Abstract
Cytokinin-Independent 1 (CKI1) belongs to a group of putative plant histidine kinases whose members do not appear to act as ethylene receptors. The deduced protein structure, combined with the observation that Arabidopsis callus cultures overexpressing CKI1 exhibit a “cytokinin-independent” cell division and greening phenotype, led to the hypothesis that CKI1 is involved in cytokinin signaling, perhaps acting as a cytokinin receptor. To test the function of CKI1, we used a reverse-genetic approach to identify plants carrying T-DNA insertions in CKI1. Two independent alleles were identified, which produce the same developmental phenotype. Analyses of populations segregating for the cki1–5 or cki1–6 T-DNA insertion alleles failed to reveal any homozygous cki1 plants, indicating that the homozygous mutant condition was lethal. Based on segregation distortion, transmission studies, a microscopy-based examination of developing female gametophytes, and mRNA expression data, we suggest that CKI1 function is required for megagametophyte development. Our work with CKI1 mutants indicates that signal transduction by means of a His/Asp phosphorelay system may play an important and previously unsuspected role in female gametophyte development in Arabidopsis.
The histidine to aspartate (His/Asp) phosphorelay is a well characterized prokaryotic signal-transduction pathway. Bacteria use this system to respond to a wide range of changes in their environment, including fluctuations in osmolarity, nutrient availability, and oxygen levels (1). Homologues of the three key proteins in a His/Asp phosphorelay, namely the histidine kinases, histidine-containing phosphotransmitters (HPts), and response regulators, have all been identified in Arabidopsis (for review see ref. 2) Aside from higher plants, the only eukaryotes in which these proteins have been found are yeast (3), Dictyostelium (4), and Neurospora (5).
The sequencing of the Arabidopsis genome has revealed 11 genes encoding histidine kinase-like proteins. Five of these proteins have been characterized as ethylene receptors (ETR1, ETR2, ERS1, ERS2, and EIN4) (6), and three are cytokinin receptors (CRE1 and its homologues AHK2 and AHK3) (7, 8). Indirect evidence implicates the remaining three histidine kinases in osmosensing (ATHK1) (9), as well as in cytokinin signaling (CKI1 and CKI2) (10).
Although plant responses to cytokinins have been studied since the discovery of kinetin in 1956 (11), the first and long-sought-after cytokinin receptor (CRE1) was identified only little more than 1 year ago (7). This major breakthrough in the field of cytokinin signaling occurred amid a renewed interest in identifying proteins that may participate in the cytokinin-signaling pathway (8, 12–15). It has now been established that an His/Asp phosphorelay plays a role in cytokinin signaling (for model see ref. 16), although the particular histidine kinases, response regulators, and HPts that mediate specific cytokinin responses have not yet been identified. The growing body of research linking Arabidopsis His/Asp phosphorelay signaling proteins to cytokinin signal transduction was initiated by Kakimoto's discovery of CKI1 (Cytokinin-Independent 1) (10).
The first nonethylene receptor histidine kinase described was CKI1. Overexpression of CKI1 was found to confer cytokinin-inducible responses on Arabidopsis callus tissue in the absence of applied cytokinin (10). Thus, CKI1 was initially implicated in hormone signaling. To explore the function of CKI1, we used a reverse-genetic approach to identify plants containing null alleles of CKI1. A genome-wide method to obtain null alleles by means of T-DNA insertions is well established in Arabidopsis thaliana (17). Here, we describe two independent T-DNA insertion alleles of CKI1 that have the same effect on plant development. Based on segregation distortion, transmission studies, a microscopy-based analysis of mutant ovules, and mRNA expression data, we have identified a role for CKI1 in megagametogenesis in A. thaliana.
Materials and Methods
Plant Materials and Growth Conditions.
Seeds of A. thaliana, ecotypes Wassilewskija (Ws) and Landsberg erecta (Ler), were germinated on plates containing half-strength Murashige and Skoog salts (18), 1% (wt/vol) sucrose, and 0.8% (wt/vol) washed agar (MS plates). Seedlings were transferred to soil after ≈8 days. The plants were then subjected to the following growth conditions: either 21°C under constant light, or 22°C with a light regime of 16 h light/8 h dark. Both T-DNA insertion lines studied were backcrossed at least twice before genetic transmission studies, CAPS (cleaved amplified polymorphic sequences) analysis, and microscopy. For transmission studies, reciprocal crosses were performed between mutant and wild-type plants. Seeds were harvested from individual siliques and germinated on MS plates containing 50 μg/ml kanamycin. The T-DNA construct used (pD991) carries a selectable marker gene that renders transgenic plants resistant to kanamycin (KanR).
Screening for T-DNA Insertion Lines.
T-DNA insertion alleles were obtained by screening the α population of 60,480 independent T-DNA insertion lines, provided by the Arabidopsis Knockout Facility at the University of Wisconsin (www.biotech.wisc.edu/Arabidopsis/). Details of the screening procedure are described in ref. 17. The CKI1 gene-specific primers used were CRFF (5′-CGCAGCCAAACTATTATTTTACCACAGAC-3′) and CR1M1 (5′-ATCGAGCCATTGGAGATGAAGAAAGAATC-3′). The T-DNA-specific primers used were left border (5′-CATTTTATAATAACGCTGCGGACATCTAC-3′) and right border (5′-TGGGAAAACCTGGCGTTACCCAACTTAAT-3′). The location of the T-DNA insert relative to the CKI1 genomic sequence was determined by sequencing PCR products containing the T-DNA/plant genomic DNA junction. The genotype of individual plants grown on MS plates was determined by two PCRs. One PCR contained both gene-specific primers, identifying the presence of a wild-type allele. The other PCR contained the appropriate combination of a gene-specific and a T-DNA-specific primer, identifying the presence of a mutant allele.
CAPS Analysis.
Two sets of primers were selected from the CAPS marker table on the Arabidopsis Information Resource (TAIR) web site (www.arabidopsis.org/aboutcaps.html). Primers were selected based on their ability to yield a polymorphism between the ecotypes Ws and Ler after PCR and digestion of the amplified product with the appropriate restriction enzyme. The primers chosen for CAPS analysis were 5′-GGGATTTGATGAAGGAGAAC-3′ and 5′-ATTCCTTGGTCTCCATCATC-3′, corresponding to the marker GPA1a on chromosome 2, as well as 5′-ACTCCTTTGTCATCTCCCGAATC-3′ and 5′-CCAACAACATGCATGATAGTTCAG-3′, corresponding to the marker 17D8LE on chromosome 3. Genomic DNA was prepared from the leaf tissue of KanR progeny resulting from crosses between wild-type (CKI1/CKI1) Ler and heterozygous (cki1/CKI1) Ws plants. DNA isolation, PCR, and restriction digests were carried out as described (19, 20). The ecotype (Ws/Ws, Ler/Ler, or Ws/Ler) of KanR plants was determined by the size of digested PCR products visualized on 1% (wt/vol) agarose gels.
Confocal Laser Scanning Microscopy (CLSM).
Tissue preparation, microscopy, image capture, and figure preparation were performed as described (21, 22).
In Situ Expression Analysis.
Nonradioactive in situ hybridization experiments were carried out as described (23) with the following modification: tissue was fixed and embedded as described (24). CKI1 antisense probes were prepared to exons 1–4 and exon 6 of CKI1. A FILAMENTOUS FLOWER [member of the YABBY gene family (25)] probe was used as a positive control.
Molecular Complementation.
After amplification of Ws genomic DNA with PCR primers 5′-AATAATTGGGAAAACATGTGATAAAAGTCTGA-3′ and 5′-GGCGCGCCCACTGGTTTCATTTGCCTACAT-3′, and subsequent restriction digests with PacI and AscI (NEB, Beverly, MA), an ≈6.5-kb genomic fragment containing the CKI1 gene, 1,300 bases upstream of the start codon, and 185 bases downstream of the stop codon, was ligated into pCAMBIA3300S, a spectinomycin-resistant derivative of pCAMBIA3300 (26). Plasmids containing the CKI1 gene were introduced by electroporation into Agrobacterium tumefaciens, and used to transform cki1/CKI1Arabidopsis plants by means of the floral dip method (27). Transformed plants were selected on MS plates containing 25 μg/ml ammonium glufosinate (Sigma–Aldrich), the active ingredient of the herbicide BASTA. The T2 generation of transformed plants (the plant that was dipped was designated generation T0) was analyzed for complementation of the cki1 phenotype by kanamycin segregation as described. Complementation was confirmed by three PCRs. The first PCR contained the gene-specific primer 5′-AATAGGCTTTCGACCGGTACGCACTGACT-3′ and the T-DNA-specific primer 5′-TTTCTCCATATTGACCATCATACTCATTG-3′, identifying the presence of the disrupted cki1 allele. The second PCR contained the gene-specific primer 5′-CCTATGGAGATGCGTAAGTCGGTATTTGA-3′ and T-DNA-specific primer 5′-GTCATGCCAGTTCCCGTG-3′, identifying the presence of the transgene. The third PCR contained the gene-specific primers 5′-GAACGGGTCAGAACATTAAAACATACATT-3′ and 5′-TCTTCCCGCTTTCGATTTTGCTCAT-3′, identifying the presence of the endogenous wild-type CKI1 allele. A complemented plant was defined as a plant lacking the endogenous wild-type allele of CKI1, yet viable because of the presence of the transgene.
Results
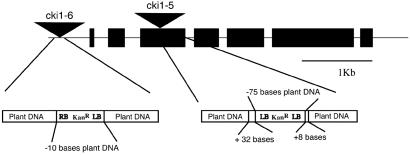
We identified Arabidopsis plants containing two independent T-DNA insertion alleles of CKI1. cki1-5 contains an insert within the third exon, and cki1-6 contains an insert 587 bases upstream of the translation start site (Fig. 1). For each allele, both T-DNA/plant genomic DNA junctions were sequenced to rule out the possibility of chromosomal translocations or large deletions (28). Analysis of the progeny resulting from self-fertilization of plants heterozygous for the cki1-5 or cki1-6 alleles did not yield a plant homozygous for the insertion in either allele. This observation indicates that the homozygous condition is lethal for some aspect of plant growth and development. Furthermore, the percent transmission of the mutant allele following the self-fertilization of plants heterozygous for the cki1-5 or cki1-6 alleles was ≈50%, which is consistent with a gametophytic defect (ref. 29; Table 1). Results were the same whether the plant genotypes were determined by using PCR or by kanamycin selection (data not shown).
Fig 1.
Description of the CKI1 T-DNA insertion alleles. Black boxes represent exons and triangles represent T-DNA integration sites. The T-DNA insertion allele cki1-6 contains an insert 587 bases upstream of the ATG, as well as a deletion of 10 bases at the integration site. The T-DNA insertion allele cki1-5 contains an insert 257 bases into the third exon, as well as a deletion of 75 bases and an addition of 40 bases of unknown origin at the integration site. LB and RB, left and right borders of the T-DNA, respectively; KanR, kanamycin resistance gene (neomycin phosphotransferase).
Table 1.
Genetic analysis of insertion alleles cki1-5 and cki1-6
| Cross performed | Resultant progeny | |||
|---|---|---|---|---|
| Female parent | Male parent | cki1/CKI1:CKI1/CKI1 | KanR:KanS | T-DNA transmission, % |
| cki1-5/CKI1 | cki1-5/CKI1 | 162:178 | ND | 47.6 |
| cki1-6/CKI1 | cki1-6/CKI1 | 145:144 | ND | 50.2 |
| cki1-5/CKI1 | CKI1/CKI1 | ND | 27:1089 | 2.4 |
| cki1-5/CKI1 | CKI1/CKI1 | ND | 5:160 | 0.0 |
| cki1-6/CKI1 | CKI1/CKI1 | ND | 21:210 | 9.1 |
| cki1-6/CKI1 | CKI1/CKI1 | ND | 12:173 | 0.0 |
| CKI1/CKI1 | cki1-5/CKI1 | ND | 589:549 | 51.3 |
| CKI1/CKI1 | cki1-6/CKI1 | ND | 143:160 | 47.2 |
ND, not determined.
Ecotype Wassilewskija.
Percent T-DNA transmission was calculated as 100 × KanR/(KanR + KanS).
Ecotype Landsberg erecta.
Percent T-DNA transmission was determined by a CAPS analysis.
To determine whether T-DNA transmission through the male and/or female gametes was reduced, reciprocal crosses were performed between cki1-5/CKI1 or cki1-6/CKI1 plants and wild-type plants. Our results show that mutant and wild-type alleles are transmitted equally well although the pollen (Table 1), but the transmission of mutant alleles through the female gametophyte is severely reduced relative to the wild-type allele (Table 1).
To determine whether transmission through the female gametophyte is completely eliminated, wild-type plants of the ecotype Ler were used as pollen donors in crosses with cki1-5/CKI1 or cki1-6/CKI1 plants (ecotype Ws), and a CAPS analysis was performed on all KanR progeny. This procedure allowed us to distinguish between KanR progeny that resulted from a successful cross, and those that were the result of self-fertilization because of imperfect emasculation. Our results indicated that cki1 T-DNA insertion alleles were never transmitted through the female gametophyte (Table 1), yielding 100% penetrance for this aspect of the mutant phenotype.
We noticed reduced seed set, in the form of empty spaces, within siliques of self-fertilized flowers from cki1-5/CKI1 and cki1-6/CKI1 plants (data not shown). Combined with the segregation distortion and transmission studies, this semisterile phenotype supported the idea that disrupting CKI1 results in a defect in female gametophyte development (29). To determine the nature of the defect in a cki1 female gametophyte, we analyzed mutant female gametophytes by using confocal laser scanning microscopy (CLSM). In this procedure, nucleoli appear white, cytoplasm appears gray, and vacuoles appear black (Fig. 2B) (21).
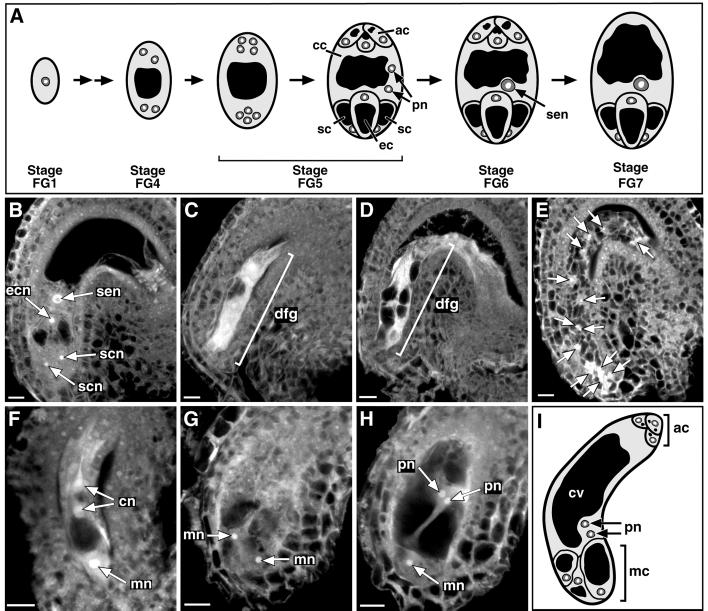
Fig 2.
Phenotypic analysis of cki1-5 female gametophytes. (A) Depiction of wild-type female gametophyte development. Megagametogenesis has been described and divided into seven stages (21). Before megagametogenesis, a diploid megaspore mother cell undergoes meiosis and produces four haploid megaspores, three of which degenerate. The surviving megaspore defines stage FG1. The megaspore then undergoes three rounds of mitosis without cytokinesis, giving rise to an eight-nucleate cell (early stage FG5). Immediately following the third mitosis, cell walls form and partition the nuclei into cellular compartments (late stage FG5). The central cell inherits two nuclei, called the polar nuclei, which fuse to form a homodiploid nucleus (stage FG6). Finally, the antipodal cells degenerate. The mature female gametophyte (stage FG7) consists of one central cell, one egg cell, and two synergid cells (22). (B–E) CLSM images of female gametophytes at the terminal developmental stage (stage FG7). (B) A wild-type Arabidopsis female gametophyte. This image is a projection of two 1.0-μm slices. (C) A degenerated cki1-5 female gametophyte. This image is a projection of five 1.5-μm optical sections. (D) A partially degenerated cki1-5 female gametophyte. This image is a single 1.0-μm optical section. (E) A cki1-5 female gametophyte that contains an excessive number of nuclei (indicated by arrows). This image is a single 1.5-μm optical section. (F) CLSM image of a cki1-5 female gametophyte from an ovary at stage FG4 (i.e., the ovary contains wild-type female gametophytes at stage FG4). This image is a projection of two 1.5-μm optical sections and shows two degenerating chalazal nuclei and a single intact micropylar nucleus. A second intact micropylar nucleus was present but not projected in this image. (G and H) CLSM images from the same cki1-5 female gametophyte from an ovary at stage FG5/6 (i.e., the ovary contains wild-type female gametophytes at stage FG5/6). Because of the abnormal cell morphologies, distinct cellular identities could not be assigned for the cells at the micropylar end. The nuclei are thus labeled as micropylar nuclei (mn). The presumed polar nuclei are unfused. Three presumed antipodal cells were observed at the chalazal end but are not projected in these images. Both images are separate projections of two 1.0-μm optical sections each. (I) Depiction of a cki1-5 female gametophyte at the FG5/6 stage illustrating misshapen vacuoles and the irregular placement of micropylar nuclei. All female gametophytes are oriented with their chalazal poles up and their micropylar poles down. ac, antipodal cells; cc, central cell; cn, chalazal nucleus; cv, central cell vacuole; dfg, degenerated female gametophyte; ec, egg cell; ecn, egg cell nucleus; mc, micropylar cells; mn, micropylar nucleus; pn, polar nucleus; sc, synergid cell; scn; synergid cell nucleus; sen, secondary nucleus. (Bar, 10 μm.)
We first determined the terminal phenotypes of the cki1 mutants by examining the female gametophytes after allowing megagametogenesis within the ovaries of heterozygous plants to progress to the terminal developmental stage, female gametophyte stage 7 (FG7) (21). Both mutants had morphologically abnormal female gametophytes at stage FG7, indicating that the cki1-5 and cki1-6 mutations similarly affect megagametogenesis. With both mutants, approximately half of the female gametophytes we observed exhibited mutant phenotypes (Table 2). This proportion is consistent with the observed 100% penetrance of cki1-5 and cki1-6 mutations in the female gametophyte (Table 1).
Table 2.
Summary of CLSM analysis of the cki1-5 and cki1-6 mutants
| Genotype | FG stage | No. of pistils analyzed | No. of normal FGs observed | No. of abnormal FGs observed | Abnormal, % |
|---|---|---|---|---|---|
| cki1-5/CKI1 | FG0–FG3 | 7 | 69 | 0 | 0 |
| FG4 | 8 | 58 | 6 | 9 | |
| FG5/6 | 13 | 74 | 73 | 50 | |
| FG7 | 32 | 173 | 204 | 54 | |
| cki1-6/CKI1 | FG7 | 19 | 89 | 117 | 56 |
FG, female gametophyte.
Embryo sacs of cki1-5 and cki1-6 mutants had similar abnormal phenotypes. In both cases, mutant female gametophytes fell into two phenotypic categories. In the first and most frequent (153/204 for cki1-5 and 114/117 for cki1-6) category, female gametophytes appeared to be in the process of degeneration. The defects ranged from partial (Fig. 2D) to complete degeneration (Fig. 2C). The embryo sac cavity of partially degenerated female gametophytes was collapsed, yet displayed evidence of cellularization (Fig. 2D). The embryo sac cavity of completely degenerated female gametophytes was collapsed and filled with brightly fluorescent material (Fig. 2C). In the second phenotypic category (51/204 for cki1-5 and 3/117 for cki1-6), the embryo sac cavity was filled with a matrix of cytoplasmic strands connecting many small vacuoles (>100), as well as a greater than normal number of nuclei (≥16) (Fig. 2E).
To determine the developmental stage at which megagametogenesis in the mutants first departs from wild type, we analyzed megagametogenesis at all developmental stages in cki1-5/CKI1 ovaries. Wild-type female gametophyte development is shown in Fig. 2A. As summarized in Table 2, abnormal female gametophytes were not observed during the one-nucleate (stage FG1) or two-nucleate stages (FG2 and FG3). The earliest stage at which abnormal female gametophytes were observed was the four-nucleate stage (FG4). At this stage, a small proportion of embryo sacs contained two normal nuclei and two degenerated nuclei (Fig. 2F). Generally, the two degenerated nuclei were those at the chalazal pole (Fig. 2F). By the eight-nucleate stage (beginning at stage FG5), half of the female gametophytes within the ovary of a heterozygous plant were abnormal, indicating full penetrance at these stages. Abnormal female gametophytes had pleiotropic defects in cell morphology and nuclear position (Fig. 2 G and H). With reference to the cell polarity characteristic of a wild-type female gametophyte, nuclei were positioned inappropriately, both within the cells and relative to the other cells. Cell vacuoles were also misshapen and out of place. Taken together, these data indicate that the CKI1 gene product is essential for normal cell morphology, as well as control over the number of nuclear divisions, during megagametogenesis.
The defect in megagametogenesis of cki1 mutants strongly indicates that the CKI1 gene is expressed within the female gametophyte. To test this prediction, we carried out RNA in situ hybridization experiments with developing ovules. The CKI1 probe produced a weak signal that was ≤1/10 that of the control FILAMENTOUS FLOWER probe, indicating that CKI1 RNA is present at low levels in developing ovules. Within mature female gametophytes, the CKI1 probe consistently produced a strong signal in the central cell nucleus and a weaker signal in the egg cell nucleus (Fig. 3 A–C). CKI1 RNA was not detected in the cytoplasm of these two cells (Fig. 3 B and C). Occasionally, a weak signal was associated with the synergid cells (seen in Fig. 3B but not in A). Although the phenotypic abnormalities in a cki1 female gametophyte are apparent by stage FG5, we did not detect a CKI1 signal in developing embryo sacs at stages before FG7 (data not shown), indicating lower RNA levels were present during those stages.
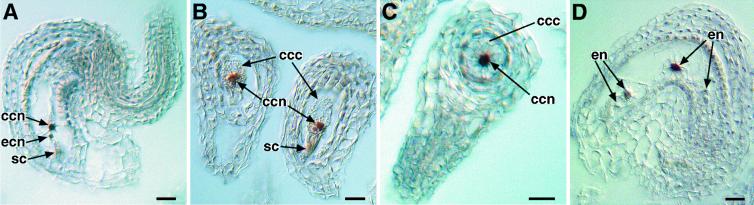
Fig 3.
CKI1 expression in the female gametophyte and endosperm. (A–C) Unfertilized ovules. A strong signal is associated with the nucleus of the central cell, and a weak signal with the nucleus of the egg cell. Note that no signal is associated with the synergid cells in A but a weak signal is associated with the synergid cells in B. (D) Fertilized ovule. A strong signal is associated with some but not all endosperm nuclei. ccc, central cell cytoplasm; ccn, central cell nucleus; ecn, egg cell nucleus; en, endosperm nuclei; sc, synergid cells. (Bar, 10 μm.)
We also analyzed CKI1 expression in fertilized ovules at 24–48 h after pollination. In fertilized ovules, a strong signal was present in the endosperm nuclei (Fig. 3D). Not all endosperm nuclei within a given ovule displayed a signal. This observation indicates that CKI1 expression may be under cell cycle control (30). CKI1 RNA was not detected in embryos by means of in situ hybridization.
We were able to detect CKI1 RNA by performing RT-PCR on RNA isolated from pistils containing mature female gametophytes or young siliques (data not shown). We were not usbable to detect a CKI1 transcript by performing RT-PCR on RNA isolated from 10- to 12-day-old seedlings, callus tissue, or shoot apical meristems. Additionally, we saw a very weak signal within the entire embryo sac of plants expressing a CKI1 promoter:GFP reporter fusion (data not shown).
The mRNA expression data, combined with the availability of two independent insertion alleles of cki1 with identical phenotypes, provides strong evidence that the defect in a cki1 female gametophyte is caused by a disruption in the CKI1 gene. To further establish a causal link between genotype and phenotype, we used molecular complementation to identify plants homozygous for an insertion in CKI1, yet viable because of the presence of a CKI1 transgene. A wild-type copy of CKI1 was introduced, along with a gene conferring BASTA resistance, into KanR cki1-5/CKI1 and cki1-6/CKI1 plants (generation T0) by means of T-DNA-mediated transformation. BASTA-resistant plants were recovered, and their progeny (generation T2) was screened for evidence of complementation. First, we analyzed the segregation of kanamycin resistance in the T2 generation. As described, the progeny resulting from self-fertilization of a cki1/CKI1 plant showed a 1:1 ratio of KanR:KanS. If the female gametophyte lethality is successfully complemented by the introduced CKI1 gene, the cki1 mutant allele should be transmitted through the female gametophyte as well as the male gametophyte. Therefore, we would expect to recover KanR progeny at a higher frequency in these lines. In most cases, the CKI1 transgene will segregate independently of the CKI1 locus, and if one copy of the transgene is present, we would expect to rescue the cki1 defect 50% of the time. Thus, we were looking for a bias toward KanR plants on the order of 2:1 KanR:KanS in the T2 generation. As seen in Table 3, several independent transgenic lines originating from both cki1-5/CKI1 and cki1-6/CKI1 T0 plants show a KanR:KanS ratio consistent with successful complementation of the cki1 homozygous lethal phenotype. Four lines were selected for further analysis. In each case, PCR genotyping confirmed the existence of individual plants homozygous for either the cki1-5 or cki1-6 insertion allele, yet viable because of the presence of the CKI1 transgene (Table 3).
Table 3.
Molecular complementation of the cki1 phenotype
| T0 genotype | KanR:KanS | T-DNA transmission, % | No. of complemented plants |
|---|---|---|---|
| cki1-5/CKI1 | 14:9 | 60.9 | 4 (13) |
| cki1-5/CKI1 | 17:7 | 70.8 | 5 (17) |
| cki1-5/CKI1 | 14:9 | 60.9 | ND |
| cki1-5/CKI1 | 19:11 | 63.3 | ND |
| cki1-5/CKI1 | 14:6 | 70.0 | ND |
| cki1-5/CKI1 | 15:6 | 71.4 | ND |
| cki1-5/CKI1 | 21:13 | 61.8 | ND |
| cki1-5/CKI1 | 18:11 | 62.1 | ND |
| cki1-5/CKI1 | 20:11 | 64.5 | ND |
| cki1-5/CKI1 | 20:8 | 71.4 | ND |
| cki1-5/CKI1 | 19:9 | 67.9 | ND |
| cki1-5/CKI1 | 19:8 | 70.4 | ND |
| cki1-5/CKI1 | 24:11 | 68.6 | ND |
| cki1-5/CKI1 | 15:9 | 62.5 | ND |
| cki1-5/CKI1 | 20:11 | 64.5 | ND |
| cki1-5/CKI1 | 19:11 | 63.3 | ND |
| cki1-6/CKI1 | 16:7 | 69.6 | 3 (16) |
| cki1-6/CKI1 | 13:6 | 68.4 | ND |
| cki1-6/CKI1 | 14:7 | 66.7 | ND |
| cki1-6/CKI1 | 16:7 | 69.6 | 5 (15) |
| cki1-6/CKI1 | 20:9 | 69.0 | ND |
| cki1-6/CKI1 | 21:9 | 70.0 | ND |
Each row describes an independent transgenic line. ND, not determined.
T0 refers to the plant whose progeny was transformed.
Kanamycin segregation data for the T2 generation of transformed plants.
Percent T-DNA transmission was calculated as 100 × KanR/(KanR + KanS).
Complemented plants were determined by PCR to have the genotype cki1/cki1 + CKI1 transgene. Numbers in parentheses indicate the total number of KanR plants genotyped.
Discussion
We described the isolation of two independent T-DNA insertion alleles that disrupt the function of a specific histidine kinase gene, CKI1. We provided several lines of evidence that, taken together, support a role for CKI1 in megagametogenesis: failure to recover a plant homozygous for the mutation, distortion from a 3:1 Mendelian segregation to a 1:1 segregation, evidence that mutant alleles are transmitted through the male gametophyte but not the female gametophyte, a ratio of 1:1 wild-type to mutant female gametophytes based on a CLSM analysis of megagametophytes within cki1/CKI1 pistils, localization of CKI1 mRNA expression in the developing female gametophyte and endosperm, and complementation of the cki1 homozygous lethality with a wild-type copy of CKI1.
The earliest stage at which phenotypic abnormalities could be detected in a cki1 female gametophyte is stage FG4, the four-nucleate stage. By this stage, meiosis, haploid megaspore degeneration, and two rounds of mitosis have occurred (21), which indicates that CKI1 function is not required for these processes. From the completion of stage FG4 through stage FG5, the stage at which the cki1 phenotype is completely penetrant, several important developmental steps occur: a final round of mitosis, cellularization, vacuole formation, and the establishment of cell identities (21). Because of the pleiotropic nature of the cki1 defect, we are unable at this time to limit the importance of CKI1 function to one of these processes. Megagametophyte-specific promoter:reporter gene fusions may be useful in assigning cellular identities to the abnormal cell types seen in FG5/6 female gametophytes; however, megagametophyte-specific promoters are uncharacterized at this time.
The same phenotypic abnormalities, most commonly, the collapse and degeneration of the embryo sac, were seen in both cki1-5 and cki1-6 female gametophytes at the terminal developmental stage. Degeneration of the embryo sac has also been described in another female gametophyte-specific mutant that is nonallelic to cki1, the ethyl methanesulfonate-generated fem1 mutant (22). Although the gene affected in the fem1 mutant is not known, we can rule out the possibility that FEM1 is allelic to CKI2, another histidine kinase-encoding gene that was identified in Kakimoto's (10) T-DNA activation screen for cytokinin independent mutants, based on mapping information (22). Whether or not CKI1 and/or CKI2 actually plays a role in cytokinin signaling, and what role cytokinins may play in female gametophyte development, remains to be seen. Currently, neither cytokinin levels nor the expression of cytokinin-inducible genes in the Arabidopsis embryo sac is known.
Given that specific molecules directing or modulating female gametophyte development have not yet been discovered, it is particularly exciting that a protein kinase with a putative extracellular ligand-binding domain and cytosolic signaling domain has now been implicated in this pathway. Elucidation of the ligand that interacts with CKI1 is an important objective for future study. Cytokinin is unlikely to be the ligand for CKI1 for several reasons. The lack of homology between the putative extracellular domain of CKI1 and that of the cytokinin receptor CRE1, and its two close homologues AHK2 and AHK3 (7), argues against a role for CKI1 as a cytokinin receptor. Membranes isolated from fission yeast expressing CKI1 do not bind radiolabeled cytokinin (8). Furthermore, the expression of CKI1 in Escherichia coli lacking the histidine kinase RcsC (8) and in Arabidopsis protoplasts coexpressing a cytokinin-inducible reporter gene (31) has been shown, in both cases, to activate histidine-kinase-signaling pathways in a constitutive, rather than cytokinin-dependent, manner. Thus, it seems plausible that the overexpression of CKI1 in Arabidopsis callus tissue created promiscuous “crosstalk,” i.e., allowed the protein to interact with other components of the cytokinin-signaling pathway with which it normally would not interact. Further studies are clearly needed to resolve this question.
Yeast two-hybrid studies by Urao et al. (13) have shown that CKI1 can interact with the HPt homologues ATHP1 and ATHP2 in vitro. In addition, work by Nakamura et al. (32) has shown that the response-regulator domain of CKI1 can act as a phosphatase when incubated with purified, radioactively phosphorylated HPts ATHP1 and ATHP3. Thus, the potential for a His/Asp phosphorelay initiated by CKI1 and involving other His/Asp phosphorelay components certainly exists. The T-DNA insertion mutants cki1-5 and cki1-6 can provide the framework for elucidating the in situ mechanisms of this first known signal-transduction pathway that operates in the female gametophyte.
Acknowledgments
We thank Jason Young for technical assistance. This work was supported by a National Science Foundation Graduate Research Fellowship (to M.S.P.); by research funds provided by the University of Wisconsin Graduate School, the University of Wisconsin College of Agricultural and Life Sciences, the National Science Foundation, and the U.S. Department of Energy (DE-FG02ER13938) (to M.R.S.); and by research funds provided by the National Science Foundation (IBN-9630371) and Ceres, Inc. (to G.N.D.).
Abbreviations
CAPS, cleaved amplified polymorphic sequences
CKI1, Cytokinin-Independent 1
CLSM, confocal laser scanning microscopy
FGn, female gametophyte stage n
HPt, histidine-containing phosphotransmitter
KanR, kanamycin resistant
KanS, kanamycin sensitive
References
- 1.Parkinson J. S. (1993) Cell 73 857-871. [DOI] [PubMed] [Google Scholar]
- 2.Urao T., Yamaguchi-Shinozaki, K. & Shinozaki, K. (2000) Trends Plant Sci. 5 67-74. [DOI] [PubMed] [Google Scholar]
- 3.Ota I. M. & Varshavsky, A. (1993) Science 262 566-569. [DOI] [PubMed] [Google Scholar]
- 4.Schuster S. C., Noegel, A. A., Oehme, F., Gerisch, G. N. & Simon, M. I. (1996) EMBO J. 15 3880-3889. [PMC free article] [PubMed] [Google Scholar]
- 5.Alex L. L., Borkovich, K. A. & Simon, M. I. (1996) Proc. Natl. Acad. Sci. USA 93 3416-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hua J. & Meyerowitz, E. M. (1998) Cell 94 261-271. [DOI] [PubMed] [Google Scholar]
- 7.Inoue T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K. & Kakimoto, T. (2001) Nature 409 1060-1063. [DOI] [PubMed] [Google Scholar]
- 8.Yamada H., Suzuki, T., Terada, K., Takei, K., Ishikawa, K., Miwa, K., Yamashino, T. & Mizuno, T. (2001) Plant Cell Physiol. 42 1017-1023. [DOI] [PubMed] [Google Scholar]
- 9.Urao T., Yakubov, B., Satoh, R., Yamaguchi-Shinozaki, K., Seki, M., Hirayama, T. & Shinozaki, K. (1999) Plant Cell 11 1743-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kakimoto T. (1996) Science 274 982-985. [DOI] [PubMed] [Google Scholar]
- 11.Miller C. O., Skoog, F., Okumura, F. S., Saltza, M. H. V. & Strong, F. M. (1956) J. Am. Chem. Soc. 78 1375-1380. [Google Scholar]
- 12.Suzuki T., Miwa, K., Ishikawa, K., Yamada, H., Aiba, H. & Mizuno, T. (2001) Plant Cell Physiol. 42 107-113. [DOI] [PubMed] [Google Scholar]
- 13.Urao T., Miyata, S., Yamaguchi-Shinozaki, K. & Shinozaki, K. (2000) FEBS Lett. 478 227-232. [DOI] [PubMed] [Google Scholar]
- 14.Sakai H., Honma, T., Aoyama, T., Sato, S., Kato, T., Tabata, S. & Oka, A. (2001) Science 294 1519-1521. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki T., Ishikawa, K., Yamashino, T. & Mizuno, T. (2002) Plant Cell Physiol. 43 123-129. [DOI] [PubMed] [Google Scholar]
- 16.Sheen J. (2002) Science 296 1650-1652. [DOI] [PubMed] [Google Scholar]
- 17.Krysan P. J., Young, J. C. & Sussman, M. R. (1999) Plant Cell 11 2283-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murashige T. & Skoog, F. (1962) Physiol. Plant. 15 473-497. [Google Scholar]
- 19.Krysan P. J., Young, J. C., Tax, F. & Sussman, M. R. (1996) Proc. Natl. Acad. Sci. USA 93 8145-8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konieczny A. & Ausubel, F. M. (1993) Plant J. 4 403-410. [DOI] [PubMed] [Google Scholar]
- 21.Christensen C. A., King, E. J., Jordan, J. R. & Drews, G. N. (1997) Sex. Plant Reprod. 10 49-64. [Google Scholar]
- 22.Christensen C. A., Subramanian, S. & Drews, G. N. (1998) Dev. Biol. 202 136-151. [DOI] [PubMed] [Google Scholar]
- 23.Klucher K., Chow, H., Reiser, L. & Fischer, R. (1996) Plant Cell 8 137-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vielle-Calzada J.-P., Thomas, J., Spillane, C., Coluccio, A., Hoeppner, M. A. & Grossniklaus, U. (1999) Genes Dev. 13 2971-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegfried K. R., Eshed, Y., Baum, S. F., Otsuga, D., Drews, G. N. & Bowman, J. L. (1999) Development (Cambridge, U.K.) 126 4117-4128. [DOI] [PubMed] [Google Scholar]
- 26.Krysan P. J., Jester, P. J., Gottwald, J. R. & Sussman, M. R. (2002) Plant Cell 14 1109-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clough S. J. & Bent, A. F. (1998) Plant J. 16 735-743. [DOI] [PubMed] [Google Scholar]
- 28.Tax F. E. & Vernon, D. M. (2001) Plant Physiol. 126 1527-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore J. M., Calzada, J. P. V., Gagliano, W. & Grossniklaus, U. (1997) Cold Spring Harbor Symp. Quant. Biol. 62 35-47. [PubMed] [Google Scholar]
- 30.Boisnard-Lorig C., Colon-Carmona, A., Bauch, M., Hodge, S., Doerner, P., Bancharel, E., Dumas, C., Haseloff, J. & Berger, F. (2001) Plant Cell 13 495-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang I. & Sheen, J. (2001) Nature 413 383-389. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura A., Kakimoto, T., Imamura, A., Suzuki, T., Ueguchi, C. & Mizuno, T. (1999) Biosci. Biotechnol. Biochem. 63 1627-1630. [DOI] [PubMed] [Google Scholar]