Abstract
The pathogen Brucella suis resides and multiplies within a phagocytic vacuole of its host cell, the macrophage. The resulting complex relationship has been investigated by the analysis of the set of genes required for virulence, which we call intramacrophagic virulome. Ten thousand two hundred and seventy-two miniTn5 mutants of B. suis constitutively expressing gfp were screened by fluorescence microscopy for lack of intracellular multiplication in human macrophages. One hundred thirty-one such mutants affected in 59 different genes could be isolated, and a function was ascribed to 53 of them. We identified genes involved in (i) global adaptation to the intracellular environment, (ii) amino acid, and (iii) nucleotide synthesis, (iv) sugar metabolism, (v) oxidoreduction, (vi) nitrogen metabolism, (vii) regulation, (viii) disulphide bond formation, and (ix) lipopolysaccharide biosynthesis. Results led to the conclusion that the replicative compartment of B. suis is poor in nutrients and characterized by low oxygen tension, and that nitrate may be used for anaerobic respiration. Intramacrophagic virulome analysis hence allowed the description of the nature of the replicative vacuole of the pathogen in the macrophage and extended our understanding of the niche in which B. suis resides. We propose calling this specific compartment “brucellosome.”
Interactions between microorganisms and their hosts extend from acute infections to persistent infectious diseases or symbiosis. This type of interaction can result from a million years of coevolution and coadaptation of the two organisms. Thus, the biology of the interaction can be read, at least partially, in the genome of the microorganism. In the specific case of pathogenic bacteria, deciphering of the genes involved in the interaction and analysis of their functions will shed light on the environment encountered by the parasite in the host and will contribute to the understanding of the complex relationship between two organisms. As a name for the whole set of genes required for virulence, i.e., involved in the invasion of the host by the bacteria and their adaptation to the environment provided by this host, we propose virulome. In this study, we will perform a thorough analysis of the intramacrophagic virulome of Brucella suis.
Brucella spp. is an α proteobacteriaceae that induces a persistent disease in some mammals, resulting in abortion. In humans, initial septicemia may be followed by a subacute or a chronic infection (1). Brucella spp. is phyletically related as well to plant symbionts such as rhizobiaceae, as to rickettsiae, which generate an acute infectious disease (2). In terms of virulence, brucellae occupy an intermediate position where the adaptation results in a mild disease that allows them to persist in mammal hosts. It is usually considered that, for a facultative intracellular bacterium such as Brucella spp., which multiplies in trophoblasts or macrophages (3), one of the challenges is to rapidly adapt to the intracellular settings but also to resist the harsh conditions generated by the immune system, including activation of the phagocytes. However, the status of Brucella spp. and of some other bacteria such as Mycobacterium spp. or Legionella spp. is more complex. Their persistence in nature depends only on their ability to infect animal cells (or protists for Legionella), because they do not survive for protracted periods of time outside their hosts. These pathogens should therefore rather be considered mainly as intracellular, facultatively extracellular organisms (4). The phagosome, to which they have adapted during evolution, is their natural niche.
The identification of the whole set of genes involved in the multiplication of Brucella spp. inside the macrophage is a prerequisite to the understanding of the relation with the macrophage. Signature-tagged mutagenesis and differential fluorescence induction approaches were recently used to identify genes required for virulence in a murine model of infection and in human and murine macrophages (5–9). A large proportion of these genes encode enzymes of various metabolic pathways. However, the small number of mutants screened resulted in a limited output, making identification of the majority of the virulence genes impossible. As a consequence, we decided to design an alternative method. We screened 10,272 individual Tn5 transposon mutants of B. suis for their inability to multiply in a macrophage model of infection, and 131 attenuated mutants were detected. This analysis allowed us to define the environment the bacteria encountered in the macrophage and to better understand the nature of the phagosome.
Materials and Methods
Bacterial Strains and Media.
B. suis 1330 (American Type Culture Collection 23444) and all derived miniTn5 mutant strains were grown in tryptic soy broth at 37°C. The Escherichia coli donor strain for conjugation SM10 λ pir containing a tagged miniTn5 Km2 transposon in pUT (7, 10) was cultivated in LB broth. Antibiotics were used at 50 μg/ml (kanamycin) and 25 μg/ml (nalidixic acid).
Acceptor Strain Construction and Mutagenesis.
A constitutively expressed copy of gfp was inserted by allelic exchange into bcsp31 on chromosome I (BMEI0796) of B. suis 1330. This gene is not involved in bacterial virulence (11). To this end, a previously described strong promoter isolated from B. suis (8) was cloned upstream of the gfp-cat reporter tandem into pBBR1-GFP (12), reisolated in fusion with gfp-cat, and inserted into the unique EcoRV site of bcsp31 cloned in pUC18. The final construct, which is a suicide vector in brucellae, was electroporated into B. suis 1330 (13), and clones that were fluorescent were further verified, confirming the insertion of gfp into chromosomal bcsp31. A spontaneous NalR mutant of this strain was used as B. suis acceptor strain for transposon mutagenesis performed as described (14). Five hundred B. suis clones were isolated per conjugation, and a total of 22 conjugations were performed, yielding 10,272 mutants that were analyzed individually.
Screening of B. suis Tn5 Mutants for Attenuation in Human THP-1 Cells.
Macrophage-like THP-1 cells were infected by individual B. suis mutants and the acceptor strain as a control at a multiplicity of infection of 50 in 96-well plates (15). At 24 and 48 h postinfection, the plates were examined by using fluorescence microscopy. Wells containing attenuated mutants showed no intracellular fluorescence, allowing their rapid identification. Attenuation of the selected mutants was validated in THP-1 infection experiments performed in duplicates in 24-well plates where the number of intracellular bacteria was determined at 90 min and 7, 24, and 48 h postinfection (15).
Analysis of Transposon Insertion Sites.
Chromosomal DNA was isolated from the attenuated mutants, and the sequence from the region flanking the transposon insertion site was obtained directly by using a PCR-based approach before automated sequence analysis (Genaxis, Nîmes, France) by using primers P6 and P7 (10). Sequence database searching was performed by using the blastn and blastx algorithms (16) and by comparison with the annotation for the genome of Brucella melitensis (17). The presence of the identified genes in the genome of B. suis was verified by using the TIGR database (http://tigrblast.tigr.org/ufmg/). This annotation was confirmed in several ways: search of functional motifs by alignment with other sequences, similarity of secondary or tertiary structures of the homologous proteins, presence of the gene in an operon, and strong homologies with phenotypically characterized genes in related bacteria such as rhizobiaceae or Agrobacterium.
Results and Discussion
We reasoned that transposon mutagenesis was the best means to target genes, provided that enough individual clones are analyzed. To perform this task, we labeled B. suis 1330 with constitutively expressed gfp encoding GFP by direct insertion into the genome, by using bcsp31 (11) as target. The resulting fluorescent B. suis was the acceptor strain for miniTn5 Km2 transposon (10), and 10,272 mutants were obtained. Statistical analysis showed that, based on the assumption that B. suis 1330 possesses ≈3,200 genes, the maximal probability to pick up a gene at least once was 96.5%. The resulting mutants were screened for their participation in virulence by infection of the macrophage cell line THP-1, as described (15, 18), and by individual analysis of intracellular multiplication by using fluorescence microscopy, as described in Materials and Methods. The disrupted genes were identified by sequencing the chromosomal DNA flanking Tn5 and homology search by using blast.
One hundred thirty-one mutants unable to multiply in THP-1 cells were selected. These 131 mutations affected 59 different genes. Many of them were drawn several times, confirming that most of the genes were probably interrupted at least once by the transposon. However, a number of previously described genes involved in virulence (5–7) were not picked up. Furthermore, the distribution was not isomorphic with the expected Poisson's distribution if we consider each gene 1,000 nucleotides in size. In fact, the distribution is closer to one that discloses only ≈60% of the screened genes. We should interpret with caution the hypothesis that most (≈95%) of the genes were targeted by miniTn5. The discrepancy between statistical analysis and the experimental results may be explained by a high threshold of detection, a nonhomogeneous size of the genes, or a nonrandom insertion of Tn5.
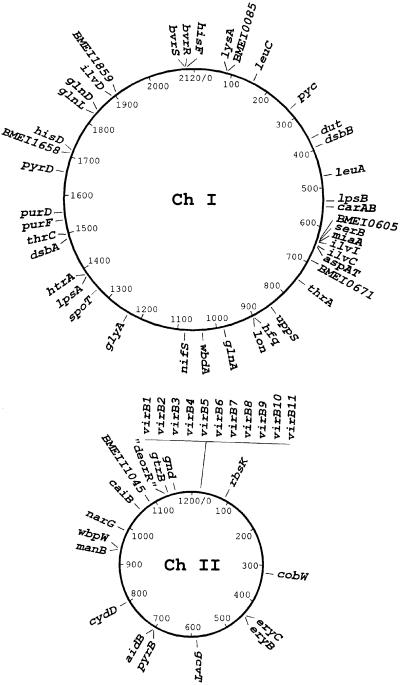
We can ascribe a function to 53 of the 59 genes identified with good confidence. The results are summarized in Table 1. Genes were classified according to the putative functions they encode. Six genes did not yield homologous sequences in microbial databases. The identified virulence genes/operons are distributed evenly on the two chromosomes with no major clustering (Fig. 1).
Table 1.
B. suis genes essential for intramacrophagic multiplication
| Functional groups | Mutated genes and putative functions | Attenuation at 48 h postinfection (log) | SpoT regulation | ORF number |
|---|---|---|---|---|
| Transporter | cobW | 2 | II0308 | |
| virB (type IV secretion system) | 5 | II0025–0035 | ||
| Amino acid synthesis | thrA, thrC (T) | 3 | + | I0725, I1450 |
| ilvC, ilvD, ilvI (I, L, V) | 2.7; 5; 5 | + | I0624, I1848, I0617 | |
| leuA, leuC (L) | 2 | I0451, I0157 | ||
| hisD, hisF (H) | 3; 1.7 | + | I1668, I2041 | |
| glyA, serB (G, S) | 2.4 | I1192, I0615 | ||
| lysA (K) | 2.7 | + | I0084 | |
| carAB (R) | 2.8 | I0526 | ||
| gcvT (S) | 2 | II0559 | ||
| aspB? (aminotransferase) | 2 | I0626 | ||
| Sugar metabolism | pyc (anaplerotic function) | 3.9 | I0266 | |
| eryB, eryC (erythritol) | 2.3; 1.4 | II0429, II0428 | ||
| gnd (pentose phosphate pathway) | 1.8 | II1124 | ||
| gtrB? (glycosyl transferase) | 2.7 | II1101 | ||
| rbsK (ribokinase) | 1.9 | II0089 | ||
| DNA/RNA metabolism | carAB (pyrimidines) | 2.8 | I0526 | |
| pyrB, pyrD (pyrimidines) | 2.4; 3 | − | II0670, I1611 | |
| purD, purF (purines) | 2.1; 2.5 | − | I1519, I1488 | |
| dut (dUTPase) | 2.1 | I0358 | ||
| miaA (tRNA isopentenyl transferase) | 2 | I0616 | ||
| Lipid metabolism | aidB (acyl-CoA dehydrogenase) | 1 | II0671 | |
| Regulation | SpotT/rsh (stringent response) | 2.1 | I1296 | |
| bvrR, bvrS (two component system) | 2; 4.5 | I2036, I2035 | ||
| deoR (transcriptional regulator) | 4.3 | II1093 | ||
| Stress proteins | hfq (host factor I) | 1.8 | I0872 | |
| htrA (protease) | 2.8 | I1330 | ||
| lon (protease) | 1.8 | + | I0876 | |
| Membrane structure | uppS (peptidoglycan synthesis) | 1.2 | − | I0827 |
| Lipopolysaccharide | wbdA (mannosyl transferase) | 0.8 | I0997 | |
| biosynthesis | lpsA, lpsB (glycosyl transferases) | 1.7; 5.9 | I1326, I0509 | |
| manB (phosphomannomutase) | 4.2 | II0899 | ||
| wbpW (phosphomannose isomerase) | 2 | II0900 | ||
| Oxidoreduction | cydD (cytochrome oxydase) | 2.5 | II0762 | |
| narG (nitrate reductase) | 2 | II0950 | ||
| caiB (carnitine dehydratase) | 1.2 | II1019 | ||
| dsbA, dsbB (disulfide bond formation) | 2.5; 5 | I1440, I0384 | ||
| Nitrogen metabolism | glnA (glutamine synthetase) | 3.3 | + | I0979 |
| glnD (PII uridylyl transferase) | 3.2 | I1804 | ||
| glnL (NRII) | 1.3 | I1786 | ||
| nifS (nitrogenase cofactor synthesis) | 1.8 | I1043 | ||
| Unknown functions | BMEI0085 | 1.5 | I0085 | |
| BMEI0605 | 2 | I0605 | ||
| BMEI0671 | 2.5 | I0671 | ||
| BMEI1658 | 1.9 | I1658 | ||
| BMEI1859 | 2.3 | I1859 | ||
| BMEII1045 | 2.2 | II1045 |
As calculated from the comparison to the number of the intracellular B. suis control strain.
Genes reported to be under positive (+) or negative (−) control during the stringent response in E. coli (19).
According to the nomenclature of the B. melitensis genome in GenBank (accession nos. NC003317 and NC003318).
In parentheses, amino acid single-letter code of the final pathway products.
Fig 1.
Physical map of the two chromosomes of B. suisdisplaying the distribution of the genes identified by Tn5 mutagenesis.
The most intriguing result is the absence of virulence factors such as toxins or other exported proteins able to interfere with host cell functions, except the already known VirB secretion system, whose operon was targeted 33 times. None of the mutants was affected in adherence or entry under the experimental conditions used. The majority of the mutations resulting in intracellular growth defect affected genes acting in housekeeping functions, confirming the results previously obtained by signature-tagged mutagenesis or differential fluorescence induction.
Adaptation of the Bacteria to the Phagosome.
Four mutations affecting genes necessary for a global adaptation to an intracellular environment were detected, and an attenuated mutant of B. suis showed an insertion of Tn5 in a gene encoding a SpoT homolog involved in the stringent response (Table 1). In E. coli, the two proteins RelA and SpoT regulate the concentration of (p)ppGpp, a global transcriptional activator of numerous metabolic pathways under starvation conditions (19). RelA is important for the virulence of Listeria monocytogenes (20), for the activation of quorum sensing in Pseudomonas aeruginosa (21), and for long-term survival of Mycobacterium tuberculosis (22). In Brucella spp. and Sinorhizobium meliloti, only the gene coding for a regulator closely related to SpoT exists and may be called rsh for rel spo homolog. In S. meliloti, the rsh mutant does not induce nodulation and fails to grow in the host plant (23). In Legionella pneumophila, in contrast, RelA is not involved in intracellular replication (24). The attenuation of the B. suis mutant, unable to induce these metabolic pathways, suggests that the phagosomal environment lacks the necessary precursors for these molecules, a hypothesis confirmed by the analysis of other mutants (see below).
Another attenuated mutant of B. suis was affected in the characterized virulence gene hfq encoding Host Factor I. Mutation of hfq in Brucella abortus results in a lowered resistance to stress induced by peroxides and acidic conditions during stationary phase (25). In Salmonella typhimurium, the transcription factor RpoS, which is required for the transcription of many genes expressed during the onset of stationary phase and for virulence, is regulated by Host Factor I. Adaptation to nutrient limitation and starvation in E. coli depend on RpoS. However, there is no gene homologous to rpoS in B. suis. The attenuation of hfq mutants in Brucella can be explained either by the existence of a stationary-phase regulating σ factor different from RpoS or by its regulation of unknown virulence factors.
We have isolated an attenuated Tn5 mutant affected in htrA. The protease HtrA has been shown to be involved in the removal of proteins exposed to reactive oxygen intermediates. Contradictory results on the role of htrA in virulence of B. abortus were reported: A htrA null mutant is not attenuated in a mouse model of infection (26). The same authors nevertheless confirm the attenuation of the mutant in isolated murine macrophages, obtaining results similar to ours. However, it has been shown that brucellae penetrate the phagocytic cells without an oxidative burst being detected (18). The biological role of HtrA in the interaction with the host therefore remains obscure.
The stress-induced protease Lon was also found to be essential for multiplication inside macrophages (Table 1). This protein has been described as necessary for virulence in B. abortus (27) and in S. typhimurium by controlling genes required for adherence and invasion (28). In S. meliloti, Lon, which shows 86% homology with the B. suis gene, regulates the synthesis of exopolysaccharides and is required for nodulation (29).
All together, these results suggest that the stress response is crucial for the adaptation of brucellae to the intraphagosomal life style.
Genes Involved in Amino Acid Synthesis.
The most striking feature of the results obtained is the number of genes involved in the biosynthesis of amino acids (Table 1). Bacterial attenuation after disruption of these genes signifies that the corresponding metabolites are not accessible to the bacteria inside the phagosome, and that they are essential for the intramacrophagic growth. Of the 15 mutated genes described, 11 were isolated two to five times in independent experiments. Synthesis of amino acids starts from the five metabolites α ketoglutarate, oxalacetate, pyruvate, 3-phosphoglyceric acid, and chorismate. The first four pathways have been identified during mutagenesis but not the pathway for the synthesis of aromatic amino acids from chorismate. Furthermore, mutants affected in the synthesis of alanine from pyruvate, of methionine from oxalacetate, and of proline from α ketoglutarate were never isolated by this screening method. There are two possible explanations: (i) the screening has not targeted all of the genes, and (ii) only some pathways are essential for intracellular brucellae. The second hypothesis is unlikely, because (i) there is no evident rational means to draw this conclusion; (ii) only a few mutations were found on genes encoding enzymes for a same specific pathway (histidine or threonine synthesis); and (iii) we and others have identified genes involved in biosynthesis of amino acids not isolated in this screening. AroC, for instance, necessary for the synthesis of aromatic amino acids, has been shown to be required for intracellular multiplication (30). We conclude that not all pathways were targeted by the method used. Our results allow us to conclude that the synthesis of amino acids is absolutely necessary for the intracellular multiplication of Brucella spp., implying that amino acids are not available inside the phagosome. This finding confirms the need for a functional spoT gene for intracellular multiplication, with (p)ppGpp inducing most of the amino acid synthesis pathways identified as essential in our virulence screening (Table 1). A schematic presentation of these pathways with the genes identified as essential for intracellular B. suis is shown in Fig. 2, which is published as supporting information on the PNAS web site, www.pnas.org.
Genes Involved in Nucleotide Synthesis.
Five genes engaged in the synthesis of purines (purD, purF) and pyrimidines (carAB, pyrB, pyrD) are required for intracellular multiplication of B. suis (Table 1; Fig. 2, which is published as supporting information on the PNAS web site), confirming previous results on the attenuation of purE mutants from B. melitensis (31). This observation suggests that inside the phagosome, Brucella spp. cannot access the bases or the nucleotides necessary for its growth and has to synthesize them.
Genes Involved in Sugar Metabolism.
Only a few genes involved in sugar metabolism have been identified as necessary for intramacrophagic multiplication (Table 1; Fig. 2, which is published as supporting information on the PNAS web site). Among them is the operon encoding enzymes involved in erythritol degradation (32). We identified two genes, eryB and eryC, necessary for growth inside the macrophage. However, erythritol has not been described in human macrophages, making clear that the role of these enzymes in virulence is rather complex and awaits further analysis. We cannot rule out the possibility that EryB, homologous to glycerol-3-phosphate dehydrogenases, and EryC, showing partial homology to hydrogenases (32), catabolize additional substrates present in human macrophages.
The gnd gene encoding 6-phosphogluconate dehydrogenase is involved in the branching of sugar metabolism to the pentose phosphate pathway (Fig. 2, which is published as supporting information on the PNAS web site). Brucella is known to have lost pfk and fda genes and thus is not able to use the Embden–Meyerhof pathway (glycolysis) to degrade glucose (17). The inactivation of gnd leaves the Entner–Doudoroff pathway as a shunt for sugar degradation (the homologues of the edd and eda genes are present on the Brucella chromosome II), explaining the slight attenuation in the macrophage model (1.8 log).
Another remarkable gene involved in intracellular survival of B. suis is pyc encoding pyruvate carboxylase. This enzyme is involved in the synthesis of oxalacetate from pyruvate, restocking the tricarboxylic acid (TCA) cycle. The anaplerotic function of this enzyme confirms that metabolites of the TCA cycle are used for synthesis of amino acids and bases. However, the anaplerotic gene homologous to icl encoding isocitrate lyase from the glyoxylate cycle is not involved in virulence of B. suis (S. K., unpublished work), in contrast to results published for M. tuberculosis (33). With Icl, acetyl-CoA is used, whereas in the reaction catalyzed by Pyc, CO2 and ATP are involved. In M. tuberculosis, Icl is necessary for persistence and is involved in the direct binding of acetyl-CoA obtained from the β oxidation of fatty acids. According to our results, Brucella spp. seems to behave differently and probably does not use mainly fatty acids as an energy source in the macrophage.
The carbon source of brucellae inside the macrophage has to be uncovered, but the necessity of a functional ribose kinase (Table 1) suggests that brucellae import ribose and thus may use pentoses as a source of carbon and energy. The pentose phosphate pathway has already been recognized earlier as a major sugar degradation pathway in Brucella spp. (34).
Oxidoreduction and Electron Transport.
Functional analysis of the genes necessary for virulence inside the macrophage reveals the presence of several genes involved in oxidoreduction, among which is cydD, part of the operon cydDCAB. CydD participates in the assembly of the cytochrome bd oxidase encoded by cydAB, which is a terminal electron donor to oxygen under low oxygen concentration conditions. The respiratory chain of B. suis contains two terminal oxidases, cytochrome bd and cytochrome bo oxidase (17). In E. coli, cytochrome bo oxidase is predominantly expressed when oxygen is not limited, and cytochrome bd oxidase, which has a higher affinity for oxygen, is used under microaerophilic conditions (35). The importance of cydD is corroborated by the seminal discovery of cydB as a virulence gene in the mouse model of infection (36). The cydB mutant shows an increased sensitivity to oxygen radicals and to pH ≤3.6, favoring the hypothesis that cytochrome bd oxidase participates in the resistance of Brucella spp. to the harsh conditions of the phagosome (36). It has been shown for B. abortus that the switch from cytochrome bo oxidase to cytochrome bd oxidase can be explained by lowered oxygen tension inside the macrophage (37). A functional cytochrome bd oxidase is also required for intracellular survival of Shigella flexneri (38).
The genome of B. melitensis contains the genetic information necessary for the use of nitrate as electron acceptor, with production of nitrite and nitrogen (17). Among the genes involved in intramacrophagic multiplication, we have identified narG, coding for an essential component of the dissimilatory nitrate reductase complex (Table 1). This complex is encoded by the narGHIJ locus, which is present in the B. suis genome together with the gene of the nitrite extrusion protein, narK. The narG mutant was unable to produce nitrite from nitrate (not shown). The genes nir and nor necessary for nitrate respiration are also present in the B. suis genome. The requirement of a functional narG gene for intracellular growth suggests that the vacuole containing B. suis is devoid of oxygen and can be interpreted as the ability of B. suis to use nitrate for respiration during intracellular growth. The impact of a mutation in caiB strengthens this hypothesis. In anaerobiosis, carnitine is degraded to butyrylbetaine, which can be used as an electron transporter. The three genes encoding the enzymes required for the carnitine metabolism, caiB, caiC, and caiD, are present as an operon in the genome of B. suis.
Nitrogen Metabolism.
B. suis has to synthesize all of the amino acids and bases to multiply inside the phagosome. Thus, the nitrogen source cannot be imported as complex molecules, but B. melitensis contains genes encoding a putative assimilatory nitrate reductase and a copper-containing nitrite reductase (17). On the other hand, our results show that at least three enzymes involved in NH utilization (GlnA, GlnD, and NRII; Table 1) are necessary for intracellular multiplication, the first being regulated by (p)ppGpp during the stringent response (see Table 1). glnA is also required for virulence of S. typhimurium (39), demonstrating that Brucella spp. uses NH
utilization (GlnA, GlnD, and NRII; Table 1) are necessary for intracellular multiplication, the first being regulated by (p)ppGpp during the stringent response (see Table 1). glnA is also required for virulence of S. typhimurium (39), demonstrating that Brucella spp. uses NH , which may be obtained either by nitrate reduction or by direct uptake from the phagosome, for amino acid or nucleotide synthesis.
, which may be obtained either by nitrate reduction or by direct uptake from the phagosome, for amino acid or nucleotide synthesis.
Genes Involved in Lipid Metabolism.
The degradation of fatty acids most likely does not play a crucial role for the growth of B. suis inside the macrophage, as only one slightly attenuated mutant has been obtained in a gene encoding a putative acyl-CoA dehydrogenase. In total, eight enzymes with this potential function are encoded in the genome of B. melitensis. If we associate this observation to the result that pyruvate carboxylase instead of isocitrate lyase is crucial in anaplerotic reactions, it can be inferred that fatty acids from the host cell are not available for degradation by the pathogen.
Regulation.
The sequence of 27 two-component regulation systems can be detected in the genome of Brucella. The only attenuated mutant selected by our technique and affected in such a function is the bvrS-bvrR mutant. BvrR and BvrS are known to be required for intracellular multiplication (40).
DsbAB Proteins, Assembly, and Exportation of Virulence Proteins.
DsbAB proteins are involved in disulphide bond formation in the periplasm of Gram-negative bacteria, hence stabilizing the interactions of many proteins (41). DsbA has been lately described as essential for the secretion of pertussis toxin (42), and it is conceivable that attenuated B. suis dbsAB mutants may be affected by the unfolding or lack of assembly of yet-to-be-identified virulence factors.
Role of Smooth Lipopolysaccharide in Intracellular Multiplication.
Although the two species Brucella ovis and Brucella canis are naturally rough and respectively infect rams and dogs, it has been shown that rough mutants from other species are avirulent (43, 44). Among the attenuated mutants, five were affected in genes required at different stages of the lipopolysaccharide biosynthesis (Table 1). Microscopic examination of infected macrophages confirms eradication of the rough bacteria, which is in agreement with results obtained in intracellular survival assays (not shown), and which could be due to their sensitivity to defensin-like factors (43).
Analytical Conclusion: The Nature of the B. suis Niche in the Macrophage.
This work, which was aimed at the definition and analysis of the intramacrophagic virulome of B. suis, reveals an extended picture of the niche in which B. suis resides in the macrophage. The pathogen remains enclosed in a vacuole throughout its intracellular life, but the nature of the phagosome is poorly defined, and its modifications during the activation of the phagocyte are totally unknown. In macrophages, it has been described that B. suis prevents the fusion of the phagosome with lysosomes and that phagocytosis inhibits TNFα production and apoptosis of the cell (45–47) .
Our results allow us to describe the environment encountered by the bacterium in the phagosome and, together with previously published data, the strategy it uses to pervert the functions of the phagocyte. Complete genomic sequences of two Brucella species have confirmed the absence of “classical” virulence factors and types I, II, and III protein secretion systems (17). Indeed, this bacterium enters the cell by rafts, using a lectin-like mechanism, and not by means of dedicated proteins such as internalin or invasin (48). Previous work has demonstrated that an acid phagosomal pH of 3.5–4 is required for early steps of infection within the cell, and neutralization of the phagosome after 6 h of infection has no effect on intracellular growth of B. suis (49). Synthesis of stress proteins such as DnaK, Lon, or Host Factor I are probably required at this stage (13, 25, 27).
The 59 genes (virB operon = 1) described here are necessary for intracellular replication. Among the 131 attenuated mutants, the virB operon, composed of 12 genes with a total size of 12 kb, was targeted 33 times. The number of virB mutants was higher than expected, possibly due to an additional hot spot for Tn5 insertion within the operon. The requirement of an acid pH for effective intracellular multiplication only during the first 6 h and the demonstration that virB is induced under acidic conditions and in the phagosome (50) suggest that VirB might be important only during the early phase of cellular infection. VirB may allow the bacterium to reach a particular cellular compartment by a yet-undiscovered mechanism and perhaps after several steps of maturation, such as transit by multilamellar autophagosome-like structures, as claimed earlier (51).
Our observations are of importance in defining the environment of the replicative niche containing Brucella spp., i.e., at 48 h postinfection. Before our work, the nature of the bacteria-containing phagosome was studied essentially by using cellular biology approaches (52). The characterization of the vacuoles is mainly performed by colocalization with specific cellular markers. Results obtained with this approach showed that in HeLa cells (nonprofessional phagocytes), B. abortus resides in a structure that exhibits markers of the endoplasmic reticulum (ER) (51, 53, 54). A functional ER should contain proteins and peptides synthesized by the host cell, which would not be consistent with the shortage in amino acid availability revealed by our work. Furthermore, synthesis of proteins with a rapid turnover is not altered in cells infected by B. abortus (54), indicating that the brucellae are probably not localized in the same functional compartment. It could be put forward that the compartment containing Brucella spp. is different in HeLa cells and macrophages. Indeed, results obtained on intracellular trafficking of B. abortus in J774 macrophages report that only a few bacteria are localized in autophagosomes and ER-like structures (55). However, partial analysis of the virulome of B. melitensis has been performed in HeLa cells, and some amino acid biosynthesis pathways have been shown to be necessary (6, 56). In addition, recent observations that ER contributes to phagosome formation (57) and that ER markers colocalize with phagosomes containing latex beads (58) make it clear that a vacuole cannot be defined only by the presence of specific proteins. The description of the nature of the replicative compartment of B. suis in the late phase of infection can be complemented by our analysis of the intramacrophagic virulome: it is poor in nutrients, as shown by the requirement of functional genes of various biosynthesis pathways; it most likely has a neutral pH; it is also characterized by low oxygen tension and, alternatively, nitrate ions are available for anaerobic respiration. In this compartment, B. suis multiplies actively, until occupying the whole cytoplasm. During infection, the main functions of the cell seem to be unaffected, as suggested by inhibition of apoptosis (47).
Altogether, these results demonstrate that brucellae actively build a pathogen-specific niche for multiplication within the cells, different from all naturally existing cellular organelles, including those playing a vital physiological role to the uninfected host. We therefore propose calling the vacuole in which B. suis multiplies “brucellosome.” B. suis seems to behave furtively, disturbing the cell functions as little as possible. It multiplies within the macrophage because of its resistance to the different stress conditions encountered in the phagosome and because of its ability to synthesize all of the metabolites necessary for its survival and to use anaerobic respiration in the replicative compartment it creates.
Supplementary Material
Acknowledgments
This work was partly supported by Grant QLK2-CT-1999-00014 from the European Union.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Young E. J. (1983) Rev. Infect. Dis. 5 821-842. [DOI] [PubMed] [Google Scholar]
- 2.Yanagi M. & Yamasato, K. (1993) FEMS Microbiol. Lett. 107 115-120. [DOI] [PubMed] [Google Scholar]
- 3.Harmon B. G., Adams, L. G. & Frey, M. (1988) Am. J. Vet. Res. 49 1092-1097. [PubMed] [Google Scholar]
- 4.Moreno E. & Moriyon, I. (2002) Proc. Natl. Acad. Sci. USA 99 1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong P. C., Tsolis, R. M. & Ficht, T. A. (2000) Infect. Immun. 68 4102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lestrate P., Delrue, R. M., Danese, I., Didembourg, C., Taminiau, B., Mertens, P., De Bolle, X., Tibor, A., Tang, C. M. & Letesson, J. J. (2000) Mol. Microbiol. 38 543-551. [DOI] [PubMed] [Google Scholar]
- 7.Foulongne V., Bourg, G., Cazevieille, C., Michaux-Charachon, S. & O'Callaghan, D. (2000) Infect. Immun. 68 1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Köhler S., Ouahrani-Bettache, S., Layssac, M., Teyssier, J. & Liautard, J. P. (1999) Infect. Immun. 67 6695-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eskra L., Canavessi, A., Carey, M. & Splitter, G. (2001) Infect. Immun. 69 7736-7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hensel M., Shea, J. E., Gleeson, C., Jones, M. D., Dalton, E. & Holden, D. W. (1995) Science 269 400-403. [DOI] [PubMed] [Google Scholar]
- 11.Halling S. M., Detilleux, P. G., Tatum, F. M., Judge, B. A. & Mayfield, J. E. (1991) Infect. Immun. 59 3863-3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouahrani-Bettache S., Porte, F., Teyssier, J., Liautard, J. P. & Köhler, S. (1999) Biotechniques 26 620-622. [DOI] [PubMed] [Google Scholar]
- 13.Köhler S., Teyssier, J., Cloeckaert, A., Rouot, B. & Liautard, J. P. (1996) Mol. Microbiol. 20 701-712. [DOI] [PubMed] [Google Scholar]
- 14.Michaux-Charachon S., Bourg, G., Jumas-Bilak, E., Guigue-Talet, P., Allardet-Servent, A., O'Callaghan, D. & Ramuz, M. (1997) J. Bacteriol. 179 3244-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekaza E., Guilloteau, L., Teyssier, J., Liautard, J. P. & Köhler, S. (2000) Microbiology 146 1605-1616. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215 403-410. [DOI] [PubMed] [Google Scholar]
- 17.DelVecchio V. G., Kapatral, V., Redkar, R. J., Patra, G., Mujer, C., Los, T., Ivanova, N., Anderson, I., Bhattacharyya, A., Lykidis, A., et al. (2002) Proc. Natl. Acad. Sci. USA 99 443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caron E., Liautard, J. P. & Köhler, S. (1994) J. Leukocyte Biol. 56 174-181. [DOI] [PubMed] [Google Scholar]
- 19.Cashel M., Gentry, D. R., Hernandez, V. J. & Vinella, D. (1996) in Escherichia coli and Salmonella, ed. Neidhardt, F. C. (Am. Soc. Microbiol. Press, Washington, DC), Vol. 1, pp. 1458–1496. [Google Scholar]
- 20.Taylor C. M., Beresford, M., Epton, H. A., Sigee, D. C., Shama, G., Andrew, P. W. & Roberts, I. S. (2002) J. Bacteriol. 184 621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Delden C., Comte, R. & Bally, A. M. (2001) J. Bacteriol. 183 5376-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Primm T. P., Andersen, S. J., Mizrahi, V., Avarbock, D., Rubin, H. & Barry, C. E., III (2000) J. Bacteriol. 182 4889-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wells D. H. & Long, S. R. (2002) Mol. Microbiol. 43 1115-1127. [DOI] [PubMed] [Google Scholar]
- 24.Zusman T., Gal-Mor, O. & Segal, G. (2002) J. Bacteriol. 184 67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robertson G. T. & Roop, R. M., Jr. (1999) Mol. Microbiol. 34 690-700. [DOI] [PubMed] [Google Scholar]
- 26.Phillips R. W. & Roop, R. M., II (2001) Infect. Immun. 69 5911-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robertson G. T., Kovach, M. E., Allen, C. A., Ficht, T. A. & Roop, R. M., II (2000) Mol. Microbiol. 35 577-588. [DOI] [PubMed] [Google Scholar]
- 28.Takaya A., Tomoyasu, T., Tokumitsu, A., Morioka, M. & Yamamoto, T. (2002) J. Bacteriol. 184 224-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Summers M. L., Botero, L. M., Busse, S. C. & McDermott, T. R. (2000) J. Bacteriol. 182 2551-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foulongne V., Walravens, K., Bourg, G., Boschiroli, M. L., Godfroid, J., Ramuz, M. & O'Callaghan, D. (2001) Infect. Immun. 69 547-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drazek E. S., Houng, H. S., Crawford, R. M., Hadfield, T. L., Hoover, D. L. & Warren, R. L. (1995) Infect. Immun. 63 3297-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sangari F. J., Aguero, J. & Garcia-Lobo, J. M. (2000) Microbiology 146 487-495. [DOI] [PubMed] [Google Scholar]
- 33.McKinney J. D., Honer zu Bentrup, K., Munoz-Elias, E. J., Miczak, A., Chen, B., Chan, W. T., Swenson, D., Sacchettini, J. C., Jacobs, W. R., Jr. & Russell, D. G. (2000) Nature 406 735-738. [DOI] [PubMed] [Google Scholar]
- 34.Robertson D. C. & McCullough, W. G. (1968) Arch. Biochem. Biophys. 127 263-273. [DOI] [PubMed] [Google Scholar]
- 35.Cotter P. A., Melville, S. B., Albrecht, J. A. & Gunsalus, R. P. (1997) Mol. Microbiol. 25 605-615. [DOI] [PubMed] [Google Scholar]
- 36.Endley S., McMurray, D. & Ficht, T. A. (2001) J. Bacteriol. 183 2454-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rest R. F. & Robertson, D. C. (1975) J. Bacteriol. 122 139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Way S. S., Sallustio, S., Magliozzo, R. S. & Goldberg, M. B. (1999) J. Bacteriol. 181 1229-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klose K. E. & Mekalanos, J. J. (1997) Infect. Immun. 65 587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sola-Landa A., Pizarro-Cerda, J., Grillo, M. J., Moreno, E., Moriyon, I., Blasco, J. M., Gorvel, J. P. & Lopez-Goni, I. (1998) Mol. Microbiol. 29 125-138. [DOI] [PubMed] [Google Scholar]
- 41.Bardwell J. C., McGovern, K. & Beckwith, J. (1991) Cell 67 581-589. [DOI] [PubMed] [Google Scholar]
- 42.Stenson T. H. & Weiss, A. A. (2002) Infect. Immun. 70 2297-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen C. A., Adams, L. G. & Ficht, T. A. (1998) Infect. Immun. 66 1008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Godfroid F., Taminiau, B., Danese, I., Denoel, P., Tibor, A., Weynants, V., Cloeckaert, A., Godfroid, J. & Letesson, J. J. (1998) Infect. Immun. 66 5485-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naroeni A., Jouy, N., Ouahrani-Bettache, S., Liautard, J. P. & Porte, F. (2001) Infect. Immun. 69 486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caron E., Gross, A., Liautard, J. P. & Dornand, J. (1996) J. Immunol. 156 2885-2893. [PubMed] [Google Scholar]
- 47.Gross A., Terraza, A., Ouahrani-Bettache, S., Liautard, J. P. & Dornand, J. (2000) Infect. Immun. 68 342-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naroeni A. & Porte, F. (2002) Infect. Immun. 70 1640-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Porte F., Liautard, J. P. & Köhler, S. (1999) Infect. Immun. 67 4041-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boschiroli M. L., Ouahrani-Bettache, S., Foulongne, V., Michaux-Charachon, S., Bourg, G., Allardet-Servent, A., Cazevieille, C., Liautard, J. P., Ramuz, M., et al. (2002) Proc. Natl. Acad. Sci. USA 99 1544-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pizarro-Cerda J., Meresse, S., Parton, R. G., van der Goot, G., Sola-Landa, A., Lopez-Goni, I., Moreno, E. & Gorvel, J. P. (1998) Infect. Immun. 66 5711-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinai A. P. & Joiner, K. A. (1997) Annu. Rev. Microbiol. 51 415-462. [DOI] [PubMed] [Google Scholar]
- 53.Pizarro-Cerda J., Moreno, E., Sanguedolce, V., Mege, J. L. & Gorvel, J. P. (1998) Infect. Immun. 66 2387-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pizarro-Cerda J., Moreno, E. & Gorvel, J. P. (2000) Microbes Infect. 2 829-835. [DOI] [PubMed] [Google Scholar]
- 55.Arenas G. N., Staskevich, A. S., Aballay, A. & Mayorga, L. S. (2000) Infect. Immun. 68 4255-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delrue R. M., Martinez-Lorenzo, M., Lestrate, P., Danese, I., Bielarz, V., Mertens, P., De Bolle, X., Tibor, A., Gorvel, J. P. & Letesson, J. J. (2001) Cell Microbiol. 3 487-497. [DOI] [PubMed] [Google Scholar]
- 57.Gagnon E., Duclos, S., Rondeau, C., Chevet, E., Cameron, P. H., Steele-Mortimer, O., Paiement, J., Bergeron, J. J. M. & Desjardins, M. (2002) Cell 110 119-131. [DOI] [PubMed] [Google Scholar]
- 58.Garin J., Diez, R., Kieffer, S., Dermine, J. F., Duclos, S., Gagnon, E., Sadoul, R., Rondeau, C. & Desjardins, M. (2001) J. Cell Biol. 152 165-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.