Abstract
Plants have a unique transdifferentiation mechanism by which differentiated cells can initiate a new program of differentiation. We used a comprehensive analysis of gene expression in an in vitro zinnia (Zinnia elegans L.) culture model system to gather fundamental information about the gene regulation underlying the transdifferentiation of plant cells. In this model, photosynthetic mesophyll cells isolated from zinnia leaves transdifferentiate into xylem cells in a morphogenic process characterized by features such as secondary-wall formation and programmed cell death. More than 8,000 zinnia cDNA clones were isolated from an equalized cDNA library prepared from cultured cells transdifferentiating into xylem cells. Microarray analysis using these cDNAs revealed several types of unique gene regulation patterns, including: the transient expression of a set of genes during cell isolation, presumably induced by wounding; a rapid reduction in the expression of photosynthetic genes and the rapid induction of protein synthesis-associated genes during the first stage; the preferential induction of auxin-related genes during the subsequent stage; and the transient induction of genes closely associated with particular morphogenetic events, including cell-wall formation and degradation and programmed cell death during the final stage. This analysis also revealed a number of previously uncharacterized genes encoding proteins that function in signal transduction, such as protein kinases and transcription factors that are expressed in a stage-specific manner. These findings provide new clues to the molecular mechanisms of both plant transdifferentiation and wood formation.
A plant body is composed of >40 types of cells with different shapes, sizes, cellular contents, and locations, each of which has an intrinsic developmental program. Moreover, plants have a unique transdifferentiation mechanism by which differentiated cells can initiate a new program of differentiation. Such cell differentiation programs in plants are not yet fully understood. The zinnia in vitro xylem-cell differentiation system is an excellent model with which to examine the whole transdifferentiation program. In this system, single isolated mesophyll cells transdifferentiate into xylem cells, tracheary elements (TEs), without cell division (reviewed in refs. 1–3). The system has provided various lines of data on transdifferentiation, foremost of which is the concept that the transdifferentiation process falls into three stages: stage 1 (first 24 h), corresponding to the functional dedifferentiation process during which mesophyll cells lose their photosynthetic capacity and acquire a new multidifferentiation potency; stage 2 (next 24 h), corresponding to the process of differentiation from procambial initials into the precursors of TEs; and stage 3 (final 24–48 h), corresponding to the process of morphogenesis that characterizes TE formation and includes secondary-wall formation and programmed cell death (PCD) (reviewed in refs. 1 and 4). Using this system, stage-dependent differentiation-regulating factors have been identified, such as the combination of auxin and cytokinin (5), brassinosteroids (6, 7), and xylogen (8). However, the molecular basis of each stage is not yet understood. Comprehensive gene transcript profiles have the potential to reveal the molecular basis of each stage, providing new insights into the transdifferentiation process and leading to the discovery of novel genes involved in differentiation.
Elucidation of the process of xylem differentiation is extremely important to our understanding of the genetic control of wood formation in trees. Indeed, the large-scale generation of ESTs from xylem tissues has progressed for such woody plants as the loblolly pine (9) and the poplars (10) and also in the model plant, Arabidopsis (11). Furthermore, the transcriptional profiling using microarray technology of nearly 3,000 poplar genes in the different developmental stages of wood-forming tissues has identified genes encoding lignin and cellulose biosynthetic enzymes and other potent regulators of xylogenesis (12). Milioni et al. (13) have also reported in their review article that expression analysis using PCR-amplified fragment-length polymorphisms during xylem-cell differentiation in zinnia culture has allowed the identification of many cell-wall-associated genes expressed during xylogenesis. Therefore, comprehensive transcript profiling throughout the transdifferentiation process of mesophyll cells into xylem cells in zinnia culture, which includes the formation of primary xylem, should provide fundamental information on the gene regulation underlying xylem formation and the commitment of cell fates. This information will enable us to compare the genes expressed during primary and secondary xylem formation, giving new insight into the common and different regulation mechanisms in primary and secondary xylem formation.
In this study, therefore, we performed comprehensive microarray analysis of gene expression during transdifferentiation in the zinnia system, using >8,000 zinnia cDNA clones isolated from the equalized cDNA library of differentiating xylem cells. Our study describes the unique gene expression patterns underlying the dramatic changes in cell fate during transdifferentiation and a number of developmental stage-specific genes. These include genes involved in auxin function, signal transduction, cell-wall formation and degradation, and PCD.
Materials and Methods
Seeds of zinnia (Zinnia elegans L. cv. Canary bird) were sown in vermiculite and grown for 2 weeks. Zinnia mesophyll cells were cultured as described in ref. 14. Differentiation-inducing medium (D-med) containing 0.1 mg/liter of naphthylacetic acid (NAA) and 0.2 mg/liter of benzyladenine and a control medium containing 0.1 mg/liter of NAA were used. Isolation of poly(A)+ RNA and total RNA, in situ hybridization, and RNA gel blot analysis were performed as described (15).
The equalized cDNA library was prepared as described (16), with some modification. cDNAs were prepared from poly(A)+ RNA isolated from cells cultured for 48 or 60 h in D-med, amplified by PCR, mixed, and equalized by three cycles of hydroxyapatite column chromatography by using 20 μm of MacroPrep Ceramic Hydroxyapatite Type II (Bio-Rad) at 30°C, followed by PCR amplification. Aliquots of equalized cDNAs were subcloned nondirectionally into pGEM-T Easy plasmid vector (Promega). Plasmid DNA was prepared by using 96-well MultiScreen NA and FB plates (Millipore), and partial sequences of the cDNA inserts were determined from one or both strands by using universal M13 forward and reverse primers and a CEQ2000 sequencer (Beckman Coulter). A semiautomatic program was developed for sequence analysis that included trimming (removal of vector, linker, and primer sequences); clipping from the 3′ end to obtain high quality sequences with <5% unidentifiable bases; the collection of ESTs; the assembly of individual ESTs into clusters; and comparisons of sequences with the GenBank nonredundant protein database by using blastx (S.S., T.D., and H.F., unpublished data). Sequences determined were deposited in the DNA Data Base in Japan (accession nos. –AU294769).
The marker genes for stage 1, ZePR, ZePI1, ZePI2, ZeRPP0, ZeRPS3a, and ZeRPS3, were isolated by differential screening of zinnia first true leaves and mesophyll cells cultured for 12 h in D-med (N.M., T.D., and H.F., unpublished data). The genes for autolysis [ZCP4, encoding a Cys peptidase similar to p48h-17 (17)] and lignification (ZePAL1, -2, and -3) were isolated by the screening of a zinnia cDNA library prepared from cells cultured for 48 h in D-med (5) by using PCR-amplified cDNA fragments of conserved domains as probes. Sequences determined were deposited in the DNA Data Base in Japan (accession nos. AB091070–AB091078).
To prepare the cDNA microarray, inserts of 9,120 cDNA clones and marker genes for each stage of transdifferentiation were PCR-amplified with an M13 universal primer set. The PCR products were purified by using 96-well MultiScreen PCR plates (Millipore). Spotting, labeling, hybridization, and scanning were performed as described (18). The fluorescence intensity for each spot was captured by using microarray suite (Scanalytics, Fairfax, VA), and the mean value for each spot was analyzed in genespring 4.02 (Silicon Genetics, Redwood City, CA). Because each cDNA was spotted in duplicate, the average value for the duplicated spots was used in genespring. The 50th percentiles of all of the averaged values obtained from each hybridization were used as the first synthetic positive controls for each cDNA, and each value for each cDNA was divided by these synthetic positive controls. Subsequently, the 50th percentiles of the normalized values for each cDNA were used as their own synthetic positive controls, and each normalized value for each cDNA was divided by its own synthetic positive control. After this double normalization, cDNAs with a >8-fold change in expression over the time-course of transdifferentiation (i.e., the maximum normalized value divided by the minimum normalized value was >8) were chosen. These were then filtered to eliminate those cDNAs with average values before normalization <50 in all hybridizations, or cDNAs with ratios between the mean values of the duplicated spots of <1/3 or >3. Finally, 523 cDNAs were selected for hierarchical clustering by using the “Clustering Gene Tree” function with the “Measure Similarity by Standard Correlation” in genespring.
Results and Discussion
Zinnia ESTs in the in Vitro Transdifferentiation of Mesophyll Cells into TEs.
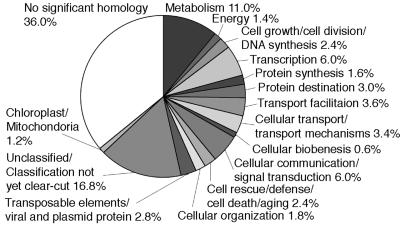
We equalized cDNAs prepared from cultured cells differentiating into TEs. The average length of the equalized cDNAs was estimated to be 0.8 kb. The cDNAs were subcloned nondirectionally into plasmid vector, and 9,120 clones were picked randomly for partial sequencing. The average read-length was 0.5 kb. The final numbers of ESTs sequenced using universal M13 forward or reverse primers were 8,182 and 1,533, respectively. The final number of cDNAs analyzed was 8,496, which formed 7,402 clusters. A total of 6,481 clusters consisted of singleton cDNAs, and the other clusters contained two (782 clusters) to eight (one cluster) cDNAs. To estimate the distribution of the putative functions of these cDNA clusters, we assigned putative functions to 500 randomly selected cDNA clusters on the bases of blastx searches (Fig. 1). About 64% of clusters were significantly similar [an expected value (E-value) <1.0 E-3] to at least one protein sequence previously described in the GenBank database. The compositions and ratios of the functional classifications of the functionally assigned cDNA clusters were similar to those for the Arabidopsis genome (19) and the tomato unigenes (20). In contrast, 36% of clusters showed no significant similarity to previously published sequences (E-value >1.0 E-3). Full-length sequences of the corresponding genes will be required to determine precisely whether they represent novel genes or nonconserved or noncoding regions of previously described genes.
Fig. 1.
Distribution of putative functions of zinnia cDNAs. Five hundred zinnia cDNA clusters were randomly picked, and their putative functions were assigned and categorized according to the Munich information Center for Protein Sequences (http://mips.gsf.de).
Of the 8,496 cDNAs analyzed in this study, only nine corresponded to previously reported zinnia cDNAs, although 42 cDNAs were registered in the GenBank database (Z115 and Z2791, TED4, D30802; Z3261, ZCAD1, D86590; Z3685, hb2, AJ312054; Z4247, TED2, D30800; Z5366, ZRNaseII, U19924; Z5563, nucZe1, U90265; Z7217, ZeHB9, AB042768; and Z2657, CesA-1, AF323039). These results suggest that most of the cDNAs represent previously undescribed zinnia genes that encode a great variety of proteins.
Zinnia cDNA Microarray.
A total of 9,113 PCR-amplified cDNA inserts were isolated from 9,120 cDNA clones. In addition to these cDNA inserts, we prepared several cDNA inserts as positive controls that had been cloned as marker genes for stage 1 (ZePR, ZePI1, ZePI2, ZeRPP0, ZeRPS3a, and ZeRPS3); stage 2 [TED2, -3, and -4 (refs. 5, 21); ZeTubB1, -2, and -3 (ref. 22)], and stage 3 [ZCP4; ZEN1 (ref. 23); ZRNaseI (ref. 24)], and for lignification (ZePAL1, -2, and -3). These PCR-amplified cDNAs were spotted twice onto each slide to increase the reliability of the microarray data. To monitor gene expression patterns during the in vitro transdifferentiation of mesophyll cells into TEs, a Cy5-labeled cDNA population was prepared from freshly isolated mesophyll cells and cells cultured for 24–96 h in D-med containing auxin and cytokinin. This population was hybridized to the microarray. To determine the changes in their expression levels during the preparation of mesophyll cells, we examined the expression levels in the first true leaves from which the mesophyll cells were isolated. After scanning, the mean values of the signal intensities for each spot were used as raw data (Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). The expression patterns of most of the marker genes revealed by our microarray analysis (Fig. 5, which is published as supporting information on the PNAS web site) were consistent with the patterns obtained by RNA gel blot analyses (5, 6, 21–24), confirming the accuracy of the microarray data presented in this paper.
Hierarchical Clustering of Developmentally Regulated Genes.
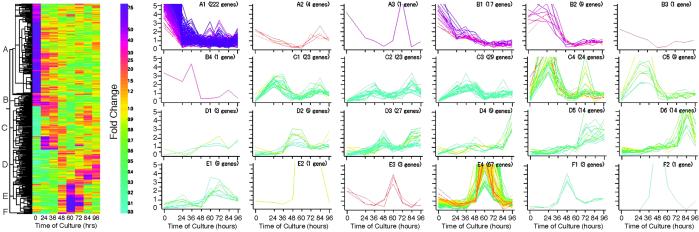
To visualize typical gene expression patterns during transdifferentiation, the 523 genes that showed an 8-fold change in expression over the transdifferentiation time-course were selected and clustered (Fig. 2). These genes were divided into six groups (A–F) with two to six subgroups (A1–F2) (Fig. 2, and Table 2, which is published as supporting information on the PNAS web site). Groups A and B mainly included genes that were down-regulated during stage 1, and group C mainly included genes that were up-regulated during stage 1. Groups D–F included genes that were up-regulated during stage 2 or 3 (Fig. 3A). Of these genes, those expressed specifically or highly preferentially during stage 1, 2, or 3 were clustered into subgroup C4, subgroups F1 and F2, or subgroups E2 and E4, respectively. Because it usually takes about 1 h to prepare isolated mesophyll cells, wound-induced events occur in association with the isolation process during cell preparation. Therefore, to identify the early gene expression profiles that are associated with cell isolation, we compared the expression levels of these clustered genes with the corresponding levels in the first true leaves and in freshly isolated mesophyll cells just before culture (Table 2, and Fig. 6, which is published as supporting information on the PNAS web site). The results show that the expression of 6% (32/523) of these cDNAs was down-regulated and the expression of 19% (97/523) was up-regulated (4-fold differential expression) during the preparation of mesophyll cells.
Fig. 2.
Hierarchical clustering of developmentally regulated genes. On the basis of the constructed tree (Left), the 523 cDNAs whose corresponding genes showed an 8-fold change in expression over the transdifferentiation time-course were clustered into 24 subgroups (subgroups A1–F2) (Right). The color scale indicates the degree of change (fold change) in expression. The expression pattern of each cDNA is colored according to the level of fold change in freshly isolated cells (0 h).
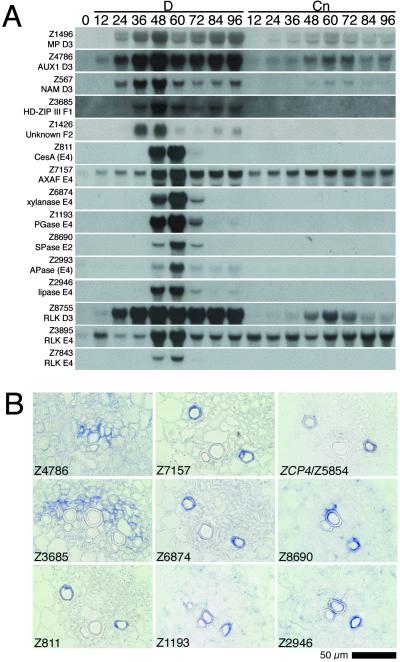
Fig. 3.
Expression patterns of several stage-2 or -3 specifically or preferentially expressed genes in vitro (A) and in vivo (B). (A) RNA gel blot analysis comparing freshly isolated cells (0) and cells cultured for 12–96 h in a D-med (D) or a control medium containing only auxin (Cn). (B) In situ hybridization in the vasculature around the shoot apical region of 14-day-old zinnia seedlings.
New Insights into the Transdifferentiation Process
(i) Wound-Induced Events During Stage 1.
We have previously suggested that stage 1 involves the expression of wound-induced genes (1). About 40% (88/222) of genes clustered in subgroup A1 showed rapidly up-regulated expression during cell preparation, followed by down-regulation during stage 1 (Table 2, Figs. 2 and 6). Because the phytohormones auxin and cytokinin were not administered to cells during cell preparation, the up-regulated expression of these genes cannot be attributed to their induction by phytohormones but was more likely induced by wound stress. These genes encode a variety of proteins including protein kinases (Z156, Z1923, Z3260, Z3908, Z4458, and Z8114) and transcription factors (Z1866, Z3739, and Z7431) (Table 2). Because wound stress is essential for transdifferentiation into xylem cells (1), genes induced by wound stress, especially those involved in signal transduction and transcriptional regulation, may play pivotal roles in switching cell fate during early stage 1.
(ii) Down-Regulation of Photosynthetic Activity During Stage 1.
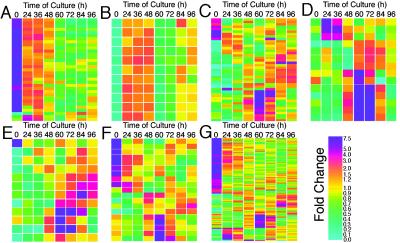
In the 523 cDNAs regulated developmentally, we found 33 cDNAs encoding proteins predicted to function in chloroplasts, such as plastid ribosomal proteins and chlorophyll a/b-binding proteins (Table 2, Fig. 4A). Of these, 29 cDNAs were clustered in subgroup A1, which includes genes down-regulated during stage 1 (Figs. 2 and 4A). This result provides direct evidence at the molecular level to confirm the previous assumption that isolated mesophyll cells lose their potential to function as photosynthetic cells during stage 1 (1). Moreover, the expression of almost all these phytosynthesis-associated genes was not down-regulated during the preparation of mesophyll cells (Table 2, Fig. 6), suggesting that the reduction of synthetic activity by the photosynthetic machinery is not a rapid response to wound stress.
Fig. 4.
Expression patterns of genes with particular functions. (A) Photosynthesis-associated proteins (33 genes). (B) Cytoplasmic ribosomal proteins (11 genes). (C) Proteins associated with cell-wall metabolism (43 genes). (D) Lignin biosynthetic proteins (14 genes). (E) Proteases, nucleases, and lipases (12 genes). (F) Protein kinases (19 genes). (G) Proteins with unknown function (126 genes). The color scale indicates the degree of change in expression (fold change).
(iii) Activation of Protein Synthesis from Stage 1 to Stage 2.
Eleven cDNAs coding cytoplasmic ribosomal proteins were clustered in subgroups C1–5, with up-regulated expression during stage 1 (Fig. 4B). Stage 1 marker genes encoding cytoplasmic ribosomal proteins also showed similar expression patterns (Fig. 5B). Expression of these genes was induced by incubating strips of the first leaves floating in a culture medium containing no phytohormone (Co-med) for 24 h, and only transient expression of these genes was induced by culturing isolated mesophyll cells in Co-med, suggesting that these genes are wound-induced (M.N., T.D., and H.F., unpublished data). From these data, together with the fact that protein synthesis is elevated from stage 1 to stage 2 (25), we can infer that the coordinated expression of genes encoding components of the translational machinery from stage 1 to stage 2 may be essential for the early transdifferentiation process.
(iv) Auxin Signal Transduction During Stage 2.
The expression pattern of subgroup D3 genes is similar to those of the stage 2 marker genes, TED2, TED3, and TED4 (Fig. 5E). Indeed, the cDNAs for TED2 (Z4247) and TED4 (Z2791) clustered in subgroup D3. Both auxin and cytokinin, which induce TE differentiation, are necessary for the expression of the stage 2 marker genes (5, 21), suggesting the presence of a hormonal signaling pathway involved in the progression of stage 2. In subgroup D3, we found genes encoding proteins similar to the Arabidopsis auxin response factor 5/monopteros (Z1496) (26); an auxin permease, AUX1 (Z4786) (27); and an Arabidopsis putative no apical meristem (NAM)-like protein belonging to the NAC plant-specific transcription factor family (Z567), several members of which have recently been found to mediate auxin signaling in the root (NAC1, ref. 28) and in the embryonic shoot apex (CUC1 and -2, ref. 29). These genes were expressed much more strongly in cells cultured in D-med containing both auxin and cytokinin than in cells cultured in control medium containing only auxin (Fig. 3A). This result suggests that the expression of these genes that is associated with auxin signal transduction is regulated by a xylem-differentiation-dependent program that proceeds in the presence of auxin and cytokinin. Immature-xylem-specific expression of the zinnia AUX1 homolog (Z4786) shown in Fig. 3B supports this idea.
Our microarray contained nine cDNAs with similarity to Arabidopsis auxin-response factors (ARFs) (Z174, Z214, Z1496, Z3670, Z6796, Z7090, Z7433, Z8416, and Z8828), of which only Z1496, the putative ARF5/MP (MP, monopteros) homolog, was expressed predominantly during stage 2. Five cDNAs with similarities to a class of auxin response genes, Aux/IAAs, were included in our microarray (Z824, Z1689, Z2172, Z3615, and Z5079), of which only one (Z2172), which is very similar to Arabidopsis IAA8 and IAA9, exhibited a stage 2-preferential expression pattern (Fig. 7A, which is published as supporting information on the PNAS web site). These results suggest that the zinnia Aux/IAA homolog (Z2172) and the zinnia monopteros homolog (Z1496) may function together in auxin signaling during stage 2.
(v) Transition from Stage 2 to Stage 3.
Four genes clustered in subgroups F1 (Z634, Z3685, and Z9472) and F2 (Z1426) showed transient expression at the transition between stage 2 and stage 3 (Fig. 2). RNA gel blot analysis of Z3685 and Z1426 indicated that their transient expression is specific for cells cultured under differentiation-inducing conditions (Fig. 3A). In situ hybridization of Z3685 also indicated that its expression is differentiating-xylem-cell specific (Fig. 3B). Z3685, identical to hb2 (GenBank accession no. AJ312054), is most similar to Athb-15, a homeodomain–leucine zipper class III protein gene (see Fig. 1 in ref. 30). We have recently identified three other distinct zinnia HD-ZIP III genes (ZeHB-10, -11, and -12) with expression patterns similar to that of Z3685 (30). Expression of these genes is regulated by brassinosteroids that trigger transdifferentiation from stage 2 to stage 3. Therefore, these genes clustered in subgroups F1 and F2 may be involved in the transition from stage 2 to stage 3.
(vi) Formation and Degradation of Cell Wall During Stage 3.
Fig. 4C presents the expression profiles of genes encoding proteins that may function in cell wall metabolism. Approximately one-third of these belong to subgroup E4 and are expressed specifically in early stage 3. Because the specialized thickening of the secondary cell walls of TEs occurs early in stage 3 (after ≈60 h of culture), the cell-wall-related genes clustered in subgroup E4 may act in secondary-wall formation. Z2657, which encodes part of the zinnia CesA-1 gene (31), is most similar to the cotton cellulose synthase (CesA) gene, GhCesA-1, which is highly expressed at the onset of secondary-wall synthesis (32). Seven other members with similarities to the CesA genes were included in our microarray (Z632, Z749, Z811, Z1737, Z2480, Z4847, and Z6118). Of these, Z811 is most similar to the Arabidopsis AtCesA-7/irregular xylem 3 gene, mutations of which cause severe deficiency in secondary-cell-wall cellulose deposition that leads to collapsed vessels (33). Z811 displays an early stage 3-specific expression pattern similar to that of the subgroup E4 genes (Figs. 3A and 7B) and also differentiating-TE-specific expression in vivo (Fig. 3B). However, the other six transcripts are expressed more or less constitutively, suggesting a role in primary-wall formation. One cDNA (Z3164) has a high degree homology to the Arabidopsis gene for the membrane-anchored endo-1,4-β-glucanase (EGase), korrigan, previously suggested to be a part of the cellulose synthesizing complex (34). The expression of Z3164 was also up-regulated transiently during early stage 3 (Fig. 7C). These results suggest the involvement of specific CesA and EGase genes in cellulose synthesis in the secondary cell walls of TEs in zinnia.
A rapid increase in degradative activity of the primary cell wall takes place during early stage 3 (35). Several cDNAs showing significant similarity to O-glycosyl hydrolases were expressed transiently during early stage 3: three cDNAs for β-xylosidase (Z6245, Z7690, and Z9107); one for arabinoxylan arabinofuranohydrolase (AXAF) (Z7157); one for β-galactosidase (Z7361); one for β-glucosidase (Z9060); one for β-xylanase (Z6874); and two for α-mannosidase (Z2422 and Z9354) (subgroup E4 in Fig. 2; Fig. 7 D and E). Furthermore, the expression of three cDNAs (Z1193, Z3465, and Z5557) that are similar to the polygalacturonase (PG) gene was up-regulated in early stage 3. RNA gel blot and in situ hybridization analyses for the transcripts corresponding to AXAF (Z7157), β-xylanase (Z6874), and PG (Z1193) revealed that the expression of these O-glycosyl hydrolases is induced specifically, or at least predominantly, in differentiating TEs both in vitro and in vivo (Fig. 3), suggesting their coordinated expression and specific role in primary-cell-wall degradation in TEs.
(vii) Lignification of Secondary Cell Wall.
Fig. 4D presents the expression profiles of genes involved in lignin biosynthesis. Six cDNAs encoding putative laccases (Z116, Z741, Z832, Z3811, Z4959, and Z8149) and one cDNA encoding a putative peroxidase (Z2977), all of which may function in the polymerization of lignin monomers, displayed stage 3-specific expression (Fig. 2, subgroup E4). In contrast, genes for the synthesis of lignin monomers, such as cinnamyl alcohol dehydrogenase [Z3261 identical to ZCAD1 (ref. 36)], phenylalanine–ammonia lyase (Z7345 and Z7975), and 4-coumarate/CoA ligase (Z9331), did not cluster in subgroup E4, which contains the putative laccases and peroxidases, but did cluster in subgroup E1. Expression of the genes in this subgroup continues into late stage 3, when autolysis of the TEs is completed. Hosokawa et al. (37) recently found that nondifferentiated zinnia cells provide lignin monomers to the lignifying secondary walls of TEs through the culture medium. Taken together, our microarray data strongly suggest the specific expression of genes encoding lignin-monomer-polymerizing enzymes in differentiating TEs and of genes encoding lignin-monomer-synthesizing enzymes in both differentiating TEs and nondifferentiating cells, probably parenchyma-like cells of the xylem.
(viii) Autolytic Process of PCD During Stage 3.
Stage 3 involves the processes of PCD during which cell components are autolyzed (reviewed in ref. 38). In the 523 cDNAs regulated developmentally, we found 12 cDNAs encoding proteases, nucleases, or lipases (Fig. 4E). Of these, two cDNAs for Cys peptidase (CPase) (Z5854 identical to ZCP4, and Z8955) and one for Ser peptidase (SPase) (Z8690) were expressed specifically during early stage 3 in vitro, and in immature TEs in vivo (subgroups E2 and E4 in Fig. 2, and Fig. 3). Our microarray contained another cDNA for CPase and eight other cDNAs for SPase, of which only Z1485 and Z4223 (both encoding SPase) were also expressed predominantly during early stage 3 (Fig. 7 F and G). Of these stage 3-specific peptidases, ZCP4/Z5854 share a high degree of homology with the Arabidopsis xylem-specific CPase, XCP1, and Z8690, Z1485, and Z4223 share a high degree of homology with the Arapidopsis xylem-specific SPase, XSP1 (39). Furthermore, three cDNAs encoding aspartic peptidase were included in our microarray, of which only Z2993 displayed the stage 3-specific expression (Figs. 3A and 7H). One cDNA (Z2946) encoding a putative lipase (lipolytic acyl hydrolase) was also expressed specifically during early stage 3 and in differentiating TEs in vivo (Fig. 3). Thus, we have identified many proteins that may be involved in autolysis, suggesting a sophisticated autolytic process involving PCD-specific hydrolytic enzymes with different tasks.
(ix) Identification of Stage-Specific Protein Kinase Genes.
Genes encoding putative protein kinases, including leucine-rich receptor protein kinases, serine/threonine kinases, and mitogen-activated protein kinase (MAP-K) kinase kinase showed unique expression patterns (Fig. 4F). Of these, several genes share a high degree of similarity to the putative Arabidopsis receptor-like protein kinases. One of these is predominantly expressed in stage 2 (Z8755), and two are predominantly expressed in stage 3 (Z3895, Z7843), when cultured in D-med (Fig. 3A). This is to our knowledge the first report to suggest the involvement of cell–cell interactions in the late stage of xylem differentiation at the molecular level.
(x) Genes for Unknown Proteins.
Of the 523 cDNAs clustered in Fig. 2, 126 display similarity only to genes for proteins with no known function, designated putative proteins, hypothetical proteins, or unknown proteins. Moreover, 85 cDNAs show no significant similarity to any known sequence. Therefore, the functions of the proteins encoded by these genes are completely unknown. However, all showed very specific expression profiles (Fig. 4G). For example, many genes displayed a peak of expression after 60 h of culture, which suggests that they are involved in TE morphogenesis, as discussed above. Fine analysis of the functions of these proteins in relation to TE morphogenesis will cast new light on xylem formation mechanisms.
Conclusion
In this study, we constructed a comprehensive profile of gene expression during transdifferentiation from mesophyll cells into xylem cells, using an in vitro zinnia culture system. Analysis of >8,000 zinnia cDNA clones revealed unique differentiation-stage-dependent patterns in gene regulation, which will enable us to pursue the master gene that controls each stage. This analysis also identified a number of previously undescribed genes that may function in the transdifferentiation process, as judged from their expression patterns. Some of these genes suggest the presence of new steps involved in differentiation, such as an auxin signaling pathway and cell–cell interactions. The discovery of >70 stage 3-specific genes, many of which encode biosynthetic or degradative enzymes, implies the involvement of many previously unrecognized factors in the morphogenesis of TEs.
Microarray analysis of poplar wood-forming tissues has shown that more than 500 genes are differentially expressed during secondary xylem development (12). Comparison of the data sets of zinnia and poplar has identified many specific genes with similar sequences and similar expression profiles in both genera (Table 2, “Poplar” column, and Fig. 8, which is published as supporting information on the PNAS web site). For instance, several zinnia sequences clustered in subgroup E4 and poplar sequences that are strongly up-regulated in a zone of secondary-wall formation (zone D) are most similar to the same Arabidopsis proteins, such as putative phytochelatin synthetase (Z466 and AI16060), a hypothetical protein (Z739 and AI161572), and an arabinogalactan protein (Z2943, AI165289, and AI166182). This fact implies that the microarray data collected here on zinnia differentiation in vitro can be used to understand the gene regulation involved in wood formation, although such analyses are often very difficult to undertake directly with trees. Recently, an Arabidopsis cDNA library was constructed from isolated xylem (39), and the EST database generated from it represents a partial gene expression profile for the xylem of Arabidopsis (11). Efforts are now underway to determine the specific gene expression profiles induced by various stage-dependent differentiation-inducing factors such as cytokinin (14), brassinosteroids (6, 7), and xylogen (8). Comparison of this data set with the ESTs and gene expression profiles of the xylem tissues of Arabidopsis and woody plants will greatly contribute to an elucidation of the common gene regulation mechanism underlying xylem differentiation.
Supplementary Material
Acknowledgments
We thank Ayumi Ihara and Tatsuya Horiguchi for technical assistance and Jun Ito (Graduate School of Science, University of Tokyo) for providing cDNA clones that encode the full length of ZEN1 and ZRNaseI. This work was supported in part by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Rice Genome Project MA-2203) to H.F., and in part by Grants-in-Aid from the Ministry of Education, Science, Sports, and Culture of Japan (14036205 and 10219201 to H.F.) and the Japan Society for Promotion of Science (12740433 to T.D. and 13440236 to H.F.).
Abbreviations
TE, tracheary element
D-med, differentiation-inducing medium
PCD, programmed cell death
References
- 1.Fukuda H. (1997) Plant Cell 9 1147-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCann M. C. (1997) Trends Plant Sci. 2 333-338. [Google Scholar]
- 3.Ye Z.-H. (2002) Annu. Rev. Plant Biol. 53 183-202. [DOI] [PubMed] [Google Scholar]
- 4.Kuriyama H. & Fukuda, H. (2001) J. Plant Growth Regul. 20 35-51. [Google Scholar]
- 5.Demura T. & Fukuda, H. (1993) Plant Physiol. 103 815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamoto R., Demura, T. & Fukuda, H. (1997) Plant Cell Physiol. 38 980-983. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto R., Fujioka, S., Demura, T., Takatsuto, S., Yoshida, S. & Fukuda, H. (2001) Plant Physiol. 125 556-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motose H., Sugiyama, M. & Fukuda, H. (2001) Plant Cell Physiol. 42 129-137. [DOI] [PubMed] [Google Scholar]
- 9.Allona I., Quinn, M., Shoop, E., Swope, K., St. Cyr, S., Carlis, J., Riedl, J., Retzel, E., Campbell, M. M., Sederoff, R. & Whetten, R. W. (1998) Proc. Natl. Acad. Sci. USA 95 9693-9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sterky F., Regan, S., Karlsson, J., Hertzberg, M., Rohde, A., Holmberg, A., Amini, B., Bhalerao, R., Larsson, M., Villarroel, R., et al. (1998) Proc. Natl. Acad. Sci. USA 95 13330-13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beers E. P. & Zhao, C. (2001) in Molecular Breeding of Woody Plants, eds. Morohoshi, N. & Komamine, A. (Elsevier, Amsterdam), pp. 43–52.
- 12.Hertzberg M., Aspeborg, H., Schrader, J., Andersson, A., Erlandsson, R., Blomqvist, K., Bhalerao, R., Uhlén, M., Teeri, T. T., Lundeberg, J., et al. (2001) Proc. Natl. Acad. Sci. USA 98 14732-14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milioni D., Sado, P.-E., Stacey, N. J., Domingo, C., Roberts, K. & McCann, M. C. (2001) Plant Mol. Biol. 47 221-238. [PubMed] [Google Scholar]
- 14.Fukuda H. & Komamine, A. (1980) Plant Physiol. 65 57-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishitani C., Demura, T. & Fukuda, H. (2001) Plant Cell Physiol. 42 1210-1218. [DOI] [PubMed] [Google Scholar]
- 16.Kohchi T., Fujishige, K. & Ohyama, K. (1995) Plant J. 8 771-776. [DOI] [PubMed] [Google Scholar]
- 17.Ye Z.-H. & Varner, J. E. (1996) Plant Mol. Biol. 30 1233-1246. [DOI] [PubMed] [Google Scholar]
- 18.Endo M., Matsubara, H., Kokubun, T., Masuko, H., Takahata, Y., Tsuchiya, T., Fukuda, H., Demura, T. & Watanabe, M. (2002) FEBS Lett. 514 229-237. [DOI] [PubMed] [Google Scholar]
- 19.The Arabidopsis Genome Initiative (2000) Nature 408 796-815. [DOI] [PubMed] [Google Scholar]
- 20.Van der Hoeven R., Ronning, C., Giovannoni, J., Martin, G. & Tanksley, S. (2002) Plant Cell 14 1441-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demura T. & Fukuda, H. (1994) Plant Cell 6 967-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshimura T., Demura, T. & Fukuda, H. (1996) Plant Cell Physiol. 37 1167-1176. [DOI] [PubMed] [Google Scholar]
- 23.Aoyagi S., Sugiyama, M. & Fukuda, H. (1998) FEBS Lett. 429 134-138. [DOI] [PubMed] [Google Scholar]
- 24.Ye Z.-H. & Droste, D. L. (1996) Plant Mol. Biol. 30 697-709. [DOI] [PubMed] [Google Scholar]
- 25.Fukuda H. & Komamine, A. (1983) Plant Cell Physiol. 24 603-614. [Google Scholar]
- 26.Hardtke C. S. & Berleth, T. (1998) EMBO J. 17 1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchant A., Bhalerao, R., Casimiro, I., Eklöf, J., Casero, P. J., Bennett, M. & Sandberg, G. (2002) Plant Cell 14 589-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie Q., Frugis, G., Colgan, D. & Chua, N.-H. (2000) Gene Dev. 14 3024-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aida M., Vernoux, T., Furutani, M., Traas, J. & Tasaka, M. (2002) Development (Cambridge, U.K.) 129 3965-3974. [DOI] [PubMed] [Google Scholar]
- 30.Ohashi-Ito K., Demura, T. & Fukuda, H. (2002) Plant Cell Physiol. 43 1146-1153. [DOI] [PubMed] [Google Scholar]
- 31.Haigler C. H., Ivanova-Datcheva, M., Hogan, P. S., Salnikov, V. V., Hwang, S., Martin, K. & Delmer, D. P. (2001) Plant Mol. Biol. 47 29-51. [PubMed] [Google Scholar]
- 32.Pear J. R., Kawagoe, Y., Schreckengost, W. E., Delmer, D. P. & Stalker, D. M. (1996) Proc. Natl. Acad. Sci. USA 93 12637-12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor N. G., Scheible, W.-R., Cutler, S., Somerville, C. R. & Turner, S. R. (1999) Plant Cell 11 769-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicol F., His, I., Jauneau, A., Vernhettes, S., Canut, H. & Höfte, H. (1998) EMBO J. 17 5563-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohdaira Y., Kakegawa, K., Amino, S., Sugiyama, M. & Fukuda, H. (2002) Planta 215 177-184. [DOI] [PubMed] [Google Scholar]
- 36.Sato Y., Watanabe, T., Komamine, A., Hibino, T., Shibata, D., Sugiyama, M. & Fukuda, H. (1997) Plant Physiol. 113 425-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hosokawa M., Suzuki, S., Umezawa, T. & Sato, Y. (2001) Plant Cell Physiol. 42 959-968. [DOI] [PubMed] [Google Scholar]
- 38.Fukuda H. (2000) Plant Mol. Biol. 44 245-253. [DOI] [PubMed] [Google Scholar]
- 39.Zhao C., Johnson, B. J., Kositsup, B. & Beers, E. P. (2000) Plant Physiol. 123 1185-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.