Abstract
Cyanobacterial clock proteins KaiA and KaiC are proposed as positive and negative regulators in the autoregulatory circadian kaiBC expression, respectively. Here, we show that activation of kaiBC expression by kaiA requires KaiC, suggesting a positive feedback control in the cyanobacterial clockwork. We found that robust circadian phosphorylation of KaiC. KaiA was essential for in vivo KaiC phosphorylation and activated in vitro KaiC autophosphorylation. These effects of KaiA were attenuated by the kaiA2 long period mutation. Both the long period phenotype and the abnormal KaiC phosphorylation in this mutant were suppressed by a previously undocumented kaiC mutation. We propose that KaiA-stimulated circadian KaiC phosphorylation is important for circadian timing.
Cyanobacteria are the simplest organisms known to generate circadian rhythms (1, 2). In the cyanobacterium, Synechococcus elongatus PCC 7942, three kai genes (kaiA, kaiB, and kaiC) are essential for circadian function and form a gene cluster (3). A transcription/translation-based autoregulatory loop by these Kai proteins is proposed as an important process in circadian kaiBC gene expression (3). This type of feedback regulation of clock genes has also been suggested as a key process to generate circadian oscillation also in eukaryotic clock systems in Drosophila, Neurospora, Arabidopsis, and mammals (4). Nevertheless, no similarity was found between the Kai proteins and eukaryotic clock proteins. We know some biochemical properties of the Kai proteins. First, all three Kai proteins interact with each other to form protein complexes (5), and KaiC also interacts with the SasA histidine kinase (6). Second, KaiC has ATP-binding and autokinase activities (7). Third, the KaiB and KaiC proteins accumulate in a circadian fashion in free-running conditions (8). However, it is unknown how these properties are related to each other and function for circadian kaiBC expression.
We report here genetic and biochemical data that provide a previously undocumented regulatory link among these properties. Combinatorial studies of genetic inactivation and overexpression of kai genes revealed coordinated functions of KaiA and KaiC for both positive and negative limbs of the circadian feedback process for kaiBC expression. We also provide genetic evidence for cooperative KaiA-KaiC functions in determining the period length by identification of a previously undocumented kaiC allele (kaiC15) that suppresses a kaiA2 long period phenotype. At the molecular level, KaiA was found to enhance in vitro KaiC autokinase activity. Moreover, we found that KaiC is phosphorylated in vivo in a robust circadian manner. KaiA was essential for this in vivo KaiC phosphorylation. Finally, we found that the level of KaiC phosphorylation is lowered in the kaiA2 mutant, whereas kaiC15 restores it. Thus, we propose that KaiA and KaiC cooperatively function in both positive and negative feedback processes in the cyanobacterial clock by controlling the state of KaiC phosphorylation in a circadian fashion.
Materials and Methods
Bacterial Strains, Media, and Culture.
We used wild-type, kaiA-inactivated and kaiC-depleted Synechococcus strains that harbor the PkaiBC:luxAB reporter gene set. The mutant reporter strains were newly generated by mutagenesis of a wild-type PkaiBC:lux luciferase reporter strain, NUC42 (9), as described (3). For overexpression of kaiA and kaiC genes, these reporter strains were transformed with pTS2KPtrc:kaiA or pTS2KPtrc:kaiC (3). For the suppressor mutant screen, we also used a kaiA2 mutant reporter strain, A30a, which harbors a PpsbAI:luxAB reporter unit (3).
Suppressor Mutant Screen.
kaiA2 mutant strain A30a was mutagenized with ethylmethane-sulfonate, and the bioluminescence profiles of mutagenized cells were monitored by a chilled charge-coupled device (CCD) camera-based multiplate bioluminescence monitoring system as described (10). Among ≈50,000 independent clones screened, we isolated one mutant that restored a period length of ≈24 h. Potential causal mutation kaiC15 was mapped to the kaiC gene by sequencing the entire kai locus. To confirm that this mutation is responsible for the kaiA2-suppressor phenotype, we generated kaiBC reporter strains that contain the kaiC15 mutation and/or the kaiA2 mutation by introduction of a mutagenized pCkaiABC vector (3) in the kaiABC-depleted reporter strain NUC42 (9) to regain a different mutant kai cluster at the original kai locus. Data in Fig. 1D illustrate bioluminescence profiles from these strains.
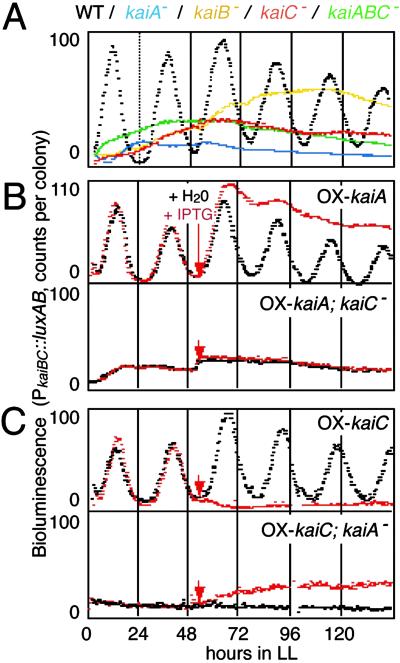
Fig 1.
(A) kaiBC promoter (PkaiBC) activity was monitored with a luciferase reporter gene set fused to PkaiBC in the wild-type (WT; black), kaiA− (blue), kaiB-inactivated (kaiB−; yellow), kaiC− (red), and kaiABC-cluster-depleted (kaiABC−; green) strains. The bioluminescence profiles of four independent clones for each mutant were measured in LL after two cycles of 12 h light:12 h dark (LD) alternations with a photomultiplier tube as described (3). Representative bioluminescence profiles normalized to that of the wild type are shown. (B) The effect of overexpression of the kaiA gene on PkaiBC activity in the wild-type and kaiC-disrupted strains. Bioluminescence from PkaiBC reporter strains carrying a Ptrc:kaiA construct (for kaiA overexpression) was monitored. Cells were grown on agar plates as described above and treated with 500 μM isopropyl β-d-thiogalactopyranoside or water after 50 h in LL (arrows). (C) The effect of overexpression of the kaiC gene on PkaiBC activity in wild-type and kaiA-disrupted strains.
Bacterial Expression and Purification of Kai Proteins.
The kaiB and kaiC ORFs were cloned into the pGEX-6P-1 vector (Amersham Pharmacia) and then transformed into Escherichia coli DH 10B. GST-fusion proteins were produced and affinity-purified in E. coli as described (5), and the GST-tag was then cleaved and removed with PreScission Protease (Amersham Pharmacia) according to the manufacturer's protocol. The kaiA ORF was cloned into pQE32 (Qiagen, Valencia, CA) and transformed with E. coli M15(pRep4) (Qiagen). The KaiA protein was produced and purified according to the manufacturer's protocol.
Protein Phosphorylation Assay.
Autokinase assay was performed at 30°C as described (7), with some modifications. Briefly, 8.6 pmol (1 μg) of KaiC was incubated with 10 μl of TD buffer (100 mM Tris⋅Cl/100 mM KCl/5 mM MgCl2,pH 7.6) in the presence or absence of KaiA (8.6 pmol), KaiB (38 pmol), or BSA (8.6 pmol) at 4°C for 40 min. ATP was supplemented to a final concentration of 0.05 mM (10,000 cpm/pmol) and incubated at 30°C. Time 0 was taken from when ATP was added (for data in Fig. 3 A and E). Reaction mixtures were subjected to SDS/PAGE on 10–12% gels followed by autoradiography as described (7). For data on Fig. 3D, KaiC was produced in E. coli and purified in the presence of 5 mM ATP. KaiC protein (1.4 μg) was incubated with 1.9 μg of KaiA in a reaction buffer (150 mM NaCl/20 mM Tris⋅Cl/0.4 mM EDTA/5 mM ATP/5 mM MgCl2, pH 8.0) at 30°C and then subjected to SDS/PAGE on 7.5% gels and Coomassie brilliant blue (CBB) staining.
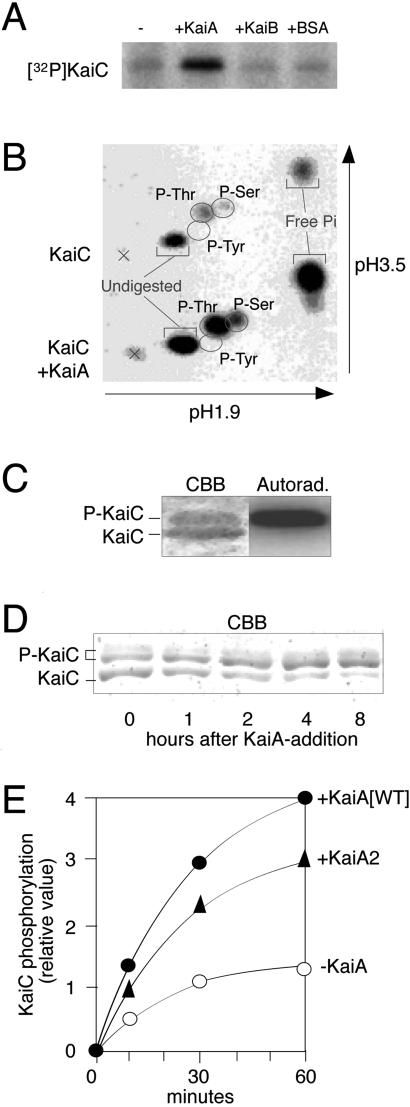
Fig 3.
KaiA-stimulated KaiC autokinase activity. (A) Autophosphorylation of the KaiC protein with [γ-32P]ATP in the presence or absence of KaiA, KaiB, and BSA. Autokinase assays were performed at 30°C for 60 min and then analyzed by SDS/PAGE and autoradiography. (B) Determination of phosphorylated aminoacyl species for KaiC autophosphorylation by the 2D thin-layer electrophoresis assay. Crosses indicate the sample origin positions. (C) Separation of phosphorylated and nonphosphorylated forms of KaiC by SDS/PAGE on 10% gels (acrylamide:N,N′-methylenebisacrylamide = 29.8:0.2). KaiC incubated with [γ-32P]ATP was subjected to SDS/PAGE followed by CBB staining (Left) and autoradiography (Right). (D) The effect of KaiA on the mobility of the KaiC protein on 7.5% SDS-polyacrylamide gels. (E) Effects of KaiA2 mutant proteins on KaiC autokinase activity. The graph shows the time course profile of KaiC autophosphorylation with or without the wild-type or kaiA2-mutation-introduced KaiA proteins (KaiA and KaiA2, respectively).
2D-TLC Analysis.
2D-TLC analysis was performed as described (11) with some modifications. KaiC (1 μg) was subjected to an autokinase assay, as described in the text, in the presence or absence of 1 μg of KaiA. After SDS/PAGE analysis, proteins were blotted onto poly(vinylidene difluoride) membranes. The phosphorylated bands were dissected from the membranes and digested with 6 M HCl at 110°C for 1 h. After centrifugation, the supernatants were lyophilized and spotted onto cellulose plates. Thin-layer electrophoresis was performed initially in 2% formic acid, 8.3% acetate (pH 1.9; 1D electrophoresis) and then in 0.5% pyridine, 10% acetate (pH 3.5; 2D electrophoresis) with the Multiphore II electrophoresis apparatus (Amersham Pharmacia). Phosphoserine, phosphothreonine, and phosphotyrosine (Sigma) were used as standard control samples.
λ Phosphatase (λPPase) Assay.
Synechococcus protein extracts were prepared with a French pressure cell press in buffer A (20 mM Hepes, pH 7.5/150 mM NaCl/1 mM EDTA/1 μg/ml PMSF/1 μg/ml pepstatin A/1 μg/ml leupeptin), and then supplemented with Triton X-100 (final 1%), sodium deoxycholate (0.5%), and SDS (0.1%). Five-hundred-microliter aliquots of the resulting protein extracts (1.4 mg/ml) were immunoprecipitated by incubation with 25 μl (bed volume) of AffiGel-Hz resin (Bio-Rad), immobilizing anti-KaiC IgG (6) at 4°C for 12 h. After washing five times with buffer A supplemented with 1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% SDS, the resin was equilibrated with λPPase buffer (50 mM Tris⋅Cl/0.1 mM EDTA/2 mM MnCl2/5 mM DTT/0.01% (wt/vol) Brij 35, pH 7.5). The resin was then incubated with 25 μl of λPPase buffer containing 100 or 1,000 units of λPPase (NEB) with or without inhibitors (50 mM NaF/20 mM Na3VO4) at 30°C for 1 h. The digested immune-complexes were resolved in an SDS sample buffer and then subjected to SDS/PAGE and Western blotting with anti-KaiC antisera (5).
Results
Cooperative Functions of KaiA and KaiC in kaiBC Expression Feedback Loops.
In the previous transcriptional feedback model, KaiA was suggested as a positive regulator for kaiBC expression (3). As described previously (3), a kaiA-inactivated (kaiA−) strain consistently reduces kaiBC promoter (PkaiBC) activity, as monitored by a luciferase reporter (Fig. 1A), and continuous kaiA overexpression enhances it and abolishes rhythmicity (Fig. 1B). On the other hand, KaiC has been proposed as a negative element for kaiBC expression (3). As shown in Fig. 1C Upper, kaiC overexpression consistently suppresses kaiBC expression (3). However, kaiC-disrupted (kaiC−) strains did not enhance but reduced kaiBC expression to ≈20% of the wild-type strain (Fig. 1A), which cannot be explained by the simple negative feedback model (1, 3). This result is in contrast to enhanced expression of the frequency (frq) gene, a negative element in the Neurospora clock, in frq-null mutants (12). Our observation in Synechococcus raises the possibility that KaiC may also function in the positive limb of the kaiBC oscillatory loop.
To examine whether the positive function of KaiA requires KaiC, we overexpressed kaiA in the wild-type and kaiC− strains under the control of an isopropyl-β-d-thiogalactopyranoside-inducible trc promoter. Although overexpression of kaiA enhanced kaiBC expression and abolished its rhythmicity in the wild-type strain (Fig. 1B; ref. 3), that of kaiA in the kaiC− strain failed to up-regulate kaiBC transcription (Fig. 1B). Thus, KaiC seems required for a positive effect of KaiA in the expression of kaiBC. Moreover, KaiC additionally participates in activation of kaiBC expression independently of KaiA function. As shown in Fig. 1C, KaiC overexpression reduces its own transcription (3), whereas in the kaiA− mutant it failed to exhibit the inhibitory effect and instead gradually enhanced kaiBC expression.
Identification of a kaiC Allele That Suppresses kaiA2 Long Period Phenotype.
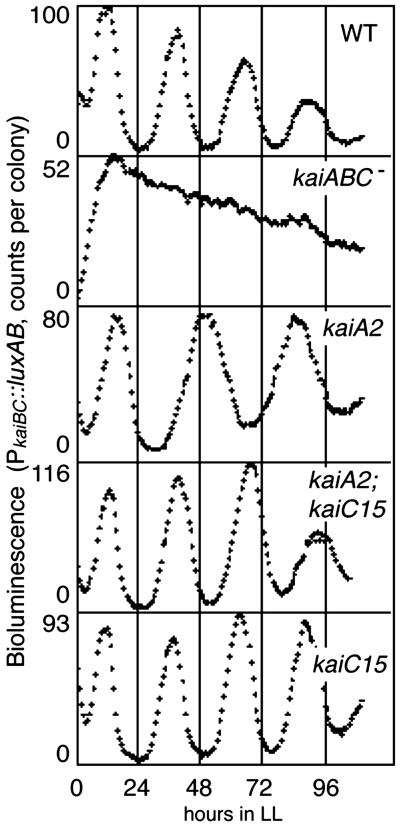
The KaiA–KaiC cooperation was further supported by identification of a previously undocumented kaiC allele named kaiC15, which suppresses a kaiA2 rhythm mutant, by a suppressor mutagenesis screen (Fig. 2). The kaiA2 mutation (Arg-249 to His in KaiA) lengthens the period by 6–7 h (3). We screened ethylmethane-sulfonate-mutagenized kaiA2 mutant strain A30a for suppressor mutants, which restore a wild-type period phenotype. The A30a strain contains a bioluminescence reporter to monitor the psbAI clock-controlled gene expression (3). Among ≈50,000 independent clones screened, we isolated a mutant that restored a period length of ≈24 h. Because most chemically mutagenized strains that impair period length have mutations in the kaiABC genes (3), we sequenced the kai locus of the genomic DNA prepared from the potential kaiA2 suppressor mutant. A mutation, designated kaiC15, was found in the kaiC ORF, which caused an amino acid substitution at the amino terminus of the KaiC (Ala-422 to Thr). To confirm that the kaiC15 mutation is responsible for the kaiA2-suppressor phenotype, we generated kaiBC reporter strains that contain the kaiC15 mutation and/or the kaiA2 mutation by site-directed mutagenesis (see Materials and Methods). As shown in Fig. 2, the kaiC15 mutation shortened the period of kaiA2 by ≈6 h. The average period length was 25.2 h in the wild-type strain, 33.0 h in the kaiA2 mutant, and 27.3 h in the kaiA2;kaiC15 strain. Note that kaiC15 mutation alone affected the period length to a lesser extent (24.7 h; Fig. 2). This finding presents genetic evidence for interactive KaiA–KaiC functions in determining the circadian period length (see Discussion).
Fig 2.
Suppression of kaiA2 long period phenotype by the kaiC15 mutation. kaiBC expression profiles in the wild-type, kaiABC−, kaiA2, kaiC2;kaiC15, and kaiC15 strains are shown. Measurement of the bioluminescence and representation of data are the same as described for Fig. 1.
KaiA Enhances KaiC Autophosphorylation Activity.
Then, how are KaiA and KaiC functions coordinated in the circadian system at the molecular level? We previously showed that all three Kai proteins interact in all possible combinations (5) and that the KaiC protein undergoes autophosphorylation in vitro (7). Thus, we examined whether the association of KaiC with KaiA (or KaiB) directly affects in vitro KaiC autokinase activity by incubation of KaiC with [γ-32P]ATP in the presence or absence of KaiA or KaiB protein. We found KaiC autokinase activity to be dramatically enhanced by KaiA (Fig. 3A). Addition of KaiB affected KaiC phosphorylation less (Fig. 3A). To examine phosphorylated aminoacyl species in KaiC, we performed 2D-TLC analysis for phosphorylated KaiC in the presence or absence of KaiA. KaiC was found to be autophosphorylated at both threonine and serine residues at a 2:1 ratio, and KaiA enhanced KaiC phosphorylation without affecting the ratio (Fig. 3B).
Circadian Rhythm in KaiC Phosphorylation in Vivo.
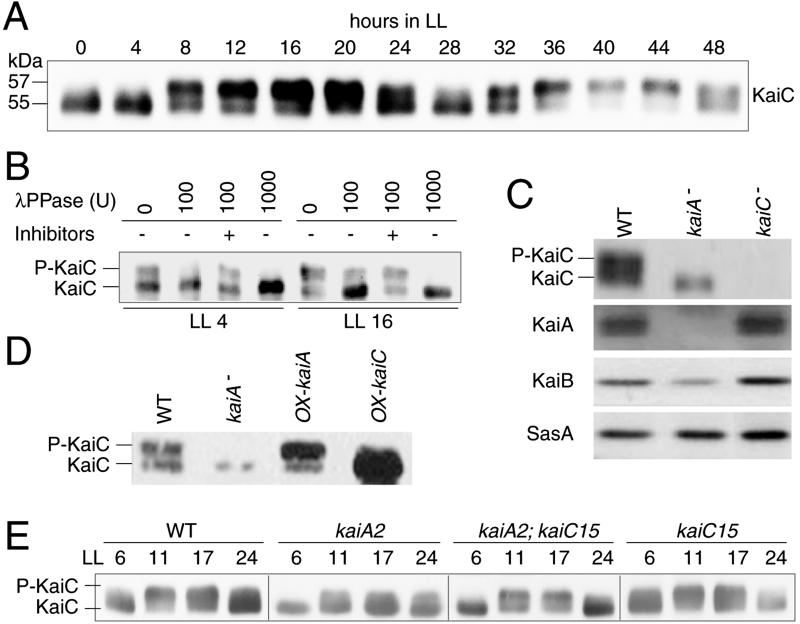
We found that KaiC is also phosphorylated in vivo. Under continuous illumination (LL) conditions, KaiB and KaiC accumulate in a circadian fashion, peaking at circadian time (CT) 16 (Fig. 4A), whereas KaiA accumulation shows much lower amplitude rhythm or even arrhythmia (8). We noted that KaiC shows circadian fluctuation in mobility shift on SDS–polyacrylamide gels (Fig. 4A). KaiC signal appeared as doublet bands, and the upper band is dominant at subjective early night (CT 12–16), whereas in subjective early day (CT 0–4), it is dramatically reduced. λPPase treatment abolished the upper band for KaiC, and this effect was inhibited by the λPPase inhibitors vanadate and fluoride (Fig. 4B). Thus, KaiC undergoes robust circadian phosphorylation, peaking at CT 16. The λPPase removes a phosphoryl group from phosphorylated Ser, Thr, Tyr, and His residues (13, 14), and our preliminary chemical stability assay suggested that KaiC is phosphorylated in vivo at Ser and/or Thr residues (T.N., unpublished results), as it is in vitro (Fig. 3B).
Fig 4.
(A) KaiC protein expression profile examined by Western blotting. Cells were cultured in a continuous culture system in BG-11 liquid medium to maintain an OD730 of 0.18 under LL (50 μE⋅m−2⋅s−1, in which E indicates an einstein, 1 mol of photons). After two LD cycles, cells were returned to LL conditions and then collected every 4 h. Total proteins were extracted by sonication of cell pellets in SDS-sample buffer and then subjected to SDS/PAGE with 10% gels (acrylamide:N,N′-methylenebisacrylamide = 29.8:0.2), followed by Western blotting assay (4.0 μg protein per lane). (B) λPPase assay. Soluble protein extracts were prepared from Synechococcus culture collected at hours 4 and 16 under LL (LL 4 and LL 16), immunoprecipitated with anti-KaiC IgG, digested with 100 or 1,000 units of λPPase in the presence or absence of λPPase inhibitors (NaF and Na3VO4), and then subjected to Western blotting with anti-KaiC antisera. (C) Expression of clock proteins in wild-type, kaiA-null, and kaiC-null mutant strains by Western blotting. Note that no circadian fluctuation was observed in both kai-gene mutants in terms of the state of KaiC phosphorylation throughout the circadian cycle (data not shown). We show here Western blots using cells collected at LL 16, when the level of KaiC phosphorylation is maximal in the wild-type strain. (D) KaiC phosphorylation profile in kaiA−, kaiA-overexpressing (ox-kaiA), and kaiC-overexpressing (ox-kaiC) cells (Western blotting). Cells were cultured in BG-11 liquid medium (100 μM isopropyl-β-d-thiogalactopyranoside) and collected at LL 16. Total proteins (8 μg) were extracted from the collected cells and subjected to 10% gels followed by Western blotting assay. (E) Circadian KaiC phosphorylation profiles in the wild-type, kaiA2, kaiC15, and kaiA2;kaiC15 strains. Total proteins (8 μg) were analyzed by Western blotting as described above.
KaiA Is Essential for in Vivo KaiC Phosphorylation.
To address whether KaiA protein also affects KaiC phosphorylation in vivo, we analyzed expression of KaiC and other clock proteins in the kaiA- and kaiC-null mutant strains (Fig. 4C). These mutants are arrhythmic (3) but did not affect the level of SasA protein. On the other hand, the Upper band in Fig. 4C (a phosphorylated form) of KaiC was evidently diminished in the kaiA− strain. Abnormality in the phosphorylation state of KaiC in the kaiA-null mutant was observed throughout the circadian cycle (data not shown). Thus, KaiA is essential for accumulation of phosphorylated KaiC protein in vivo. Decrease in the levels of KaiB and KaiC in the kaiA− cells (Fig. 4C) is most likely due to inactivation of kaiBC expression (3). The kaiA-overexpressor consistently enhanced the magnitudes of both KaiC phosphorylation and accumulation, whereas the kaiC-overexpressor accumulated a nonphosphorylated form of KaiC (Fig. 4D). Hyperphosphorylation of KaiC in the arrhythmic kaiA-overexpressor strain further supports that abolishment of KaiC phosphorylation in the kaiA-null mutant is not due to lack of circadian oscillation but to lack of the KaiA protein.
Because KaiA enhances both in vivo and in vitro KaiC phosphorylation, we tested whether activation of KaiC autophosphorylation by KaiA in vitro is accompanied by any mobility shift of the KaiC protein on SDS-polyacrylamide gels as observed in vivo. KaiC was autophosphorylated by incubation with [γ-32P]ATP, subjected to SDS/PAGE, and stained by CBB. As shown in Fig. 3C, KaiC appeared as doublet bands on 10% gels. Autoradiography of the gel revealed that the Upper band is the phosphorylated form as observed in vivo. To test the effect of KaiA on the mobility of the KaiC protein on gels, we purified KaiC protein in the presence of ATP, incubated it with KaiA, and then analyzed by SDS/PAGE. As shown in Fig. 3D, KaiC purified in the presence of ATP appeared as three bands on 7.5% gels. Incubation with KaiA attenuated the Bottom band signal, whereas it enhanced accumulation of the Top and Middle band signals. Incubation of KaiC without KaiA protein did not enhance accumulation of the Top and Middle bands (data not shown). Thus, KaiA enhances both in vitro and in vivo KaiC phosphorylation with a similar mobility shift profile of KaiC on SDS/PAGE gels.
Reduction in KaiA-Mediated KaiC Phosphorylation by kaiA2 Mutation.
To test whether KaiA-modulated KaiC phosphorylation is important for the timing mechanism in Synechococcus, we examined the effects of kaiA period mutations on the phosphorylation state of KaiC. We found that, in the kaiA2 mutant, the nonphosphorylated form of KaiC was highly accumulated throughout the circadian cycle (Fig. 4E). In contrast, the kaiA2;kaiC15 suppressor strain regained the wild-type circadian phosphorylation pattern, as did the kaiC15 single mutant strain (Fig. 4E). The kaiA2 mutation consistently reduced the enhancing effect of KaiA on in vitro KaiC autokinase activity to ≈70% of that of the wild-type protein (Fig. 3E). The abnormality in the in vivo KaiC phosphorylation state by the long period mutant thus strongly suggests a physiological relevance for the activation of KaiC phosphorylation by KaiA in determining the circadian period length in Synechococcus.
Discussion
The previously proposed simple negative feedback model for the Synechococcus clock (3) focused on KaiC function as a negative factor. In addition to this role, our genetic study (Fig. 1B) supported the idea that KaiA-mediated activation of kaiBC is KaiC dependent, and thus KaiC could be involved in the positive limb to form a positive feedback process. A certain state of KaiA–KaiC complex would activate the kaiBC promoter activity. Alternatively, KaiA may derepress kaiBC expression by affecting an inhibitory KaiC-containing complex. Moreover, it is also possible that KaiA is partly involved in KaiC-dependent repression of kaiBC expression because, in the absence of KaiA, overexpression of KaiC failed to repress kaiBC expression (Fig. 1C). In either case, the state of interaction between KaiA and KaiC determines both positive and negative feedback effects on kaiBC expression. The moderate reduction in the magnitude of kaiBC expression in the kaiC-null mutant is thus most likely due to the lack of the KaiC-dependent positive function. Note that many biological oscillations such as cell cycle control, glycolytic oscillations, and Ca2+ oscillations contain positive feedback (self-amplifying) processes (15), in addition to negative feedback regulations that are well described in circadian clock models (4). Coupling of the previously proposed negative feedback loop in the Synechococcus oscillator (3) with the positive feedback process revealed here would be important to make oscillation robust.
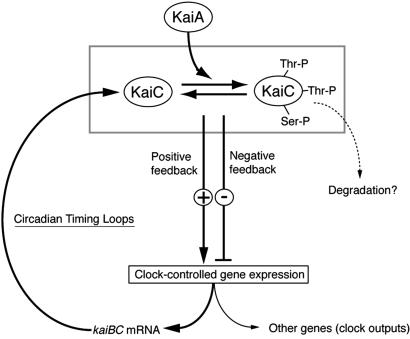
Our biochemical and genetic studies strongly suggest that KaiA-stimulated phosphorylation of KaiC is an important process in the coordination of the KaiA–KaiC functions. Phosphorylation may affect the stability of the KaiC protein (and/or KaiC-containing complexes) as proposed for some eukaryotic clock proteins (4), or it might affect the formation of different stoichiometric complexes of the Kai and/or SasA proteins (5, 6). Alternatively, circadian KaiC phosphorylation may modulate the state of chromosome condensation for global circadian gene expression (2). In any case, changes in the rate of KaiC phosphorylation by the KaiA protein could affect the rate of kaiBC repression, which is important for circadian rhythm generation (Fig. 5).
Fig 5.
A model for KaiA–KaiC functions in the Synechococcus clock. KaiA protein activates KaiC autokinase activity. The state of KaiC phosphorylation would regulate kaiBC expression. KaiC protein (complex) might affect its own promoter indirectly through controlling a clock output mechanism, such as regulation of the state of chromosome condensation (2), or by regulating SasA kinase activity (6). Phosphorylation also possibly affects the stoichiometry of clock protein complex(es) and/or the turnover of the KaiC protein (complex), which will affect the equilibrium state between nonphosphorylated and phosphorylated forms of KaiC. Because KaiC autophosphorylates at multiple residues, multiple phosphorylated forms of KaiC may be able to play different biochemical roles.
Moreover, our study revealed a similarity between in vivo and in vitro KaiC phosphorylation. Both types of phosphorylation (i) are stimulated by KaiA, (ii) occur at Ser and/or Thr residues, (iii) are accompanied by similar mobility shift profiles on SDS/PAGE gels, and (iv) are affected equivalently by the kaiA2 long period mutation. Thus, we suggest that in vivo KaiC phosphorylation is most likely due to KaiA-activated KaiC's autokinase activity, although we do not rule out the possibility that KaiC is additionally phosphorylated by other kinases, which are activated by KaiA.
Consistent changes by kaiA2 mutation, both in the KaiC phosphorylation profile and the period length, further support the physiological importance of KaiC phosphorylation for circadian timing. The kaiA2 mutation most likely reduces the rate of KaiC phosphorylation, and so it would retard the timing of KaiC degradation and/or that of activation (or derepression) of the kaiBC genes to lengthen the period. The kaiC15 mutation may compensate such effects to regain the wild-type period length. Consistently, kaiC15 has been mapped to one of two KaiA-binding domains (CKABD2) of KaiC (16) and kaiA2 to a KaiC-interacting, C-terminal domain of KaiA (amino acyl residues 180–286) that is sufficient for enhancing KaiC phosphorylation (17).
Acknowledgments
We thank Stanly B. Williams, Ioannis Vakonakis, Andy LiWang, and Susan S. Golden (Texas A&M University, College Station, TX) for sharing unpublished results and comments on the manuscript. We thank Jun Tomita, Hakuto Kageyama, and Yoriko Murayama (Nagoya University) for technical help, and our lab members for discussion. This research was partly supported by grants-in-aid from the Japanese Society for Promotion of Science (13680778 to H.I.), from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (11233203 to H.I. and T.K., and COE 13CE2005 to T.K.), and from the Kurata Memorial Hitachi Science and Technology Foundation (to H.I.), and by Inoue Research Award for Young Scientists from the Inoue Foundation (to H.I.). T.N. was supported by the Hayashi Memorial Foundation for female natural scientists, and Y.K. by the Japanese Society for Promotion of Science fellowship for young scientists (13001761).
Abbreviations
λPPase, λ phosphatase
LL, continuous illumination
CBB, Coomassie brilliant blue
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Iwasaki H. & Kondo, T. (2000) Plant Cell Physiol. 41 1013-1020. [DOI] [PubMed] [Google Scholar]
- 2.Mori T. & Johnson, C. H. (2001) Semin. Cell Dev. Biol. 12 271-278. [DOI] [PubMed] [Google Scholar]
- 3.Ishiura M., Kutsuna, S., Aoki, S., Iwasaki, H., Andersson, C. R., Johnson, C. H., Golden, S. S. & Kondo, T. (1998) Science 281 1519-1523. [DOI] [PubMed] [Google Scholar]
- 4.Young M. W. & Kay, S. A. (2001) Nat. Rev. Genet. 2 702-715. [DOI] [PubMed] [Google Scholar]
- 5.Iwasaki H., Taniguchi, Y., Ishiura, M. & Kondo, T. (1999) EMBO J. 18 1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwasaki H., Williams, S. B., Kitayama, Y., Ishiura, M., Golden, S. S. & Kondo, T. (2000) Cell 101 223-233. [DOI] [PubMed] [Google Scholar]
- 7.Nishiwaki T., Iwasaki, H., Ishiura, M. & Kondo, T. (2000) Proc. Natl. Acad. Sci. USA 97 495-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y., Mori, T. & Johnson, C. H. (2000) EMBO J. 19 3349-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishimura H., Nakahira, Y., Imai, K., Tsuruhara, A., Kondo, H., Hayashi, H., Hirai, M., Saito, H. & Kondo, T. (2002) Microbiology 148 2903-2909. [DOI] [PubMed] [Google Scholar]
- 10.Kondo T., Tsinoremas, N. F., Golden, S. S., Johnson, C. H., Kutsuna, S. & Ishiura, M. (1994) Science 266 1233-1236. [DOI] [PubMed] [Google Scholar]
- 11.Hardie D. G., (1993) Protein Phosphorylation: A Practical Approach (Oxford Univ. Press, Oxford).
- 12.Aronson B. A., Johnson, K. A., Loros, J. J. & Dunlap, J. C. (1994) Science 263 1578-1584. [DOI] [PubMed] [Google Scholar]
- 13.Cohen P. T. & Cohen, P. (1989) Biochem. J. 260 931-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhuo S., Clemens, J. C., Hakes, D. J., Barford, D. & Dixon, J. E. (1993) J. Biol. Chem. 268 17754-17761. [PubMed] [Google Scholar]
- 15.Goldbeter A., (1996) Biochemical Oscillations and Cellular Rhythms (Cambridge Univ. Press, Cambridge, U.K.).
- 16.Taniguchi Y., Yamahuchi, A., Hijikata, A., Iwasaki, H., Kamagata, K., Ishiura, M. & Kondo, T. (2001) FEBS Lett. 496 86-90. [DOI] [PubMed] [Google Scholar]
- 17.Williams S. B., Vakonakis, I., Golden, S. S. & LiWang, A. (2002) Proc. Natl. Acad. Sci. USA 99 15357-15362. [DOI] [PMC free article] [PubMed] [Google Scholar]