host resistance loci [e.g., Char4, natural resistance-associated macrophage protein-1 (Nramp1) (1, 2)] are involved in the complex response to an infection represents a largely unexplored area of research. Studies over the last 30 years have identified Nramp1 (also called Slc11a1 for solute carrier 11a1) as a major regulator of host susceptibility to antigenically unrelated intracellular pathogens, including Mycobacterium bovis bacillus Calmette–Guérin and Salmonella typhimurium (STM) (3). The absence of functional Nramp1 in mice leads to unrestrained bacterial replication in the liver and spleen, resulting in a lethal infection. Genetic polymorphisms of the human homologue, NRAMP1, have been linked to susceptibility to various bacterial diseases, including tuberculosis and leprosy, as well as diseases with a suspected bacterial etiological component, including inflammatory bowel disease and rheumatoid arthritis (4).
However, the mechanism of action of Nramp1 on intracellular pathogens at the subcellular level remains ambiguous. A late endocytic/lysosomal protein located primarily in macrophages, Nramp1 appears to function as a divalent cation transport system across the membrane of the parasitic vacuole (3). Nramp1 has been shown to transport divalent cations, including Fe2+, Mn2+, and, to a lesser extent, Zn2+, in Xenopus oocytes (5, 6), although the direction of transport of substrate cations remains controversial. More recently, Nramp1 has been proposed to play a role in iron recycling, removing iron and iron-containing compounds from the macrophage after phagocytosis of dead red blood cells (7). Despite the importance of Nramp1 in infection, surprisingly few studies have examined Salmonella virulence in Nramp1+/+ hosts. The question remains: How does a pathogen respond to the presence of this host resistance factor?
Salmonella species are the etiological agents of two major human diseases, the systemic disease typhoid fever (Salmonella typhi) and a self-limiting gastroenteritis (STM). STM is an intracellular pathogen notable for its ability to survive and replicate inside macrophages, a complex process attributed to a number of virulence genes located either within a large pathogenicity island at centisome 31 (known as Salmonella pathogenicity island 2, SPI2) or within “pathogenicity islets” scattered around the chromosome (8). These virulence genes collectively encode a discrete type three secretion system and associated secreted effectors, which function to establish the intracellular niche and maintain the bacterium in a discrete vacuole within the macrophage known as the Salmonella-containing vacuole (SCV), wherein they replicate (9).
The murine model of Salmonella infection relies on the development of a systemic disease similar to human typhoid in susceptible (i.e., Nramp1−/−) strains of mice. This model has allowed much to be elucidated about this pathogen's strategy to survive within a host. It is known that development of a systemic infection relies on the ability of STM to gain access to, survive, and replicate within host macrophages. Mutations in structural or regulatory genes associated with the SPI2 type three secretion system are dramatically attenuated in the murine typhoid model (10); such mutants disperse through the reticuloendothelial system of infected mice but do not proliferate (11). The requirement for intramacrophage survival in Salmonella disease and the colocalization of Nramp1 to the SCV within macrophages (12) suggest the possibility of crosstalk between this host resistance protein and the intracellular pathogen. Recently it was demonstrated that Nramp1 alters the maturation pattern of the SCV in cultured transfected cell lines (13). It is possible that such modulatory effects of Nramp1 on the parasitic vacuole, either at the level of trafficking or divalent cation concentration, have downstream effects on bacterial responses. Because virtually all work to date on Salmonella pathogenesis has been undertaken in Nramp1−/− cell lines or animals, this interplay has not been adequately studied and remains poorly understood. Considering the extensive characterization of Salmonella genetics as well as in vitro and in vivo infection models, the murine typhoid model represents an ideal system for investigating the interaction between Nramp1 and an intracellular pathogen in vivo. Therefore, we investigated the interplay between STM virulence genes required for intramacrophage survival and Nramp1 using both cell lines and congenic strains of resistant and susceptible mice.
Materials and Methods
Bacterial Strains, Vectors, and Growth Conditions.
Mouse virulent STM 14028s was used throughout. The ssrA:Km strain was obtained from Samuel Miller (University of Washington). Vectors used included pCRTOPO2.1 (Invitrogen) for cloning of PCR products and pFZY1 as the monocopy transcriptional fusion vector (14). LB broth and agar plates were used for growth and maintenance of bacterial strains. N minimal media (pH 7.4) were used for in vitro expression studies and were supplemented with 0.3% glycerol/0.1% casamino acids/8 μM MgCl2 (15). When required, compounds were added to the N minimal media at the final concentrations noted: dipyridyl, 250 μM; EDTA, 250 μM; EGTA, 250 μM; MnSO4, 500 μM; MgSO4, 500 μM; FeSO4, 500 μM; ampicillin, 100 μg/ml; carbenicillin, 100 μg/ml; kanamycin (Km), 50 μg/ml; tetracycline, 30 μg/ml; gentamicin, 10 or 100 μg/ml.
Cell Lines.
The Nramp1−/− murine macrophage-like cell line RAW 264.7 (American Type Culture Collection TIB 71) was used throughout. Stably transformed cell lines and their maintenance were as previously described (12).
DNA Manipulations.
Plasmid preparations, purification of PCR products, and isolation of DNA fragments from agarose gels were performed by using the appropriate Qiagen (Chatsworth, CA) kit. Transcriptional fusions to the genes of interest were created by PCR amplifying a fragment, including the upstream regulatory region of the desired genes. Oligonucleotides were designed to contain an EcoRI site at the 5′ end of the transcript and a HindIII site at the 3′ end to facilitate cloning into pFZY1. Sequences are available on request. All PCR products were amplified by using Pfx DNA polymerase (GIBCO) and verified by sequencing.
Intracellular Gene Expression and Invasion Assays.
The combined intracellular gene expression and invasion assays were carried out as described in Kehres et al. (16). Plates were read by using the luminescence detection function of the Spectrafluor Plus (TECAN, Austria) and data evaluated by using magellan software, Version 1.1 (TECAN, Austria). Assays were performed in duplicate on five independent occasions, and all results were averaged.
In Vitro Gene Expression Assay.
Bacterial cultures were grown overnight in N minimal media at 37°C with shaking. Cultures were subcultured at a 1:100 dilution to fresh N minimal media containing various supplements as described in the text. Subcultures were grown for 3 h and assayed for bacterial enumeration and β-galactosidase activity, as described above. Assays were performed in duplicate on three separate occasions, and all results were averaged.
Murine Typhoid Model.
For in vivo gene expression assays, bacterial strains were grown overnight in LB broth at 37°C with shaking at 200 rpm in an Orbit gyrorotary incubator (Lab-Line Instruments) and subcultured 1:100 for 3 h in fresh LB. Cultures were normalized to an OD600 of 0.3 and diluted 1:6,000 for inoculation. Groups of five age- and sex-matched 129svJ Nramp1−/− and Nramp1+/+ mice (17) between the ages of 6 and 18 weeks were infected i.p. with ≈2.5e5 bacteria per mouse as determined by retrospective bacterial plate counts. Mice were killed 24 h postinfection (p.i.) and their spleens harvested. Splenic tissue was homogenized in PBS containing Complete EDTA-free protease inhibitor mixture (Roche Diagnostics). Half of the homogenate was used for bacterial enumeration on selective plates and the other half for β-galactosidase assays, as described above. Background splenic β-galactosidase activity was taken into account by infecting control mice with wild-type STM lacking the lacZY plasmid. Assays were repeated on four separate occasions with groups of five mice; all data points are represented. Bacterial enumeration assays were carried out as described (18).
Results
SPI2 Is Necessary for Replication of STM in Nramp1+/+ Mice.
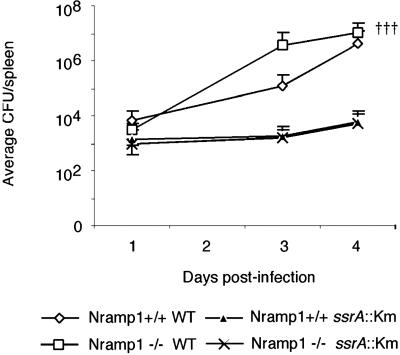
It has been established that the SPI2 type three secretion system and associated effectors are necessary for full virulence of STM in the murine typhoid model (10). However, these studies have all been carried out in Nramp1−/− mice and cultured cell lines or unrelated Nramp1+/+ and Nramp1−/− mouse strains, which makes comparison difficult (10, 11, 19–21). It is also well documented that expression of functional Nramp1 controls replication of wild-type STM both in vivo and in vitro (3). Therefore, we compared the role of SPI2 in bacterial survival and replication in congenic Nramp1+/+ and Nramp1−/− mice (Fig. 1). Previous studies in our laboratory using the same infection model determined that 100% of Nramp1−/− animals infected with wild-type STM succumbed to infection by day 5 p.i., whereas the bacterial load in the liver and spleen of Nramp1+/+ animals peaked at day 4 p.i. and plateaued thereafter, with the animals ultimately surviving the infection (ref. 22 and data not shown). Accordingly, we observed extensive replication of wild-type bacteria in the Nramp1−/− animals, such that by day 4, 30% of the animals in this group had succumbed to infection. Further, although Nramp1+/+ mice do not usually succumb to infection, we observed that wild-type bacteria replicated substantially in these animals. In contrast, the SPI2− bacteria behaved nearly identically in both strains of mice, replicating to a limited extent in both the presence and absence of functional Nramp1. This demonstrates that SPI2 is essential for bacterial replication in both Nramp1−/− and Nramp1+/+ mice.
Fig 1.
SPI2 permits replication of STM in murine spleen in the presence of Nramp1. Age- and sex-matched congenic 129sv/J mice were infected i.p. with 1e5 cfu of either wild-type or SPI2- (ssrA:Km) STM 14028s and spleens were harvested for bacterial enumeration at designated time points. Data represent the average ± the SEM of two experiments performed on experimental groups of five mice.
SPI2-Associated Loci Are Up-Regulated in Nramp1-Transfected Cell Lines.
The presence of Nramp1 on the membrane of the SCV during infection of macrophages may influence the composition of the intravacuolar environment due to its putative role as a divalent cation transporter or observed effects on vacuolar trafficking (13, 23). Considering the importance of SPI2 for replication of STM in the presence of Nramp1, we examined the transcriptional regulation of STM genes important for intramacrophage survival, in the presence and absence of Nramp1. It was possible that Nramp1 exerted its effects on STM by down-regulating expression of SPI2 genes, thereby mediating its bacteriostatic effect. Three SPI2-associated genes were selected for this study. ssrA encodes, within SPI2 itself, the sensor kinase of SsrAB, the two-component regulatory system (TCRS) responsible for SPI2 gene expression (24). sseA is the first gene of the proposed SPI2 effector operon and represents a SPI2-encoded locus. sseJ (25), a SPI2-associated locus encoded outside SPI2, is a recently characterized SPI2 effector. In addition, the locus encoding the response regulator for the PhoPQ TCRS (phoP) gene encoding the sensor for the PhoPQ TCRS and the Salmonella pathogenicity island-1 (SPI1)-encoded regulatory gene hilA were also chosen. PhoP is involved in intramacrophage survival and the control of the most extensive virulence-associated regulon known to date (15). hilA plays a central role in the expression of SPI1 loci (26) and was used as a negative control, because SPI1-associated genes are believed to be poorly expressed in the intracellular environment.
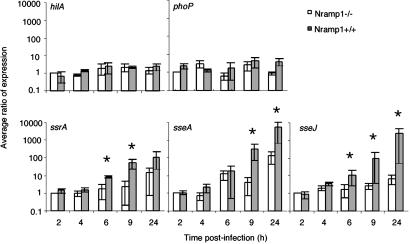
Pioneering studies have identified the expression of certain bacterial genes during infection (24, 27–29), but with one recent exception (30) they are qualitative and do not give us much information on time course of expression or quantification. To quantify virulence gene expression over a comprehensive time course, STM carrying transcriptional fusions to the genes of interest on monocopy plasmids were generated as described in Materials and Methods. The impact of Nramp1 on the expression of these genes was determined by infecting previously characterized isogenic stably transfected Nramp1+ or Nramp1− cell lines (12), with STM carrying the respective transcriptional fusion of interest over a 24-h time course. The fusion plasmids were stably maintained by the intracellular bacteria during this time course (data not shown). Although hilA expression was not affected by the presence of transfected Nramp1 (Fig. 2), different patterns of expression were observed for the other gene fusions. Basal expression levels of phoP remained relatively unchanged throughout the experiment (Fig. 2). Although a trend toward increased phoP expression in the presence of Nramp1 was observed at later time points, it did not reach significance. In contrast, the SPI2-associated genes, ssrA, sseA, and sseJ, demonstrated low expression levels for the first few hours p.i., which increased by 6 h p.i (Fig. 2). In the Nramp1− cell lines, expression of ssrA, sseA, and sseJ increased ≈15-, 125-, and 7-fold over the 24-h time course. In the Nramp1+ cell lines, this induction rose to a total of 80-, 5,000-, and 3,000-fold over 24 h for the respective genes. (Fig. 2). Further comparison of the induction ratios between the cell lines at 24 h demonstrates that ssrA, sseA, and sseJ are up-regulated 8-, 40-, and 360-fold, respectively, in the presence of Nramp1 at this final time point. This response is attributable to the alteration of intracellular conditions by Nramp1, because extracellular bacteria exposed to the Nramp1+ and Nramp1− macrophages displayed equivalent lower levels of gene expression (data not shown).
Fig 2.
Presence of Nramp1 in transfected RAW264.7 cells leads to the up-regulation of SPI2 and SPI2-associated virulence genes. Cells were infected with STM14028s carrying the plasmids containing the different transcriptional fusions. Patterns of expression over a 24-h time course are shown for the SPI1 regulatory gene hilA, the pleiotropic virulence regulatory gene phoP, the locus encoding the kinase for the two-component regulatory system that regulates SPI2, ssrA, the SPI2-encoded effector locus sseA, and the SP12 effector locus sseJ in RAW264.7 cell lines transfected with empty pCB6 vector (Nramp1−) or with pCB6 carrying a constitutively expressed wild-type Nramp1 gene (Nramp1+). These data are expressed as the average ratio of expression normalized to the initial observed level of expression at 2 h postinfection in the Nramp1− cell line and represent the mean ± the SEM of five separate experiments performed in duplicate. Statistical significance (P < 0.05) was determined by using the Wilcoxon rank sum test for unpaired samples.
Nramp1 Mediates the Up-Regulation of Virulence Genes in Vivo.
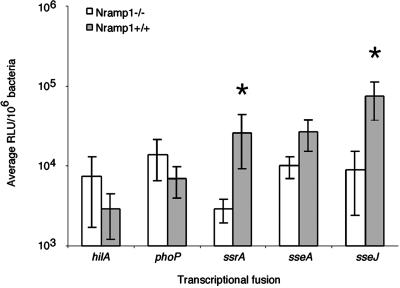
To confirm that observations obtained with the transfected cell lines were representative of events that occur during in vivo infection, we studied the expression levels of these genes in congenic Nramp1+/+ and Nramp1−/− mice. Both hilA and phoP were expressed in both strains of mice but neither was significantly affected by Nramp1 (Fig. 3). In contrast, all three SPI2 loci displayed higher expression levels in the Nramp1+/+ mice, on average 10-, 5-, and 35-fold increases for ssrA, sseA, and sseJ, respectively (ssrA and sseJ values are statistically significant) when normalized to expression levels observed in the Nramp1−/− mice. Thus in both a transfected cell line model and the murine typhoid infection model, the presence of functional Nramp1 correlates with significant up-regulation of SPI2-associated genes.
Fig 3.
Expression of SPI2 and SPI2-associated genes is up-regulated in spleens of STM-infected Nramp1+/+ 129sv mice. An average of 2.5e5 STM 14028s bearing the different transcriptional fusion plasmids were injected i.p. into age- and sex-matched 129sv mice and spleens were harvested at 24 h postinfection for evaluation of β-galactosidase activity. These data represent the mean ± the SEM of five separate experiments performed on groups of five mice per fusion of interest. Statistical significance (P < 0.05) was determined using the Wilcoxon rank sum test for unpaired samples.
Chelation of Iron Mimics the Nramp1-Associated Up-Regulation of SPI2-Associated Genes.
The specific effects of Nramp1 on the intramacrophage environment have not been established, although some reports indicate it affects divalent cation concentrations. To find an in vitro correlation between the proposed role of Nramp1 as an efflux pump of divalent cations and the results obtained in this study, we tested the effect of three different chelators, including the general chelators EGTA and EDTA and the “iron specific” chelator dipyridyl (31), on SPI2 gene expression. The basal medium in these studies was N minimal media at pH 7.4, previously defined as SPI2 expression media (15), because it is thought to mimic the intravacuolar environment. Divalent cations were also added with chelators to ensure the specificity of the chelation effects on bacterial gene expression.
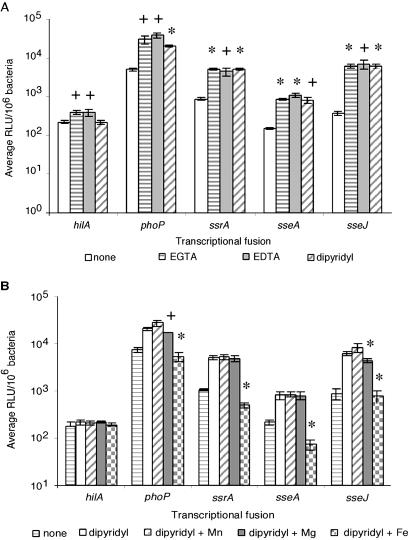
As can be seen from Fig. 4A, addition of all three individual chelators had minimal effects on hilA expression, although EGTA and EDTA had more noticeable effects than dipyridyl. phoP responded more dramatically to EGTA and EDTA than to dipyridyl. SPI2-associated genes responded as strongly to dipyridyl as to EDTA and EGTA. Addition of Mg2+ to EDTA- or EGTA-chelated cultures reduced expression of phoP to a moderate extent but had no effect on SPI2 gene expression (data not shown); addition of Mn2+, Mg2+, or Fe2+ to unchelated media also had no effect of expression levels of all three SPI2-associated loci (data not shown).
Fig 4.
(A) Chelation of divalent cations by various chelators results in up-regulation of STM virulence genes. Expression levels are shown for hilA, phoP, ssrA, sseA, and sseJ in N minimal media alone, or N minimal media supplemented with one of the following chelators: EGTA, EDTA, or dipyridyl. Statistical significance (+, P < 0.05; *, P < 0.01) was determined by using Student's t test for paired samples. (B) Chelation of divalent cations results in up-regulation of SPI2 and SPI2-associated genes in vitro. Expression levels in N minimal media alone, N minimal media supplemented with the chelator dipyridyl, or dipyridyl plus one of the following divalent cations MgSO4, MnSO4, or FeSO4, are shown for hilA, phoP, ssrA, sseA, and sseJ. Statistical analyses reflect differences in expression levels between cultures grown in dipyridyl-chelated media versus cultures grown in media supplemented with dipyridyl and the indicated excess divalent cation. Statistical significance (+, P < 0.05; *, P < 0.01) was determined by using Student's t test for paired samples.
Because there appeared to be a SPI2-restricted up-regulation of gene expression induced by dipyridyl and SPI2 expression did not appear to be affected by Mg2+ concentration, subsequent studies used dipyridyl as the chelator (Fig. 4B). Growth of STM in media supplemented with dipyridyl and excess Mn2+ had no effect on expression of any of these selected genes. The addition of excess Mg2+ repressed expression of phoP and sseJ but had no effect on expression of hilA, ssrA, or sseA. In contrast, addition of excess Fe2+ to dipyridyl-chelated media had no effect on hilA expression, yet significantly repressed expression of phoP, ssrA, sseA, and sseJ back to or below basal levels. Similar results were obtained when N minimal media at pH 4.5 were used to mimic the acidified SCV (data not shown). Iron regulation is still observed in STM carrying mutations in genes coding for regulatory proteins that respond to iron limitation, such as fur and pmrAB (32, 33), as well as in phoP (data not shown). The unique pattern of expression of phoP and sseJ was also found not to be attributable to coordinate regulation by PhoPQ (not shown). These results correlate with the hypothesis that Nramp1 is creating an iron-deprived environment within the SCV and suggest that STM has developed or acquired a specific iron-sensing regulatory mechanism that responds to iron limitation by increasing the expression of SPI2 genes necessary for intracellular survival. The gene coding for the transcriptional regulator responsible for this iron-limitation response is a matter of current investigation.
Discussion
To our knowledge, a direct comparison of the effect of SPI2 in congenic Nramp1+/+ and Nramp1−/− lineages of mice had not been undertaken. Similarly, the interplay between mammalian and pathogen resistance factors, including potential interactions between Nramp1 and SPI2, have not yet been adequately examined. Using congenic knockout mice infected i.p. with STM, we determined that minimal bacterial replication occurred in both Nramp1+/+ and Nramp1−/− mice in the absence of SPI2, reinforcing the importance of these virulence genes during infection in either model system. In Nramp1−/− mice infected with wild-type (SPI2+) STM, the bacteria replicated rapidly, and eventually the animals were overwhelmed (Fig. 1 and data not shown). In contrast, during a similar infection of Nramp1+/+ mice, significant bacterial replication in the spleen of the mice was observed, although ultimately bacterial load in this organ plateaued and mice survived the infection. Therefore, SPI2 is essential for maintaining a moderate level of bacterial replication in Nramp1+/+ animals.
It is not known at what time point or stage during the infectious process Nramp1 begins to affect intracellular pathogens. Previous studies indicate that after tail-vein injection of bacteria, bacterial replication is reduced in Nramp1+/+ animals when compared with congenic Nramp1−/− animals as early as 24 h p.i (12). In contrast, our data (Fig. 1) demonstrate no significant difference in bacterial load until day 2–3, which may reflect kinetic differences due to the i.p. route of infection used in this study. Interestingly, Nramp1 colocalizes with the phagosome and begins to extrude Mn2+ ions as early as 60 min p.i. in transfected cultured cells (23). Despite this apparent rapid activity of Nramp1, our most dramatic Nramp1-mediated effects were not observed on loci that are expressed immediately on reaching the intracellular environment (hilA, phoP), but with the SPI2-associated genes, loci that are expressed to higher levels at later stages of infection (i.e., after 6 h p.i.; ref. 24). In these experiments, the Nramp1-associated increase in SPI2-associated gene expression was maximal at 24 h p.i. We observed an up-regulation of SPI2-associated genes not only in transfected cell lines but also by isolating viable bacteria from spleens of infected Nramp1+/+ and Nramp1−/− animals 24 h after i.p. infection. The in vitro and in vivo results correlate well, such that: (i) hilA and phoP are not differentially expressed between the two lineages of animals; and (ii) the SPI2-associated genes ssrA and sseJ are significantly up-regulated in the Nramp1+/+ animals. To our knowledge, this represents the first study to demonstrate that the function of a particular host protein influences the expression pattern of STM genes required for intracellular survival.
Nramp1 has been suggested to be a H+/divalent cation antiport system such that direction of transport of divalent cations can vary depending on the magnitude of a pH gradient (5). Jabado et al. demonstrated that the phagosome reaches pH 6.5 by 1 h p.i., and that by this time, Nramp1 is responsible for the active efflux of Mn2+ from the SCV (23). Thus, because we had identified a relationship between Nramp1 and expression of SPI2 genes within the SCV, it was of interest to determine whether this proposed function of Nramp1 was related to our gene expression data. The most strongly documented transport substrates for Nramp1 and homologous transporters are Fe2+ and Mn2+, and as such these were the cations of focus in our in vitro experiments (3, 16, 23). Because the reported affinity of Nramp1 for Mg2+ does not support a role for physiological relevance of Mg2+ in this system, Mg2+ was primarily studied as a control for phoP expression (5). An in vitro expression assay based on growth media known to induce expression of SPI2-associated genes (15) was developed and used to determine that the SPI2-associated genes, and phoP to a lesser extent, were up-regulated in iron-depleted media and were repressed by addition of excess iron. There was no significant impact of Mn2+ levels on expression of these genes. To our knowledge, this is the first demonstration of phoP and SPI2-associated genes responding to iron levels. Therefore, we conclude that these virulence genes may be responding to iron limitation in the cell, as a result of Nramp1 depleting the SCV of this essential divalent cation. This response to cellular iron levels is not mediated by PhoPQ or by the global iron regulatory system Fur or the Fe3+-responsive pathogenicity-associated two-component regulatory system PmrAB (data not shown).
Initially, induction of phoP by iron levels was perplexing, because iron has not been indicated as an inducer of phoP expression in the past. However, expression of phoP has primarily been limited to studies investigating its autoregulation by PhoPQ, which does not respond to cellular iron levels as a sensory stimulus (34). Although phoP did demonstrate some tendency toward up-regulation in the presence of Nramp1 in the transfected cell line model, these levels did not reach significance, and phoP failed to be up-regulated in the in vivo model system. Therefore, it theoretically remains possible that phoP may be independently regulated by an iron-responsive regulatory system, but this effect does not appear to play an extensive role in our in vivo infection model. Thus it appears that up-regulation of the SPI2-associated genes is under the control of a distinct, and as yet undiscovered, iron-responsive regulatory system. Studies are currently underway to identify this novel regulatory system.
It would be preferable to be able to study the correlation between Nramp1-mediated depletion of iron from the SCV and STM gene expression in a more relevant model system. Ideally, these investigations would measure the ion concentration of the SCV in parallel with quantitative measurements of bacterial gene expression. The recent development of metal-sensitive fluorescent probes (23) allows for the visualization of divalent cation efflux mediated by Nramp1. Transcriptional fusions to destabilized GFP proteins have been used to quantitate STM virulence gene expression on a single cell basis (P. Cuellar-Mata, M.L.Z., and S. Grinstein, unpublished observations). Although an iron-specific probe is not currently available, the combination of these two techniques for single cell imaging offers exciting possibilities for the future study of the host–pathogen interaction on a single cell level.
Nramp1 is also associated with altering the trafficking of vacuoles containing intracellular parasites. For example, Nramp1+ vacuoles containing viable Mycobacterium avium display a lower pH than Nramp1− vacuoles due to enhanced acquisition of V-ATPase (35) and have markedly different patterns of acquisition of endosomal and lysosomal markers (36). In comparison, although the pH of the SCV was not different in Nramp1+ vs. Nramp1− cells, Nramp1+ SCVs are no longer isolated from normal cellular trafficking patterns, as determined by acquisition of M6PR (13). In addition, STM is known to alter trafficking of the SCV in a SPI2-dependent manner by diverting NADPH oxidase and inducible nitric oxide synthase (iNOS) from the SCV (37, 38). Thus it is possible that the effects we observed on STM SPI2-associated virulence gene expression are not related to Nramp1 function as a divalent cation transport system but may instead be related to the effect of Nramp1 on trafficking of the SCV.
Using Nramp1-transfected cell lines and congenic knockout mice, we have been able to study the dynamic nature of infection in two relevant model systems, which has yielded exciting insights into the STM–host interaction. Initially, finding that the presence of functional Nramp1 results in the up-regulation of genes required for intramacrophage survival was surprising. An effective strategy for halting replication of this pathogen inside host cells would have been to suppress SPI2 expression in some manner. Interestingly, we observed the opposite effect, such that in Nramp1+/+ animals, loci involved in intramacrophage survival are up-regulated. Presumably, this increase in SPI2 expression permits replication of STM, at least at early time points, but eventually Nramp1+/+ mice still control and ultimately survive an infection with wild-type STM.
This SPI2-mediated ability to replicate in the presence of Nramp1, although not to the extent of killing the host, may reflect the dynamic tug-of-war that occurs between host and pathogen during the infectious process. It is commonly held that the most successful pathogens are those that infect and colonize a host but do not kill it, allowing for prolonged shedding and thus successful transmission to new hosts. Nramp1 is found in all mammals examined to date, and functional polymorphic variants are found in human populations. Therefore, it is expected that intracellular pathogens such as STM would continuously encounter this innate host defense system “in the wild.” Thus SPI2-enhanced replication in Nramp1+/+ hosts may be essential for the development of the theoretically most “successful” pathogenic strategy, that of the production of a self-limiting disease (STM and gastroenteritis) or the establishment of a carrier state (S. typhi and typhoid fever).
In addition to Nramp1, the roles of the innate, nonspecific host defense mechanisms NADPH phagocyte oxidase (phox) and iNOS have also been studied in the murine typhoid model. This line of study is especially interesting, because SPI2 is involved in diversion of both phox and iNOS from the SCV in Nramp1−/− macrophages (37, 38), providing further insight into the function of SPI2 during infection. Phox and iNOS were determined to play significantly different roles during infection, phox contributing to killing of STM within the first few hours p.i (39) and iNOS contributing to a sustained bacteriostatic effect, which is most pronounced 7 days p.i. (40). Unfortunately, only the role of iNOS was investigated in both Nramp1+/+ and Nramp1−/− backgrounds (40), because a phox−/− Nramp1+/+ inbred lineage is not currently available. The availability of such a mouse strain would allow the determination of how these other host defense mechanisms interface with the bacteriostatic role of Nramp1 and control of an STM infection.
In summary, we have demonstrated that the presence of Nramp1 up-regulates STM genes specifically required for intramacrophage survival and that this induction can be mimicked by chelation of iron. This supports the hypothesis that Nramp1 serves to deplete the SCV of iron and potentially other essential divalent cations during infection, thereby mediating its bacteriostatic effect on intracellular pathogens by starving the bacteria for these essential nutrients. We have also identified depletion of iron as a novel stimulus for expression of SPI2. Overall, we have defined an active interplay between SPI2 and an innate genetic host defense mechanism during infection of a relevant host population (i.e., Nramp1+/+ animals). We suggest that during an STM infection of naturally resistant hosts, SPI2 needs to be up-regulated to mediate continued survival and replication sufficient to establish an infection and allow prolonged shedding and transmission to novel hosts, but does not permit sufficient replication required to kill its host.
Acknowledgments
We thank members of the Finlay laboratory for careful reading of the manuscript and Dr. Michael Maguire for advice with ion-related studies. This work was supported by grants to B.B.F. from the Canadian Institutes of Health Research (CIHR). Grant support to P.G. is from the National Institutes of Health (National Institute of Allergy and Infectious Diseases; ROI-AI35237), and from Dirección General de Asuntos del Personal Académico; Consejo Nacional de Ciencia y Tecnología to J.L.P. Support to B.A.V. is from a Medical Research Council of Canada/Canadian Digestive Diseases Foundation Fellowship; support to M.L.Z. is from a CIHR Doctoral Research Award. B.B.F., P.G., and J.L.P. are International Research Scholars of the Howard Hughes Medical Institute, and B.B.F. and P.G. are Distinguished Investigators of the CIHR. B.A.V. is an honorary Isaac Walton Killam Fellow.
Abbreviations
Nramp1, natural resistance-associated macrophage protein 1
STM, Salmonella typhimurium
SPI1/SPI2, Salmonella pathogenicity island-1 or -2
SCV, Salmonella-containing vacuole
p.i., postinfection
phox, NADPH phagocyte oxidase
iNOS, inducible nitric oxide synthase
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fortin A., Cardon, L. R., Tam, M., Skamene, E., Stevenson, M. M. & Gros, P. (2001) Proc. Natl. Acad. Sci. USA 98 10793-10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gruenheid S. & Gros, P. (2000) Curr. Opin. Microbiol. 3 43-48. [DOI] [PubMed] [Google Scholar]
- 3.Govoni G. & Gros, P. (1998) Inflamm. Res. 47 277-284. [DOI] [PubMed] [Google Scholar]
- 4.Blackwell J. M., Goswami, T., Evans, C. A., Sibthorpe, D., Papo, N., White, J. K., Searle, S., Miller, E. N., Peacock, C. S., Mohammed, H. & Ibrahim, M. (2001) Cell. Microbiol. 3 773-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goswami T., Bhattacharjee, A., Babal, P., Searle, S., Moore, E., Li, M. & Blackwell, J. M. (2001) Biochem. J. 354 511-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atkinson P. G. & Barton, C. H. (1999) Immunology 96 656-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulero V., Searle, S., Blackwell, J. M. & Brock, J. H. (2002) Biochem. J. 363 89-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcus S. L., Brumell, J. H., Pfeifer, C. G. & Finlay, B. B. (2000) Microbes Infect. 2 145-156. [DOI] [PubMed] [Google Scholar]
- 9.Gorvel J. P. & Meresse, S. (2001) Microbes Infect. 3 1299-1303. [DOI] [PubMed] [Google Scholar]
- 10.Shea J. E., Hensel, M., Gleeson, C. & Holden, D. W. (1996) Proc. Natl. Acad. Sci. USA 93 2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hensel M., Shea, J. E., Waterman, S. R., Mundy, R., Nikolaus, T., Banks, G., Vazquez-Torres, A., Gleeson, C., Fang, F. C. & Holden, D. W. (1998) Mol. Microbiol. 30 163-174. [DOI] [PubMed] [Google Scholar]
- 12.Govoni G., Canonne-Hergaux, F., Pfeifer, C. G., Marcus, S. L., Mills, S. D., Hackam, D. J., Grinstein, S., Malo, D., Finlay, B. B. & Gros, P. (1999) Infect. Immun. 67 2225-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuellar-Mata P., Jabado, N., Liu, J., Furuya, W., Finlay, B. B., Gros, P. & Grinstein, S. (2002) J. Biol. Chem. 277 2258-2265. [DOI] [PubMed] [Google Scholar]
- 14.Koop A. H., Hartley, M. E. & Bourgeois, S. (1987) Gene 52 245-256. [DOI] [PubMed] [Google Scholar]
- 15.Deiwick J., Nikolaus, T., Erdogan, S. & Hensel, M. (1999) Mol. Microbiol. 31 1759-1773. [DOI] [PubMed] [Google Scholar]
- 16.Kehres D. G., Zaharik, M. L., Finlay, B. B. & Maguire, M. E. (2000) Mol. Microbiol. 36 1085-1100. [DOI] [PubMed] [Google Scholar]
- 17.Vidal S., Tremblay, M. L., Govoni, G., Gauthier, S., Sebastiani, G., Malo, D., Skamene, E., Olivier, M., Jothy, S. & Gros, P. (1995) J. Exp. Med. 182 655-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knodler L. A., Celli, J., Hardt, W. D., Vallance, B. A., Yip, C. & Finlay, B. B. (2002) Mol. Microbiol. 43 1089-1103. [DOI] [PubMed] [Google Scholar]
- 19.Cirillo D. M., Valdivia, R. H., Monack, D. M. & Falkow, S. (1998) Mol. Microbiol. 30 175-188. [DOI] [PubMed] [Google Scholar]
- 20.Ochman H., Soncini, F. C., Solomon, F. & Groisman, E. A. (1996) Proc. Natl. Acad. Sci. USA 93 7800-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shea J. E., Beuzon, C. R., Gleeson, C., Mundy, R. & Holden, D. W. (1999) Infect. Immun. 67 213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Govoni G., Vidal, S., Gauthier, S., Skamene, E., Malo, D. & Gros, P. (1996) Infect. Immun. 64 2923-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jabado N., Jankowski, A., Dougaparsad, S., Picard, V., Grinstein, S. & Gros, P. (2000) J. Exp. Med. 192 1237-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee A. K., Detweiler, C. S. & Falkow, S. (2000) J. Bacteriol. 182 771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruiz-Albert J., Yu, X. J., Beuzon, C. R., Blakey, A. N., Galyov, E. E. & Holden, D. W. (2002) Mol. Microbiol. 44 645-661. [DOI] [PubMed] [Google Scholar]
- 26.Lucas R. L., Lostroh, C. P., DiRusso, C. C., Spector, M. P., Wanner, B. L. & Lee, C. A. (2000) J. Bacteriol. 182 1872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valdivia R. H. & Falkow, S. (1997) Science 277 2007-2011. [DOI] [PubMed] [Google Scholar]
- 28.Pfeifer C. G., Marcus, S. L., Steele-Mortimer, O., Knodler, L. A. & Finlay, B. B. (1999) Infect. Immun. 67 5690-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daigle F., Graham, J. E. & Curtiss, R., III (2001) Mol. Microbiol. 41 1211-1222. [DOI] [PubMed] [Google Scholar]
- 30.Bumann D. (2002) Mol. Microbiol. 43 1269-1283. [DOI] [PubMed] [Google Scholar]
- 31.Janakiraman A. & Slauch, J. M. (2000) Mol. Microbiol. 35 1146-1155. [DOI] [PubMed] [Google Scholar]
- 32.Foster J. W. & Hall, H. K. (1992) J. Bacteriol. 174 4317-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wosten M. M., Kox, L. F., Chamnongpol, S., Soncini, F. C. & Groisman, E. A. (2000) Cell 103 113-125. [DOI] [PubMed] [Google Scholar]
- 34.Groisman E. A. (2001) J. Bacteriol. 183 1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hackam D. J., Rotstein, O. D., Zhang, W., Gruenheid, S., Gros, P. & Grinstein, S. (1998) J. Exp. Med. 188 351-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frehel C., Canonne-Hergaux, F., Gros, P. & De Chastellier, C. (2002) Cell. Microbiol. 4 541-556. [DOI] [PubMed] [Google Scholar]
- 37.Vazquez-Torres A., Xu, Y., Jones-Carson, J., Holden, D. W., Lucia, S. M., Dinauer, M. C., Mastroeni, P. & Fang, F. C. (2000) Science 287 1655-1658. [DOI] [PubMed] [Google Scholar]
- 38.Chakravortty D., Hansen-Wester, I. & Hensel, M. (2002) J. Exp. Med. 195 1155-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vazquez-Torres A., Jones-Carson, J., Mastroeni, P., Ischiropoulos, H. & Fang, F. C. (2000) J. Exp. Med. 192 227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mastroeni P., Vazquez-Torres, A., Fang, F. C., Xu, Y., Khan, S., Hormaeche, C. E. & Dougan, G. (2000) J. Exp. Med. 192 237-248. [DOI] [PMC free article] [PubMed] [Google Scholar]