Abstract
There are very few molecules known to transport Mg2+ in eukaryotes. The membrane of Paramecium tetraurelia passes a large Mg2+-selective current and exhibits a corresponding backward swimming behavior. Both are missing in a group of mutants called eccentric. By sorting an indexed WT genomic library through microinjection into the macronucleus, we have isolated a DNA fragment that complements the eccentric mutations. The Mg2+ currents and behavior are restored fully in the transformed cells. Surprisingly, the conceptually translated protein is not homologous to any known ion channel but instead has some similarity to K+-dependent Na+/Ca2+ exchangers. Exchangers are either electrically silent or only pass very small and slow currents compared with ion-channel currents. In light of recent ion-channel crystal structures and considering the need to have narrow ion-selective filters, we speculate on how an exchanger might evolve to show channel-like activities in special circumstances. The significance of finding the molecular basis of a Mg2+-specific pathway is also discussed.
Despite the importance of Mg2+ in biology, much less is known about how eukaryotic cells regulate Mg2+ than Ca2+. Studies of Mg2+ have been slowed by the lack of convenient isotopes or other tags, and as a result, very few genes regulating or transporting Mg2+ have been cloned. Bacterial Mg2+ transporters encoded by corA, mgtA, mgtCB, and mgtE loci have been studied in detail (1), including their distant homologs in eukaryotes, some of which may be restricted to mitochondrial membranes (2–5). In eukaryotes, the intracellular Mg2+ concentration is maintained at 0.3–1 mM by the activity of a plausible Na+/Mg2+ exchanger (6–10) and other perhaps less specific pathways (8, 11–14), but they have yet to be identified in molecular terms. Recently, a Na+/Ca2+ exchanger (NCX) homolog was cloned from Arabidopsis that apparently mediates a pH-dependent Mg2+ transfer across vacuolar membranes (AtMHX, ref. 15).
The fresh-water ciliate Paramecium provides a unique opportunity to elucidate the molecular basis for Mg2+ transport. When stimulated, paramecia exhibit avoidance responses reflecting a redirection of ciliary beat, governed by changes in membrane potential and Ca2+ influx. Membrane potential is determined by various ion channels whose activities can be readily measured directly with intracellular electrodes (16). The avoidance behavior can easily be observed under a microscope, and many mutants with distinct behavioral changes have been isolated after random mutagenesis (17). These changes often indicate the loss of or changes in ion currents (18). In 1990, Preston (19) discovered a highly unusual Mg2+-specific current in Paramecium. Unlike the previously described Mg2+-specific transport mechanisms, this Mg2+ current is large (1–2 nA) and activates rapidly (<10 ms) under voltage clamp in solutions that mimic fresh water. The current normally flows inward to promote prolonged depolarization and backward swimming. This distinctive Mg2+ phenotype coupled with transmission genetics made it possible to isolate mutants that fail to exhibit the Mg2+-specific current or the associated behavior (“eccentric,” refs. 20–22). We have previously established a method to clone genes in Paramecium by sorting library fractions that effect clonal phenotype reversion when injected into the macronucleus of behavioral mutants (23–25). We have thus exploited the opportunity Paramecium offers to find a eukaryotic Mg2+-specific transporter. Here we report on the cloning and analysis of the eccentric gene, XNTA, and its conceptually translated product, XntAp.
Materials and Methods
Stocks and Cultures.
Paramecium tetraurelia stock 51s (+/+) (26), nd6 (nd6/nd6) (27), K+-resistant (K-21) {d4–521} (nd6/nd6, fA1/fA1) (28), baB {d4–596} (nd6/nd6, baB/baB) (29), xnt-A1 {d4–700} (nd6/nd6, xntA/xntA), xnt-A2 {d4–702} (nd6/nd6, xntA/xntA), xnt-A3 {d4–703} (nd6/nd6, xntA/xntA) (21), xnt-A4 {d4–710} (nd6/nd6, xntA/xntA), xnt-A5 {d4–718} (nd6/nd6, xntA/xntA), xnt-A6 {d4–719} (nd6/nd6, xntA/xntA) (R.R.P. and J. A. Hammond, unpublished work), xnt-B {d4–704} (nd6/nd6, xntB/xntB) (21), were cultured as described (25). nd6 mutants are normal in behavior but do not discharge trichocysts and therefore survive better the trauma of microinjection and intracellular electric recording.
DNA Samples and Microinjection.
Standard molecular biology techniques were used (30, 31). We used a frozen replica of an indexed genomic library of Paramecium DNA consisting of 160 separate 384-well microtiter plates (32). To generate the injected DNA, each plate was thawed and the bacteria was transferred to six duplicate LB-ampicillin plates by using a 384 pin replicator (Genetix, New Milton, U.K.) and grown overnight. The colonies were scraped from the six plates and mixed in liquid LB-ampicillin. After centrifugation, half of the pellet was then used to make a single midi-preparation of plasmid DNA (Qiagen, Chatsworth, CA). Eluates from the DNA-purification columns were filtered and precipitated. Samples were resuspended in 100 μl of 10 mM Tris, pH 8.5 and stored at either −20°C or −80°C. The purified samples ranged from 70 to 200 μg (100 ± 30 μg) so that an estimated 850–2,500 copies of each individual plasmid species could be injected per 5 pl into a macronucleus. Sample preparations, methods of injection, preinjection, and postinjection culture were as described (24, 33). On discovering a transforming plate of 384 bacterial clones, midi-preparations were made from eight separate sections, each containing 48 bacterial clones. Then 48 individual plasmid minipreparations were prepared from the single transforming section and arranged as a 6 × 8 matrix. Aliquots from the individual minipreparations were combined into 14 mixtures representing each row and each column. Transformation occurred with only one row mixture and one column mixture and allowed for the identification of a single transforming clone. For further analyses, 5–0.05 pg/pl of the single plasmid DNA were injected to effect transformation.
Behavioral Testing and Electrophysiology.
Putative clonal transformants were individually transferred to an adaptation solution of 4 mM KCl, 1 mM CaCl2, 1 mM Hepes, 0.01 mM EDTA, pH 7.2 for ≈10 min and then individually transferred into a Mg2+ test solution (10 MgCl2, replaced KCl above). The duration of backward swimming on transfer was monitored with a stereomicroscope and recorded (34). Membrane currents were recorded from cells bathed in Mg2+/tetraethylammonium solution as established (35, 36). The microelectrodes used for clamping membrane potential contained 3 M CsCl, tip resistance 10–25 MΩ. Voltage steps were commanded, and resultant currents were analyzed by using pclamp software (Axon Instruments, Foster City, CA). All currents were filtered at 1 kHz and corrected for linear leak current, estimated from averaged currents elicited by repeated 20-ms, 3- to 12-mV hyperpolarizations.
Sequence Analyses.
DNAs were amplified and tagged by using the ABI PRISM Big Dye Terminator cycle sequencing kit (Perkin–Elmer Applied Biosystems) and sequenced by the University of Wisconsin Biotech Center. We searched for homologues in current databases with blast, fasta, mpsrch; searched for domains in the Conserved Domain Database at the National Center for Biotechnology Information, which includes protein family consensus sequences (Pfam) identified through multiple alignments and hidden Markov models (Sanger Institute, Cambridge, U.K.); analyzed secondary structure with protean (DNAstar, Madison, WI); aligned sequences with megalign (DNAstar) and pileup (GCG). The dendrogram presented in this article was generated by using distances calculated by PAUP 4.0 (GCG) to generate a heuristic bootstrap 50% majority-rule consensus tree (100 replicates) from a clustalw alignment (megalign, Gonnet 250) of the entire sequences of just the following proteins: Arabidopsis (ARATH 17, GenBank accession no. AAB70411; and ARATH MHX 8, GenBank accession no. AAF14229), Caenorhabditis (CAEEL 12, GenBank accession no. CAB03047; CAEEL 16, GenBank accession no. AAK71415; and CAEEL NCX 7, GenBank accession no. CAA62913), Drosophila [DROME K+-dependent NCX (NCKX) 11, GenBank accession no. AAF07938; DROME NCX 6, GenBank accession no. AAB63464; and DROME 15, GenBank accession no. AAF59056], Escherichia (ECOLI YRGB 3, GenBank accession no. P45394), Gallus (NCKX-1 10, GenBank accession no. AAF25808), Homo (HOMO NCX 6, GenBank accession no. P32418; and HOMO NCKX-3 9, GenBank accession no. AAG12988), Loligo (LOLOP NCX 4, GenBank accession no. AAB52920), Methanococcus (METTH 2, GenBank accession no. AAB85644), Schizosaccarhomyces (SCHPO 14, GenBank accession no. CAB08751), Tetrahymena (TETPY Tcep1p 13, GenBank accession no. AAL79511), and Paramecium (PARTE XntAp 1, GenBank accession nos. AF548349 and AF548350).
Results
Isolation of a DNA Fragment by Phenotypic Complementation.
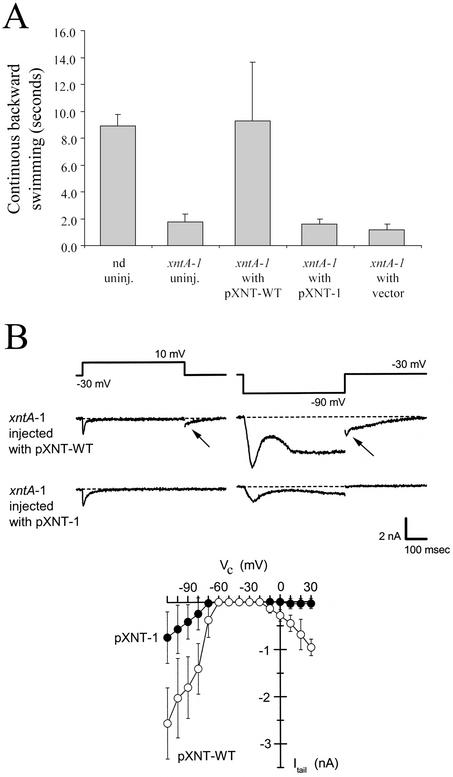
WT paramecia exhibit avoidance responses (rapidly reversing ciliary beat and changing directions) to test solutions that contain Mg2+. Eccentric mutants do not (Fig. 1A, ref. 21). After consecutive rounds of injecting plasmid midi- or mini-preparations from the library into the macronuclei of the mutants, we identified a single phenotype-transforming clone and purified a pBluescript plasmid with a 6.9-kb insert (pP76C20). Restriction-digested pP76C20 or subcloned fragments were then tested. Only those that retained a relatively long ORF found in the corresponding 5′ half of the insert were active. The behavioral phenotype was complemented with pBluescript (pXNT-WT), which contained a 2,427-bp PCR product from WT genomic DNA but not by pXNT-1, the equivalent from xntA-1 genomic DNA, or by the empty plasmid (Fig. 1A). The plasmid pXNT-WT effectively complemented eight different independently isolated xntA alleles, but not a mutant of an unlinked locus, xntB. The empty plasmid or pXNT-1 with the xntA-1 mutation did not rescue any of the mutants. Pleiotropic behavioral effects of xntA (20, 21) were also complemented by pXNT-WT.
Fig 1.
Complementation of the eccentric phenotypes. (A) Behavior response on transfer into the Mg2+ test solution of clonal descendents of various cells. Those of uninjected WT (nd) or eccentric (xntA-1) injected with the XNTA1-bearing plasmid (pXNT-WT) swim backward for ≈10 s; those of xntA-1 uninjected or injected with xntA-1-bearing (pXNT-1) or empty plasmid (vector) swim backward for only ≈2 s. Mean ± SD are shown, n = 50 = 10 × 5 clones for each entry. (B) The Mg2+-specific current that is best measured at the start of its tail current (arrow). Descendents of xntA-1 injected with pXNT-WT or pXNT-1 were placed under voltage clamp in Mg2+ solution. The membrane was held at −30 mV and then stepped for 500 ms to 10 mV or −90 mV to elicit the membrane currents shown. pXNT-WT-injected, but not the pXNT-1-injected, cells exhibit an inward tail current on returning to −30 mV, a current that has been shown previously to be Mg2+-specific. (Lower) The amplitude of such tail currents (Itail) elicited by 500-ms steps to the potentials indicated (Vc): mean ± SD (n = 4 pXNT-WT-expressing cells, n = 9 pXNT-1 cells) (19).
The membrane currents of the transformants were examined by using a two-electrode voltage clamp. As described, both depolarization and hyperpolarization can elicit the Mg2+ current in the WT but not in eccentric mutants (20–22). The xntA-1 mutant complemented with the WT plasmid pXNT-WT showed strong inward Mg2+ currents whereas those injected with the mutant plasmid pXNT-1 (Fig. 1B) or empty plasmid (not shown) did not. The Mg2+ current is most easily measured at the onset of it tail current, which allows us to examine its magnitude over a range of voltage (Fig. 1B Lower). The current restored in xntA-1 cells exhibited amplitudes, kinetics, and voltage dependence that were indistinguishable from the WT current described in ref. 19.
The XNTA ORF and Translated Product.
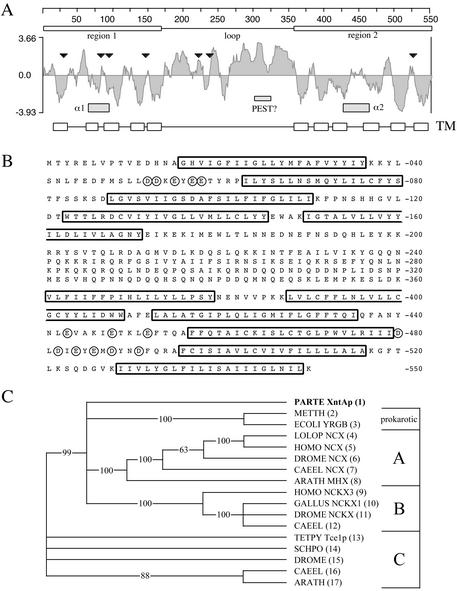
RT-PCR analyses and translated protein sequence alignments delimited an ORF that is 1,707 bp within the 2,427-bp complementing PCR fragment. The ORF includes two typical Paramecium introns (32 and 22 bp) flanked by an AT-rich 5′ and 3′ UTR. PCR products and subcloned fragments were sequenced from genomic DNA isolated from the eight eccentric mutant stocks in our collection. Each has a different mutation within the ORF (Fig. 2A, triangles). The nature and consequences of the mutations are summarized in Table 1. Many of these mutations occur in the first half of the molecule, result in truncations, and could have significant effects on membrane insertion and protein folding, transport, or function. Initial blastp searches at the National Center for Biotechnology Information with the translated product, XntAp, showed very little homology with other molecules (Fig. 2B). However, the low complexity filter masked eight stretches of residues containing 120 of the 550 predicted amino acids (61). blastp searches with the low complexity filter off reveal an alignment with Pfam 1699 [expect value (E) = 4 × 10−6, ref. 37] and significant alignments with NCKX-type exchangers along the entire 550 aa of XntAp (Fig. 2B). The most significant alignment is with a human variant of NCKX-3 (E = 2 × 10−14). The hydropathy plot (Fig. 2A) reveals two regions with potential transmembrane (TM) segments (Fig. 2 A and B, open boxes) and a long connecting hydrophilic loop. The loop has many charged residues together with a possible proteolytic signal (or PEST) sequence (38) and the transition from the loop to the second TM region also includes a number of proline residues, all being features that can be found in other exchangers. The Pfam 1699 recognition sequence (see Materials and Methods) is derived from multiple alignments by using a hidden Markov model of the two conserved regions, α1 and α2, known to be crucial in ion translocation and thought to be derived from a gene duplication. Whereas the blastp alignment scores for the Pfam sequence indicate a weak recognition of the XntAp's α1, they do not suggest a significant similarity for the corresponding α2 sequence (Fig. 2A, gray boxes). The putative α1 domain of XntAp (TM2, TM3, and surrounding loops) shows the strongest similarity to TM3 and the C-terminal half of α1 of the NCXs found in the blast searches. This includes some residues thought to be in the aqueous pathway of the ions from the accessibility studies of NCXs (39–41). clustal alignments with a large number of different transporters very consistently grouped the XntAp sequence with the NCX and NCKX family of exchangers (data not shown). However, a clustal w alignment restricted to sequences with homology to Pfam 1699 results in distance values that indicate a significant divergence from other members of this family (Fig. 2C). Whereas the function of some of the NCKX (group A) and NCX (group B) clusters has been experimentally determined, group C is of exchanger-like proteins of unknown function.
Fig 2.
XntAp, the eccentric gene product. (A) A Kyte–Doolittle hydrophilicity plot of XntAp (7-aa window). Marked are two hydrophobic regions (boxes above plot) and a connecting hydrophilic loop, the putative TM segments (TM boxes at bottom), a PEST-like sequence (slashed), and the regions corresponding to the α1 and α2 regions seen in other NCX/NCKXs (shaded boxes). Black triangles mark the location of the mutations found in the eight independently isolated mutant alleles (Table 1). (B) The XntAp sequence. Putative TM domains are outlined, and acidic residues near the possible α1 and α2 regions are circled. (C) An unrooted 50% majority-rule consensus dendrogram showing XntAp is distantly related to NCX/NCKXs. They are consistently clustered into three groups. XntAp is only slightly more related to group A (verified NCKXs and immediate relatives) and group B (NCXs and the Arabidopsis Mg2+ exchanger) than group C (putative exchangers with divergent C-terminal hydrophobic regions).
Table 1.
The mutations of eight xntA alleles
| Allele | Nucleotide | Amino acid | Ref. |
|---|---|---|---|
| xntA-1 | C111Δ | 21 | |
| xntA-2 | G1630A | G526R | 21 |
| xntA-3 | G338A | G95E | 21 |
| xntA-4 | C718G+A | Unpublished work | |
| xntA-5 | G302A | S83N | Unpublished work |
| xntA-6 | G337A | G95R | Unpublished work |
| xntA-521 | A767G | Q238(stop) | 28 |
| xntA-596 | G495A | W147(stop) | 29 |
Three independent genomic-DNA isolations followed by PCR and sequencing for each mutant stock. Nucleotide number includes intron sequences.
Deletion frameshift, peptide ends in 30VLYITFIRSICQICLKTL.
Insertion frameshift, peptide ends in 222EKKDKHF.
xntA-521 was d4-521 isolated as K+-resistance K21.
xntA-596 was Ba2+-overreactive BaB (d4-596).
Silencing.
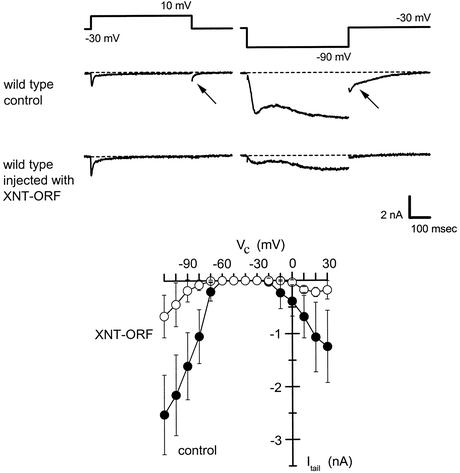
The loss of the Mg2+ current and Mg2+-induced avoidance is apparently the null phenotype of eccentric because it is seen in cells with nonsense xntA alleles that truncate the protein early (xntA-1, xntA-4, xntA-521, and xntA-596, Table 1). We used the silencing procedure to further test the relation between the XNTA product and these Mg2+ phenotypes. High concentrations of flankless ORFs silence the corresponding endogenes in the Paramecium macronucleus (42, 43). A PCR product consisting of only the 1,707-bp WT XNTA ORF, without UTRs, was concentrated to 6–8 pg/pl, and ≈5–10 pl of the sample was injected into the macronucleus of individual WT paramecia (nd6). Clonal descendants of the recipients were examined after three fissions. Transferring these cells into the Mg2+-containing test solution elicited no avoidance response. Parallel cells injected with empty plasmids at a similar concentration showed a WT avoidance response. Under a voltage clamp, these control cells showed normal inward Mg2+ currents, whereas the experimental cells had lost almost completely the Mg2+ current on both depolarization and hyperpolarization (Fig. 3).
Fig 3.
Suppression of the Mg2+-specific current in the WT by gene silencing. WT cells were injected with empty plasmid (control) or XNT-ORF and their clonal descendents were examined under voltage clamp in Mg2+ solution for evidence of a Mg2+-specific current. The membrane was held at −30 mV and then stepped for 500 ms to 10 mV or −90 mV to elicit the membrane currents including tail currents (arrows) shown. The control cells exhibited normal membrane currents that were indistinguishable from an untreated WT, but cells injected with XNT-ORF closely resembled eccentric mutant cells. (Lower) The amplitudes of tail currents (Itail) elicited by 500-ms steps to the test potentials indicated (Vc). Current amplitudes on both depolarization and hyperpolarization are reduced by ≈75% in the silenced cells. Data represent mean ± SD; currents from six cells injected with XNT-ORF (○) or seven cells injected with control plasmid (•).
Discussion
We have identified a gene that is required for a large Mg2+-specific current in Paramecium. The current has all of the hallmarks of an ion channel but the XNTA gene is distantly related to a family of NCXs. The discovery is significant because little is known about the molecules involved with Mg2+ permeation in eukaryotes. Furthermore the possibility that the observed current is passing through this molecule may provide some insights into the functional relatedness of channels and exchangers (see below).
Although it is common and very fruitful to seek homologs of an archetype, as in the case of NCX1 (44), the strength of starting from a mutant phenotype in vivo, without preconcieved notions is clearly demonstrated here. This forward-genetics approach led us to discover a gene that we would never have suspected based on the properties of the current nor would we have focused on such a distant relative of an exchanger even if a Paramecium genome sequence was available (45). The identification of XNTA demonstrates the power of the forward-genetic approach, despite the prevalence of searches based on sequence homology.
Exchanger or Channel?
At the moment, we cannot say with certainty that the Paramecium whole-cell Mg2+ current (IMg) flows through the XntAp protein. It is possible that XntAp is only a regulatory subunit of the Mg2+ flux. However, because XntAp has strong similarities to known proteins that facilitate TM fluxes, we therefore assume the simplest interpretation to be correct. This notion raises the question of how Mg2+ permeation might be achieved. The ion selectivity of the whole-cell current (Mg2+ > Co2+, Mn2+ > Ca2+, ref. 19) is without precedent in the known world of ion channels, but a bacterial transporter (CorA) does have a similar preference for Mg2+. Countering this observation is the fact that IMg is relatively large, fast-activating (Fig. 1B, ref. 19), and strongly reminiscent of a Na+ current shown to be via an ion channel in the same membrane (46). Further support for the notion that IMg is through a channel-like molecule is that the current is insensitive to changes in the concentration of extracellular Na+, K+, or Cl− or even pH. Indeed, the current is evident and can be caused to reverse direction in the absence of all three ions extracellularly (unpublished data, ref. 19). This attribute argues against XntAp cotransporting or exchanging Mg2+ for one of these three monovalents. The IMg does require a rise in intracellular Ca2+ concentration for activation, and it is formally possible that XntAp transports both Ca2+ and Mg2+, yet the channel-like characteristics of IMg are sufficiently compelling that it seems appropriate to speculate on how an exchanger molecule might be modified to pass a large ion current apparently without a counter flux.
XntAp vs. Known Ion Exchangers.
Ion exchangers use the electrochemical gradient of one ion to transport another in an opposite direction (47). NCX and NCKX contain two domains of multiple TMs (48, 49) that form the ion pathway (47, 50). These two TM domains appear to have an antiparallel arrangment (49, 51), topologically reminiscent of the Escherichia coli Cl− channel (52). The two domains contain two highly similar regions called α1 and α2 shown to govern ion permeation and specificity (48, 53). The sequence of XntAp corresponding to the α1 can be aligned but with several distinctive substitutions and the α2 is even more distant (data not shown). Some of the substitutions could reflect reciprocal substitutions whereas others may represent the natural variation seen in the sequences currently aligned to generate Pfam 1699 (37). For example, a methionine uniquely seen in the TM2 (α1) of XntAp might play a role similar to the one played by the methionine seen in NCX1's TM3 but not found in NCKX types. Another striking substitution is the absence of a highly conserved negative charge in the TM2 of both XntAp and AtMHX (15). The role of this charged residue might be shifted in both of these proteins to an aspartic acid unique to their TM3 regions. One of the most striking differences in XntAp's α1 and α2 sequences is the density of acidic residues seen at ends of these two regions. Especially the distal end of α2 where a pattern of charges spaced at every other residue alternating with hydrophobic residues (see Fig. 2B, circled residues between TM9 and TM10) is strikingly similar to sequences found in the hydrophilic portions of CorA as well as the N terminus of NCX-1 (1, 50). A disulfide bridge in NCX-1 may bring these negative charges close to the α1/α2 structure (50). This is an intriguing coincidence, because NCX1 may pass Mg2+ under certain circumstances (54).
Ion Specificity and Permeation.
Exchangers and channels share two basic biophysical features: ion specificity and use of the potential energy from ion gradients to facilitate their work. Recent crystal structures show that channel proteins narrow to a filter that coordinates the naked permeant ions that have been stripped of their hydration spheres and that this narrow filter defines the unitary conductance and ion specificity (52, 55, 56). The path and mechanism of ion translocation via exchangers is not so clear, but a filter-like narrowing seems necessary because dehydration (or partial dehydration) and coordination are basic to ion recognition (57). The positions where α1 and α2 re-entrant loops meet may well be part of a narrowing filter. The discovery that the ClC Cl− channel has an antiparallel topology (52), similar to that predicted for the transporter NCX1 (49–51, 58, 59) suggests that an exchanger could also be composed of multiple parallel subunits, each with internal antiparallel halves and the filter a multimer of the re-entrant loops. Channel filters open and close and can be viewed as a second gate. For example, the KcsA K+ channel's tube-like filter can be occluded when [K+] is low (56). The exchanger's “filter” can be envisioned to have two closed conformations as in the biochemical schemes of previous authors (50, 58) except for specifying the cyclic conformational changes of the filter. Unlike channels the filter here is never open at both ends and one exchange of the two sets of ions entails two conformational changes. No conformational change is necessary for the near-diffusion-limited flow of specific ions through open channels. This finding explains why exchangers transport only ≈103 per s (50), as opposed to ≈106–8 per s through an open channel (56, 57).
Unlike close homologs, distant relatives often reveal how a basic molecular design serves different purposes. Whereas XntAp's α1 loop has clear semblance to those of exchangers, its α2 is much less recognizable. If one imagines that an exchanger loses one of its two closed conformations, this could convert it into a tube. A Mg2+-specific filter could be constructed of noncontiguous amino acid residues from different parts of the protein as in the Cl− channel (52) and/or through arranging carbonyl oxygens like those in the KcsA (56). The prominently placed string of negative charges in XntAp, with parallels in CorA and NCX1, may play a role in the permeation of ions. Such negative charges neighboring α1 and α2 of XntAp might encourage “ion-atmosphere”-type interactions with Mg2+ as observed with several single-stranded polynucleotides (60). Such electrostatic association through secondary (outer sphere) coordination could result in faster and preferred passage of Mg2+ through the filter. At the moment, we do not know what part of XntAp could be a Mg2+ filter or whether it exists beyond freshwater protists. Yet many exchangers to date are identified only through sequence similarity and have not been tested for their electrophysiological functions. It will be interesting to see whether some of them in fact handle Mg2+ and/or pass channel-like larger ion currents. Future work should help us understand how XntAp functions and, indirectly, how exchangers and channels function in general.
Acknowledgments
We thank Derek Ho, Jean Sgro, and Lynn Hammond for technical assistance. This work was supported by National Institutes of Health Grants GM22714 (to C.K.), GM51498, and GM61716 (to R.R.P.).
Abbreviations
NCX, Na+/Ca2+ exchanger
NCKX, K+-dependent NCX
TM, transmembrane
References
- 1.Moncrief M. B. & Maguire, M. E. (1999) J. Biol. Inorg. Chem. 4 523-527. [DOI] [PubMed] [Google Scholar]
- 2.Zsurka G., Gregan, J. & Schweyen, R. J. (2001) Genomics 72 158-168. [DOI] [PubMed] [Google Scholar]
- 3.Schock I., Gregan, J., Steinhauser, S., Schweyen, R., Brennicke, A. & Knoop, V. (2000) Plant J. 24 489-501. [DOI] [PubMed] [Google Scholar]
- 4.Gregan J., Bui, D. M., Pillich, R., Fink, M., Zsurka, G. & Schweyen, R. J. (2001) Mol. Gen. Genet. 264 773-781. [DOI] [PubMed] [Google Scholar]
- 5.Graschopf A., Stadler, J. A., Hoellerer, M. K., Eder, S., Sieghardt, M., Kohlwein, S. D. & Schweyen, R. J. (2001) J. Biol. Chem. 276 16216-16222. [DOI] [PubMed] [Google Scholar]
- 6.Flatman P. W. (1991) Annu. Rev. Physiol. 53 259-271. [DOI] [PubMed] [Google Scholar]
- 7.Beyenbach K. W. (1990) Magnesium Trace Elem. 9 233-254. [PubMed] [Google Scholar]
- 8.Cefaratti C., Romani, A. & Scarpa, A. (2000) J. Biol. Chem. 275 3772-3780. [DOI] [PubMed] [Google Scholar]
- 9.Touyz R. M., Mercure, C. & Reudelhuber, T. L. (2001) J. Biol. Chem. 276 13657-13663. [DOI] [PubMed] [Google Scholar]
- 10.Tashiro M. & Konishi, M. (2000) Am. J. Physiol. 279 C1955-C1962. [DOI] [PubMed] [Google Scholar]
- 11.Rasgado-Flores H., Gonzalez-Serratos, H. & DeSantiago, J. (1994) Am. J. Physiol. 266 C1112-C1117. [DOI] [PubMed] [Google Scholar]
- 12.Rasgado-Flores H. & Gonzalez-Serratos, H. (2000) Front. Biosci. 5 D866-D879. [DOI] [PubMed] [Google Scholar]
- 13.Gunther T. & Vormann, J. (1990) Magnesium Trace Elem. 9 279-282. [PubMed] [Google Scholar]
- 14.Gunther T. & Hollriegl, V. (1993) Biochim. Biophys. Acta 1149 49-54. [DOI] [PubMed] [Google Scholar]
- 15.Shaul O., Hilgemann, D. W., de-Almeida-Engler, J., Van Montagu, M., Inz, D. & Galili, G. (1999) EMBO J. 18 3973-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naitoh Y. & Eckert, R. (1968) Z. Vergl. Physiol. 61 453-472. [Google Scholar]
- 17.Kung C. (1971) Genetics 69 29-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saimi Y., Martinac, B., Preston, R. R., Zhou, X. L., Sukharev, S., Blount, P. & Kung, C. (1994) Soc. Gen. Physiol. Ser. 49 179-195. [PubMed] [Google Scholar]
- 19.Preston R. R. (1990) Science 250 285-288. [DOI] [PubMed] [Google Scholar]
- 20.Preston R. R. & Kung, C. (1994) J. Membr. Biol. 139 203-213. [DOI] [PubMed] [Google Scholar]
- 21.Preston R. R. & Kung, C. (1994) Genetics 137 759-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preston R. R. (1998) J. Membr. Biol. 164 11-24. [DOI] [PubMed] [Google Scholar]
- 23.Haynes W. J., Ling, K. Y., Saimi, Y. & Kung, C. (1996) J. Neurogenet. 11 81-98. [DOI] [PubMed] [Google Scholar]
- 24.Haynes W. J., Vaillant, B., Preston, R. R., Saimi, Y. & Kung, C. (1998) Genetics 149 947-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haynes W. J., Ling, K. Y., Preston, R. R., Saimi, Y. & Kung, C. (2000) Genetics 155 1105-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonneborn T. M. (1970) Proc. R. Soc. London Ser. B 176 347-366. [DOI] [PubMed] [Google Scholar]
- 27.Lefort-Tran M., Aufderheide, K., Pouphile, M., Rossignol, M. & Beisson, J. (1981) J. Cell Biol. 88 301-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shusterman C. L., Thiede, E. W. & Kung, C. (1978) Proc. Natl. Acad. Sci. USA 75 5645-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shusterman C. L., (1981) Ph.D. thesis (University of Wisconsin, Madison).
- 30.Sambrook J., Fritsch, E. F. & Maniatis, T., (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 31.Ausubel F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K., (2002) Current Protocols in Molecular Biology (Wiley, New York).
- 32.Keller A. M. & Cohen, J. (2000) J. Eukaryotic Microbiol. 47 1-6. [DOI] [PubMed] [Google Scholar]
- 33.Haynes W. J., Ling, K. Y., Saimi, Y. & Kung, C. (1995) J. Eukaryotic Microbiol. 42 83-91. [DOI] [PubMed] [Google Scholar]
- 34.Kung C. (1975) Genetics 79,Suppl. 423-431. [PubMed] [Google Scholar]
- 35.Hinrichsen R. D. & Saimi, Y. (1984) J. Physiol. (London) 351 397-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preston R. R., Saimi, Y. & Kung, C. (1992) J. Gen. Physiol. 100 233-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bateman A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S. R., Griffiths-Jones, S., Howe, K. L., Marshall, M. & Sonnhammer, E. L. (2002) Nucleic Acids Res. 30 276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rechsteiner M. & Rogers, S. W. (1996) Trends Biochem. Sci. 21 267-271. [PubMed] [Google Scholar]
- 39.Doering A. E., Nicoll, D. A., Lu, Y., Lu, L., Weiss, J. N. & Philipson, K. D. (1998) J. Biol. Chem. 273 778-783. [DOI] [PubMed] [Google Scholar]
- 40.Iwamoto T., Nakamura, T. Y., Pan, Y., Uehara, A., Imanaga, I. & Shigekawa, M. (1999) FEBS Lett. 446 264-268. [DOI] [PubMed] [Google Scholar]
- 41.Nicoll D. A., Ottolia, M., Lu, L., Lu, Y. & Philipson, K. D. (1999) J. Biol. Chem. 274 910-917. [DOI] [PubMed] [Google Scholar]
- 42.Ruiz F., Vayssie, L., Klotz, C., Sperling, L. & Madeddu, L. (1998) Mol. Biol. Cell 9 931-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galvani A. & Sperling, L. (2001) Nucleic Acids Res. 29 4387-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicoll D. A., Longoni, S. & Philipson, K. D. (1990) Science 250 562-565. [DOI] [PubMed] [Google Scholar]
- 45.Dessen P., Zagulski, M., Gromadka, R., Plattner, H., Kissmehl, R., Meyer, E., Betermier, M., Schultz, J. E., Linder, J. U., Pearlman, R. E., et al. (2001) Trends Genet. 17 306-308. [DOI] [PubMed] [Google Scholar]
- 46.Saimi Y. & Ling, K. Y. (1990) Science 249 1441-1444. [DOI] [PubMed] [Google Scholar]
- 47.Saier M. H. (2000) Microbiol. Mol. Biol. Rev. 64 354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwarz E. M. & Benzer, S. (1997) Proc. Natl. Acad. Sci. USA 94 10249-10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saaf A., Baars, L. & von Heijne, G. (2001) J. Biol. Chem. 276 18905-18907. [DOI] [PubMed] [Google Scholar]
- 50.Philipson K. D. & Nicoll, D. A. (2000) Annu. Rev. Physiol. 62 111-133. [DOI] [PubMed] [Google Scholar]
- 51.Qiu Z., Nicoll, D. A. & Philipson, K. D. (2001) J. Biol. Chem. 276 194-199. [DOI] [PubMed] [Google Scholar]
- 52.Dutzler R., Campbell, E. B., Cadene, M., Chait, B. T. & MacKinnon, R. (2002) Nature 415 287-294. [DOI] [PubMed] [Google Scholar]
- 53.Nicoll D. A., Hryshko, L. V., Matsuoka, S., Frank, J. S. & Philipson, K. D. (1996) J. Biol. Chem. 271 13385-13391. [DOI] [PubMed] [Google Scholar]
- 54.Tashiro M., Konishi, M., Iwamoto, T., Shigekawa, M. & Kurihara, S. (2000) Pflügers Arch. 440 819-827. [DOI] [PubMed] [Google Scholar]
- 55.Doyle D. A., Morais Cabral, J., Pfuetzner, R. A., Kuo, A., Gulbis, J. M., Cohen, S. L., Chait, B. T. & MacKinnon, R. (1998) Science 280 69-77. [DOI] [PubMed] [Google Scholar]
- 56.Zhou Y., Morais-Cabral, J. H., Kaufman, A. & MacKinnon, R. (2001) Nature 414 43-48. [DOI] [PubMed] [Google Scholar]
- 57.Hille B., (2001) Ion Channels of Excitable Membranes (Sinauer, Sunderland, MA).
- 58.Shigekawa M. & Iwamoto, T. (2001) Circ. Res. 88 864-876. [DOI] [PubMed] [Google Scholar]
- 59.Blaustein M. P. & Lederer, W. J. (1999) Physiol. Rev. 79 763-854. [DOI] [PubMed] [Google Scholar]
- 60.Porshke D. (1995) in The Biological Chemistry of Magnesium, ed. Cowan, J. A. (VCH, New York), pp. 85–110.
- 61.Wootton J. C. & Federhen, S. (1996) Methods Enzymol. 266 554-571. [DOI] [PubMed] [Google Scholar]