Abstract
Abscisic acid (ABA) mediates plant responses to environmental stress, particularly to water status. During germination, the embryo emerges from dormancy as the ABA concentration declines. Exposure to exogenous ABA during germination arrests development rapidly, but reversibly, enabling seedlings to withstand early water stress without loss of viability. Postgermination proteolytic degradation of the essential ABI5 transcription factor is interrupted by perception of an increase in ABA concentration, leading to ABI5 accumulation and reactivation of embryonic genes. Making use of the ABA-hypersensitive hyl1 mutant of Arabidopsis, we show that the ABA signal is transmitted to the transcriptional apparatus through mitogen-activated protein kinase signaling.
The phytohormone abscisic acid (ABA) mediates many plant responses to environmental stress, particularly the ability to sense and respond to water status. The most extensively studied physiological effects of ABA are those that regulate stomatal closure to adjust the rate of water loss in aerial plant parts (1–3) and those required for seed maturation (4–6). The developing embryo enters a dormant state late in seed maturation. This state is triggered by an increase in the ABA concentration and leads to the cessation of cell division and activation of genes encoding seed storage proteins and proteins required to establish desiccation tolerance (5, 7).
ABA is thought to maintain dormancy, and germination is the process by which the embryo emerges from dormancy and resumes embryonic development (8). A decrease in the ABA concentration relative to that of the growth-promoting hormone gibberellin, together with an increase in the water content, can suffice to release the embryo from dormancy in many plant species, although others require additional signals, such as light or a period of chilling. Exposure of seeds to ABA during germination arrests development rapidly, but reversibly. ABA-arrested plants can withstand desiccation early in development without losing viability and it has been suggested that ABA-mediated postgermination arrest represents a developmental checkpoint that allows germinating seedlings to survive early water stress (9).
The ability of ABA to interfere with seed germination has been used to identify mutations that render germination both more resistant and less resistant to ABA. Analysis of such mutants has identified genes encoding protein phosphatases, transcription factors, a farnesyl transferase and three RNA-binding proteins, as well as genes belonging to other hormone-signaling pathways (6, 10). Recent studies have revealed that the ABI5 transcription factor encoded by the abscisic acid insensitive5 (ABI5) gene is a key player in ABA-triggered postgermination growth arrest (9). Germinating abi5 seeds are insensitive to growth arrest by ABA, whereas seeds of an ABI5 overexpresser are hypersensitive to ABA. Exogenous ABA interrupts the postgermination proteasome-mediated decline in the ABI5 concentration, leading to the accumulation of ABI5 protein and transcripts (9); but the ABA signaling pathway is poorly understood. ABA has been reported to activate mitogen-activated protein kinases (MAPK), but it has not been established that kinase activation mediates growth arrest (11, 12). Here we show that the ABA signal is transmitted to the transcriptional apparatus through MAPK signaling.
Materials and Methods
Plant Lines and Growth Conditions.
Arabidopsis thaliana ecotypes Nossen and Columbia were used. The hyponastic leaves 1 (hyl1) transposon insertion mutant has been described (13). Plants were grown on Murashige and Skoog (MS) medium (GIBCO/BRL, Grand Island, NY) containing 1% sucrose. Seeds were sterilized and grown as described (13). ABA (Sigma), the MAPK kinase (MAPKK)-specific inhibitor PD98059 (Sigma), and the general kinase inhibitor K252a (Calbiochem) were added to the medium where indicated. To examine ABA sensitivity, wild-type and mutant seeds were stratified for 2 days at 4°C, then transferred to room temperature under constant light. The germination rate was measured at 10 days (0 and 0.6 μM ABA) or 20 days (3 μM ABA) after stratification. To assess the effects of kinase inhibitors, wild-type and mutant seeds were plated on filter paper saturated with MS solution with or without either 100 μM PD98059 or 1 μM K252a.
RT-PCR.
Total RNA was treated with DNase I according to the manufacture's instructions (GIBCO/BRL). The first-strand cDNA was synthesized from 1.5 μg of total RNA in a volume of 20 μl containing 50 mM Tris⋅HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 5 mM DTT, 2 mM dNTP mixture, 5 μM oligo(dT) primer, 10 units of RNase inhibitor (Invitrogen), and 100 units of Moloney murine leukemia virus reverse transcriptase (Ambion, Austin, TX) for 1 hour at 42°C, then incubated at 92°C for 10 min to inactivate the enzyme. One microliter of the first-strand solution was used for the subsequent PCR in a total volume of 20 μl with 1 unit of RedTaq DNA polymerase (Sigma). The following gene specific primers were used to detect the gene expression level: ABI1-fw (5′-CCATTCCACTGAATCACTTT-3′) and ABI1-rev (5′-GATATCTCCGCCGGAGAT-3′); ABI3-fw (5′-TCCTTCCGAGGTGACCCACGT-3′) and ABI3-rev (5′-CGGAAGATTATACATTTGCAATGG-3′); ABI5-fw (5′-CCTTGACGAGTTCCAACATG-3′) and ABI5-rev (5′-ACCATAAAGCTGTTGCTGTG-3′); ANP1-fw (5′-GAAGCTTCATCACCTCCATTG-3′) and ANP1-rev (5′-TGATCACATTCTCTCCCAGTTG-3′); AtMPK1-fw (5′-ATAGGATCGATGCGTTGAGG-3′) and AtMPK1-rev (5′-TCGGTACCAACGAGTCACAA-3′); AtMPK2-fw (5′-TCTGGTCAGTCGGTTGCATA-3′) and AtMPK2-rev (5′-TCATCTCTGCTCCCAAATCC-3′); AtMPK3-fw (5′-ATGAACACCGGCGGTGGC-3′) and AtMPK3-rev (5′-CTGCTGCACTTCTAACCG-3′); AtMPK4-fw (5′-AAACCACCGCAGAGAGAGAA-3′) and AtMPK4-rev (5′-GGAGCTCGATACCAACGTGT-3′); AtMPK5-fw (5′-GACTTTGGTTTGGCAAGGAC-3′) and AtMPK5-rev (5′-GACGGATCCTCAAAATGGAA-3′); AtMPK6-fw (5′-TCTCCTCCTGAACGCAAACT-3′) and AtMPK6-rev (5′-GATGGATTGGCGAGGATAAG-3′); AtMPK7-fw (5′-TGTGATCGCGCTTAAAGATG-3′) and AtMPK7-rev (5′-TTCTGTCCCTGGAAAAATCG-3′); AtMPK8-fw (5′-GGAATACCTTCGTGGTGGTG-3′) and AtMPK8-rev (5′-TTGTCTGCTTCCTCGGATTC-3′); AtMPK9-fw (5′-GAGGCACTGGCAGATCCTTA-3′) and AtMPK9-rev (5′-TCTAGGCAAGGAAGCGTGTT-3′).
AtMPK3 cDNA Cloning.
AtMPK3 cDNA was amplified by RT-PCR using total RNA from ozone-treated (30 min at 350 ppb) 4-week-old Arabidopsis Columbia ecotype plants. The complete ORF was amplified by using the primers 5′-CATTTAAATATGAACACCGGCG-3′ and 5′-GACTAGTGCACTTCTAACCG-3′. The PCR fragment was cloned into the pT-Adv vector (CLONTECH), sequenced, then subcloned into the pFGC1008 binary vector (http://ag.arizona.edu) between the CaMV35S promoter and the ocs 3′ terminator either as an inverted repeat or as a single cDNA copy.
Plant Transformation.
The AtMPK3 cDNA vector was transferred to Agrobacterium tumefaciens strain LBA4404 by a triparental mating, and the bacteria were used to transform A. thaliana ecotype Columbia by vacuum infiltration or floral dip (14, 15). Hygromycin B-resistant (50 μg/ml) F1 plants were grown, and the presence of the transgene was verified by PCR.
Agroinfiltration.
AtMPK3 cDNA was tagged with the hemagglutinin (HA) epitope at the N terminus and inserted into the pRTL2 vector containing a CaMV 35S promoter and a NOS terminator. The 35S:HA:AtMPK3:NOS was transferred to the pCGN1547 binary vector (16) and transformed into A. tumefaciens LBA4404. Four-week-old Nicotiana benthamiana plants were agroinfiltrated as described in ref. 17. Four days after the first infiltration, the leaves were infiltrated with 0.07% EtOH or 50 μM ABA in 0.07% EtOH for 5 min or with either 20 mM H2O2 or H2O for 15 min. Protein was extracted as described below.
RNA Gel Blot Analysis.
Total RNA was isolated by using the RNeasy Plant Mini kit (Qiagen) from young seedlings ≈3–7 days after stratification. Seeds were plated on MS medium with or without ABA. Seedling samples were collected after various treatment times, total RNA was isolated, and 10 μg of RNA was fractionated on a formaldehyde agarose gel, then transferred to a nitrocellulose membrane. The ABI5-specific probe was from the cDNA coding region. All of the DNA probes were labeled with 32P-dCTP. Hybridization was performed at 65°C as described (18).
Protein Extraction.
Frozen seedlings were ground in liquid nitrogen and thawed in extraction buffer containing 100 mM Hepes (pH 7.5), 5 mM EDTA, 5 mM EGTA, 10 mM DTT, 10 mM Na3VO4, 10 mM NaF, 50 mM β-glycerophosphate, 10% glycerol, 7.5% poly(vinyl polypyrrolidone), and protease inhibitor mixture (as recommended by Sigma). The ground slurry was centrifuged at 20,000 × g for 20 min at 4°C. Supernatants were collected into new tubes, frozen in liquid nitrogen, and stored at −80°C. The protein concentration was determined by using the Bio-Rad protein assay kit with BSA as a standard.
In-Gel Kinase Assays.
The in-gel kinase assay was performed as described in ref. 19. Typically, 50-μg aliquots of protein extracted from leaf tissue were fractionated on an SDS/10% polyacrylamide gel containing 0.25 mg/ml myelin basic protein (MBP, Sigma) or purified His-tagged ABI5 protein in the resolving gel as substrate for the kinases. After electrophoresis, the gel was washed three times with 25 mM Tris, pH 7.5/0.5 mM DTT/0.1 mM Na3VO4/5 mM NaF/0.5 mg/ml BSA/0.1% (vol/vol) Triton X-100 for 30 min each at room temperature. Proteins in the gel were then renatured by incubating the gel in 25 mM Tris, pH 7.5/1 mM DTT/0.1 mM Na3VO4/5 mM NaF at 4°C overnight with three changes of buffer. The kinase reactions were then carried out by incubating the gel in 30 ml of buffer containing 25 mM Tris (pH 7.5), 2 mM EGTA, 12 mM MgCl2, 1 mM DTT, 0.1 mM Na3VO4, 200 nM ATP, and 50 μCi of [γ-32P]ATP (>4,000 Ci/mmol; 1 Ci = 37 GBq) for 60 min at room temperature. To remove free 32P, the gel was extensively washed at room temperature with several changes of 5% (wt/vol) trichloroacetic acid and 1% (wt/vol) NaPPi until 32P-radioactivity in the used wash solution was barely detectable. The gel was then dried under vacuum on Whatman 3MM paper. Labeled proteins were detected by using a PhosphorImager or x-ray film. Protein sizes were estimated by using molecular mass markers (Bio-Rad).
Immunocomplex Kinase Activity Assay.
Aliquots of protein extracts (250 μg) were incubated with 6 μg of anti-HA antibody (agarose conjugate, Santa Cruz Biotechnology) in immunoprecipitation buffer (10 mM Tris⋅HCl, pH 8.0/150 mM NaCl/1 mM Na3VO4/1 mM NaF) at 4°C on a rotator overnight. Immunoprecipitates were pelleted by centrifugation at 1,000 × g for 5 min at 4°C and washed 4 times with 1.5 ml of immunoprecipitation buffer. The beads were resuspended in 25 μl of kinase reaction buffer [25 mM Tris, pH 7.5/5 mM MgCl2/1 mM EGTA/1 mM DTT/10 μM ATP/10 μCi of [γ-32P]ATP (>4,000 Ci/mmol)/0.5 mg/ml MBP] and incubated at room temperature for 20 min. The reaction was stopped by adding SDS/PAGE sample buffer and heating at 95°C for 4 min. The 32P-labeled MBP was separated on an SDS/15% polyacrylamide gel and visualized by autoradiography or by using a PhosphorImager.
Results
Expression of the ABI5 Transcription Factor in the ABA-Hypersensitive hyl1 Mutant.
The ability of exogenous ABA to arrest the growth of germinating seedlings requires the accumulation of the ABI5 transcription factor, normally degraded soon after germination (9). Overexpression of ABI5 renders seeds ABA-hypersensitive, whereas homozygosity for the recessive abi5 mutation confers insensitivity to postgermination growth arrest by ABA (9). A number of Arabidopsis ABA-hypersensitive mutants have been described, including the pleiotropic hyl1 mutant (6). The hyl1 mutation arose by a transposon insertion into a gene encoding a nuclear double-stranded RNA (dsRNA)-binding protein. The mutation affects the response to several hormones, reducing sensitivity to auxin and cytokinin and increasing sensitivity to ABA during seed germination. An ABA concentration of 0.5 μM suffices to arrest development of germinating hyl1 seedlings. This is roughly an order of magnitude lower than the ABA concentration required to arrest growth of wild-type seedlings (13).
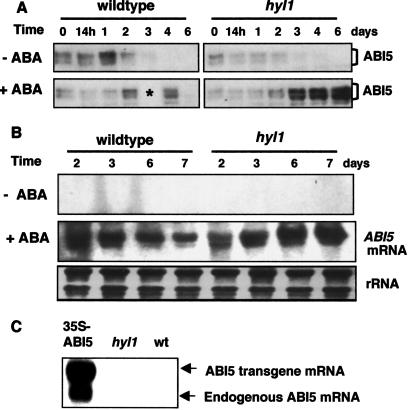
Because the ability of ABA to arrest growth declines with the disappearance of ABI5 after germination, we hypothesized that ABA-hypersensitivity during germination might result from a reduction in the concentration of ABA required to trigger ABI5 accumulation. To test this hypothesis, we germinated wild-type seeds and seeds of a plant that was homozygous for the ABA-hypersensitive hyl1 mutation on agar medium containing 0.7 μM ABA and assessed both ABI5 protein and ABI5 transcript levels over the 6-day period following germination. In the absence of ABA, the ABI5 protein levels are low and comparable in newly germinated hyl1 mutant and wild-type seedlings (Fig. 1A) and ABI5 transcript levels are too low to be detected by blot hybridization (Fig. 1B). Wild-type seedlings germinating in the presence of 0.7 μM ABA show a transient increase in the amount of both the ABI5 protein (Fig. 1A) and ABI5 transcript (Fig. 1B). At this concentration of ABA, the development of wild-type seedlings is mildly retarded, but not arrested, whereas development of hyl1 seedlings is completely arrested (13). By contrast to what is observed in wild-type seedlings, both the ABI5 protein and its transcript continue to accumulate in the growth-arrested hyl1 seedlings (Fig. 1 A and B). Because overexpression of an ABI5 cDNA transgene stimulates expression of the endogenous gene, as shown in Fig. 1C, it is likely that the ABI5 transcription factor activates its own transcription, directly or indirectly. Thus the ABA-hypersensitivity of the hyl1 mutant is correlated with accumulation of ABI5 at a lower ABA concentration in mutant than in wild-type seedlings. However, because overexpression of the ABI5 protein has no effect in the absence of ABA (9), ABI5 accumulation is necessary, but not sufficient to arrest growth, and ABA signaling must involve additional molecules or modifications.
Fig 1.
ABI5 protein and mRNA accumulation in wild-type and hyl1 seedlings. Wild-type and hyl1 seeds were stratified at 4°C for 3 days with or without 0.7 μM ABA and then transferred to constant light at 22°C. Seeds were harvested at the indicated times after transfer and total protein and RNA were isolated. (A) Western blot analysis using antibodies to ABI5. Each lane contained 10 μg protein (asterisk marks an empty lane caused by sample loss). (B) Northern blot analysis. Each lane contained 10 μg total RNA. (C) RNA was isolated from germinating seeds of transgenic plants containing a 35S-ABI5 cDNA construct, hyl1 seeds, and wild-type seeds 3 days after stratification. Each lane contained 10 μg total RNA.
ABA-Activated MAPK and ABI5 Kinase Levels Are Elevated in hyl1 Seedlings.
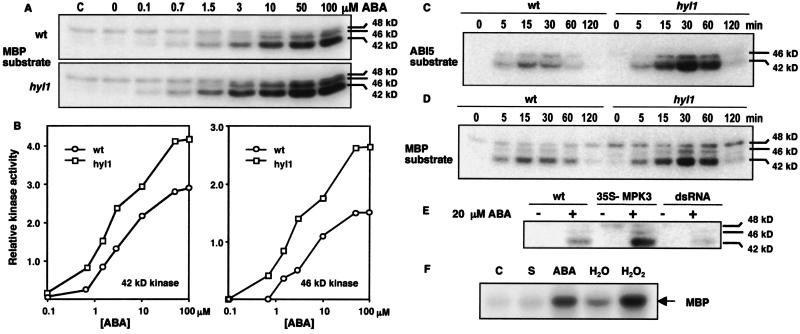
Rapid activation of MAPKs in response to exogenous ABA or a variety of abiotic stress conditions has been reported in a number of different plant species (20–22). H2O2, oxidative stress, cold, drought, salinity, touch, and wounding activate one or more of 3 Arabidopsis MAPKs, AtMPK3, -4, and -6 (23–25). Furthermore, it has been reported that ABI5 is phosphorylated (9). However, neither MAPK activation nor ABI5 phosphorylation have been causally connected with the ability of ABA to arrest seedling growth. We first asked whether the hyl1 mutation affects MAPK activation in response to ABA. We carried out in-gel MAPK assays on hyl1 mutant and wild-type seedlings germinated in liquid medium to which ABA was added at different concentrations 15 min before harvest. Fig. 2A shows that two MAPKs of ≈42 and 46 kDa are activated by ABA in both wild-type and hyl1 mutant seedlings. A third MAPK of ≈48 kDa is detected under these growth conditions, but its activity is unaffected by ABA. Strikingly, both the 42- and 46-kDa MAPKs are activated at a lower concentration of ABA in hyl1 than in wild-type seedlings (Fig. 2 A and B). In particular, markedly greater activation of both MAPKs is detected in mutant seedlings at 0.7 μM ABA, an ABA concentration sufficient for complete arrest of mutant, but not wild-type, seedling growth. Activation of both MAPKs increases with increasing ABA concentration and both exhibit higher levels of activity at all ABA concentrations in hyl1 mutant than in wild-type seedlings (Fig. 2 A and B). These observations suggest the possibility that ABA signals through a MAPK cascade to the transcriptional apparatus to arrest growth of seedlings.
Fig 2.
Kinase assays using MBP and ABI5 substrates. (A) Wild-type and hyl1 seedlings were germinated in liquid medium for 6 days. ABA (in 0.1 mM NaOH) was added at the indicated concentrations (nothing was added to the control, C) and the seedlings were harvested after 15 min. Protein extracts (30 μg) were fractionated on a 10% polyacrylamide gel containing MBP as a kinase substrate, renatured, and the gels were incubated with 32P-ATP. (B) Quantification of PhosphorImager band intensities for the 42- and 46-kDa MAPK activities detected in A. (C) Wild-type and hyl1 seedlings were germinated as in A; 10 μM ABA (in 0.1 mM NaOH) was added to the medium, and seedlings were harvested at the indicated times. Protein extracts were prepared and used for an in-gel kinase assay as described in A, but with ABI5 as substrate. (D) Wild-type and hyl1 seedlings were germinated as in A; 10 μM ABA (in 0.1 mM NaOH) was added to the medium, and seedlings were harvested at the indicated times; proteins were extracted and analyzed as described in A with MBP as kinase substrate. (E) Wild-type seeds and seeds from transgenic plants expressing a 35S-AtMPK3 cDNA or a 35S-AtMPK3 dsRNA construct were germinated as in A, then either 0.1 mM NaOH (−) or 20 μM ABA in 0.1 mM NaOH (+) was added 30 min before harvesting. Proteins were fractionated and assayed for MAPK activity as in A. (F) Leaves of Nicotiana benthamiana seedlings grown at 22°C were infiltrated with an Agrobacterium strain carrying a CaMV 35S promoter fusion expressing an HA epitope-tagged AtMPK3. After 4 days, the leaves received the following treatments: C, none; S, 0.07% EtOH for 5 min; ABA, 50 μM ABA in 0.07% EtOH for 5 min; H2O, water for 15 min; and H2O2, 20 mM for H2O2 for 15 min. The infiltrated leaves were then harvested and proteins were extracted. HA-tagged AtMPK3 was immunoprecipitated with anti-HA antibodies, incubated with 32P-ATP and MPB, then fractionated on a 15% polyacrylamide gel.
It has been reported that the ABI5 transcription factor is phosphorylated in seedlings germinated on ABA (9). To determine whether the hyl1 mutation affects the activity of the kinase or kinases that phosphorylate ABI5, we performed an in-gel kinase assay incorporating recombinant ABI5 protein into the gel as the kinase substrate. Two protein kinases of ≈42 and 46 kDa phosphorylate recombinant ABI5 protein (Fig. 2C). Both activities are present at higher levels in protein extracts from hyl1 plants than in wild-type protein extracts and both are rapidly activated by ABA (Fig. 2C). Moreover, the time course of MAPK activation after ABA treatment is very similar to that observed for the activation of the kinases that phosphorylate ABI5 (Fig. 2D). The similarity in size and ABA activation kinetics of the ABA-activated MAP and ABI5 kinases, as well as the consistent quantitative differences in these activities between the hyl1 mutant and wild-type seedlings, suggests that ABI5 is a substrate for the ABA-activated 42- and 46-kDa MAPKs.
Stress Kinase Cascade Genes Are Overexpressed in hyl1.
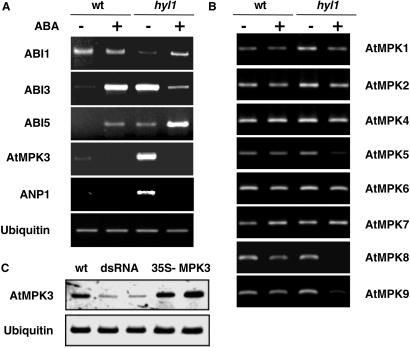
DNA microarray expression profiling experiments identified a number of genes that are overexpressed in hyl1 mutant seedlings compared with wild-type seedlings of the same age (data not shown). Among these are the ANP1 and the AtMPK3 genes, which encode component kinases of the stress-activated MAPK cascade (12), as well as the ABI3 and ABI5 transcription factor genes (6). The differences in gene expression were confirmed by quantitative RT-PCR amplification of RNA extracted from hyl1 mutant and wild-type seedlings 3 days after germination in the presence or absence of 0.7 μM ABA (Fig. 3A). Of the genes tested, the ANP1, AtMPK3, ABI3, and ABI5 genes show the most marked differences in expression level between wild-type and hyl1 plants in the absence of ABA. RT-PCR amplification of other AtMPK transcripts is shown in Fig. 3B and reveals that among the MAPK genes tested, the AtMPK3 gene is the only one whose mRNA level is markedly higher in hyl1 than in wild-type seedlings. Germination of seeds on a low concentration of ABA results in higher transcript levels for some genes and lower transcript levels for others. ABI5 transcript levels increase after ABA treatment in both wild-type and hyl1 mutant seedlings because of positive autoregulation (Fig. 1C), but the increase is more marked in the mutant than in the wild-type seedlings at the low ABA concentration used in this experiment.
Fig 3.
Relative amounts of MAPK and other gene transcripts in wild-type and hyl1 plants. Wild-type and hyl1 seeds were plated on MS medium with (+) or without (−) 0.7 μM ABA. The seeds were harvested and used for RNA isolation 3 days after stratification. Equal amounts of RNA from each sample were used for RT-PCR analysis. The sequences of primers used to detect expression of the indicated genes in A and B are given in Materials and Methods. Gene abbreviations: ABI1, -3, and -5 are the genes identified by the abi1, -3, and -5 mutations and encode a protein phosphatase and two transcription factors, respectively; AtMPK1–9 are the genes that encode A. thaliana MAP kinases 1–9; the ANP1 gene encodes a MAPKKK that activates the stress kinase cascade. Ubiquitin mRNA served as a reference. (C) RT-PCR analysis of AtMPK3 and ubiquitin mRNA levels in wild-type (wt) and transgenic A. thaliana plants expressing a CaMV 35S promoter-driven inverted repeat AtMPK3 cDNA construct (dsRNA) or a 35S-AtMPK3 cDNA construct (35S-MPK3).
The markedly elevated levels of ANP1 and AtMPK3 transcripts in hyl1 seedlings suggested that the stress-activated AtMPK3, with a predicted molecular mass of 42.7 kDa, is the major ABA-activated MAPK detected in seedlings. We confirmed this identification by in gel MAPK assays (Fig. 2E) and RT-PCR assays (Fig. 3C) of plants expressing an AtMPK3 cDNA from a 35S promoter or an inhibitory AtMPK3 dsRNA construct (see Materials and Methods). Although AtMPK3 has been reported to be activated by H2O2 (24) and flagellin (26), it has not been reported to be activated by ABA. To directly test whether AtMPK3 is activated by ABA, an HA epitope-tagged AtMPK3 protein was transiently overexpressed in Nicotiana benthamiana seedling leaves by agroinfiltration (17) from the tagged AtMPK3 cDNA expressed from a cauliflower mosaic virus (CaMV) 35S promoter. Infiltrated leaves were treated briefly with ABA (50 μM, 5 min) or H2O2 (20 mM, 15 min) before harvest. The epitope-tagged AtMPK3 was immunoprecipitated, and the immunoprecipitate was tested for its ability to phosphorylate MBP. The results show that both ABA and H2O2 activate AtMPK3 (Fig. 2F). Thus, the ABA-activated 42-kDa MAPK overexpressed in the hyl1 mutant is AtMPK3.
Overexpression of the AtMPK3 Gene Increases ABA Sensitivity.
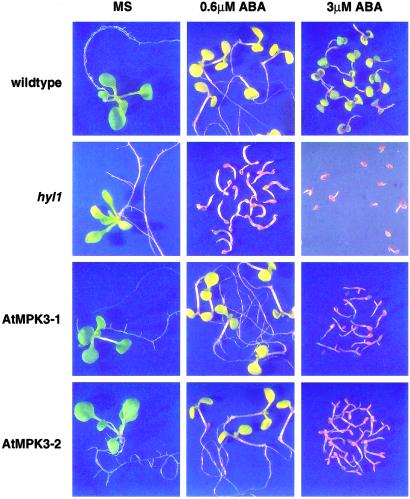
Because the hyl1 mutation is pleiotropic and changes the expression levels of a number of genes other than those encoding proteins of the stress MAPK cascade, we asked whether increased expression of just the AtMPK3 gene can alter the sensitivity of otherwise wild-type Arabidopsis seeds to postgermination arrest. We used two independently derived Arabidopsis lines containing a 35S-AtMPK3 cDNA construct, neither of which exhibited more than a 2-fold overexpression of AtMPK3 mRNA (Fig. 3C) or had a mutant phenotype under normal growth conditions. Nonetheless, as shown in Fig. 4, both of the independently derived lines were more sensitive than were wild-type plants to ABA-triggered postgermination growth arrest. Development of the hyl1 mutant is arrested at 0.6 μM ABA, whereas that of wild-type seedlings and the AtMPK3 transgenic seedlings is only mildly retarded at 10 days after germination. However, the AtMPK3 transgenic seeds are clearly more sensitive to the higher concentration of 3 μM ABA, which by 20 days after germination has only retarded the development of wild-type plants, but completely arrested development of the AtMPK3 transgenic seeds. The implication of this finding is that the ABA signal can be transmitted through the stress-activated MAPK cascade, of which AtMPK3 is a member, to trigger postgermination arrest of seedling development.
Fig 4.
Transgenic plants overexpressing MPK3 are hypersensitive to ABA. Seeds of wild-type, hyl1, and two independent 35S-AtMPK3 cDNA transgenic plants (AtMPK3–1 and –2) were stratified at 4°C for 2 days and then transferred to constant light at 22°C. Pictures were taken 10 days (0 and 0.6 μM ABA) and 20 days (3 μM) after stratification.
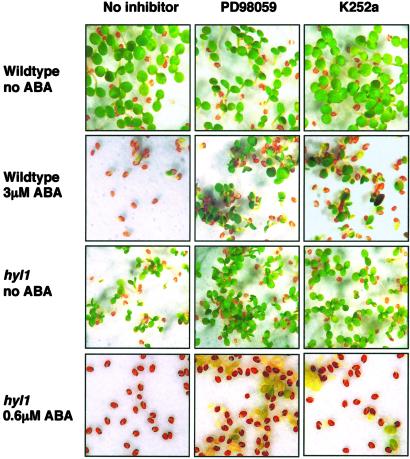
Inhibition of MAPK Signaling Decreases Sensitivity to Postgermination Growth Arrest by ABA.
If signaling through the MAPK cascade is the primary or the only route by which ABA information is transmitted to trigger postgermination growth arrest, interfering with MAPK cascade signal transmission should reduce the sensitivity of both wild-type and hyl1 mutant seeds to growth arrest by ABA. We therefore asked whether inhibition of MAPKK activity by PD98059, a widely used MAPKK-specific kinase inhibitor, would interfere with the ability of ABA to trigger postgermination growth arrest. PD98059 has been reported to show the same specificity in plants as in animals (27), and we confirmed its ability to inhibit MAPK activation by ABA and H2O2 in Arabidopsis protoplasts (data not shown). We germinated wild-type and hyl1 seeds on medium containing no ABA or either 0.6 μM or 3 μM ABA in the presence and absence of either PD98059 or the general kinase inhibitor K252a. As shown in Fig. 5, neither kinase inhibitor alone had a marked effect on the germination of either wild-type or hyl1 mutant seeds. Both PD98059 and K252a significantly reduced the sensitivity of wild-type seeds to postgermination growth arrest by 3 μM ABA. Moreover, both inhibitors supported the growth of hyl1 mutant seedlings on 0.6 μM ABA, a concentration sufficient to arrest their growth in the absence of the kinase inhibitors. We conclude from these observations that ABA signal transduction through MAPK cascade components is necessary to trigger postgermination growth arrest.
Fig 5.
Kinase inhibitors decrease sensitivity to ABA-mediated postgermination arrest. Wild-type and hyl1 seeds were plated on filter paper saturated with MS solution alone or MS containing either PD98059 (100 μM) or K252a (1 μM). ABA was present at the indicated concentrations. After 2 days at 4°C, seeds were transferred to constant light at 22°C. Pictures were taken 10 days after stratification.
Discussion
Although the ability of exogenous ABA to arrest postgermination development has been used extensively to identify genes that encode proteins involved in ABA signaling, the mechanism of developmental arrest remains poorly understood. It was reported recently that ABA stops and reverses the postgermination decline in the concentration of the ABI5 transcription factor, activating both transcription of the ABI5 gene and accumulation of the ABI5 protein by interrupting its degradation (9). The ABI5 transcription factor appears to be a necessary component of the seedling ABA response because the recessive abi5 mutation confers ABA insensitivity, whereas overexpression of an ABI5 cDNA renders seedlings hypersensitive to ABA. However, it is not known how the ABA signal is transmitted to ABI5.
We showed here that both ABI5 protein and ABI5 mRNA levels are elevated in hyl1 mutant, but not wild-type seedlings, at the low concentration of ABA necessary to arrest development of the mutant. This observation suggests the existence of a quantitative relationship between ABA concentration and some as yet unidentified signal transducers. Because ABA has been reported to activate MAPKs, we examined the relationship between ABA concentration and MAPK activation in wild-type and hyl1 mutant seedlings. We detected the ABA concentration-dependent activation of both a 42-kDa and a 46-kDa MAPK and showed that both activated kinases are detected at higher levels in hyl1 mutant than in wild-type seedlings over the entire concentration range from 0.1 to 100 μM ABA. We further showed that two ABA-activated protein kinases of essentially the same electrophoretic mobilities as the ABA-activated MAPKs phosphorylate recombinant ABI5 and that both ABI5 kinases are present at higher levels in hyl1 mutant than in wild-type seedlings. We showed that the AtMPK3 gene, as well as the ANP1 gene encoding the MAPKKK of the stress-activated kinase cascade, are overexpressed in the hyl1 mutant. We presented evidence that the major 42-kDa MAPK is AtMPK3 and that it is activated by ABA in vivo.
Taken together, these observations suggest that the ABA hypersensitivity of the hyl1 mutant is attributable to the elevated expression of stress MAPK signaling components. This, in turn, bears the implication that the ABA signal is transduced through the stress MAPK cascade to ABI5 (and possibly other transcription factors, such as ABI3) to arrest seedling growth. However, because the hyl1 mutation is both physiologically pleiotropic and causes misexpression of a number of genes other than those encoding stress MAPK cascade components ANP1 and AtMPK3, we sought direct evidence that MAPK signaling is involved in transmitting the ABA signal to arrest development of germinating seedlings.
To determine whether there is a causal connection between the elevated expression of the AtMPK3 gene and hypersensitivity to growth arrest by ABA, we asked whether overexpression of an AtMPK3 cDNA in an otherwise wild-type plant would increase seedling sensitivity to growth arrest by ABA. Although we expressed the AtMPK3 cDNA from the strong 35S promoter, we did not recover plants with very high levels of AtMPK3 transcripts or MAPK activity. Nonetheless, plants showing even a modest 2-fold overexpression of AtMPK3 mRNA produced seeds that were more sensitive to postgermination growth arrest by ABA than were wild-type seeds, suggesting that even a small increase in endogenous MAPK activity affects ABA sensitivity. A similar inference can be drawn from the ABA concentration curves for MAPK activation (Fig. 2). These observations lead us to conclude that the ABA signal that triggers postgermination growth arrest can be transmitted through the stress kinase cascade of which the AtMPK3 is a component.
However, these experiments do not address the importance of the MAPK cascade route in ABA signal transmission to trigger growth arrest. If MAPK cascade signaling is either the obligatory or the primary route, then disrupting MAPK cascade signaling should make seedlings insensitive to growth arrest by ABA. Moreover, it should rescue the hyl1 mutant, allowing it to germinate and grow on ABA. By contrast, if ABA signaling is degenerate in the sense of using alternative routes that do and do not include a MAPK cascade with equal efficiency, then disruption of MAPK signaling should have little or no effect on ABA-triggered growth arrest. These predictions were tested by plating wild-type and mutant seeds on a medium containing the MAPKK inhibitor PD98059. PD98059 both decreased the sensitivity of wild-type seeds to ABA and rescued the ABA hypersensitive phenotype of the hyl1 mutant, though both wild-type and mutant seedlings grow somewhat more slowly in the presence of both ABA and the inhibitor than in the presence of just the inhibitor. Nonetheless, the presence of MAPKK inhibitor permitted both wild-type and mutant seedlings to develop on ABA concentrations which fully arrest their development in the absence of the inhibitor. We conclude that MAPK cascade components are the predominant, if not the exclusive, transmission route for the ABA signal that triggers postgermination growth arrest.
Although the ABI5 transcription factor is likely to be an important target of the ABA signal, its activation mechanism is not well understood. ABA signaling stabilizes the ABI5 protein and activates the gene (9). We showed here that expression of an ABI5 cDNA transgene elevates expression of the endogenous gene, suggesting positive autoregulation. However, overexpression of the protein does not itself lead to growth arrest, although it does render seedlings hypersensitive to ABA (9). The ABI5 protein is phosphorylated (9) and we showed here that recombinant ABI5 protein is a substrate for two ABA-activated protein kinases, that both ABI5 kinases are present at higher levels in the hyl1 mutant than in wild-type seedlings, and that the two kinases have the same electrophoretic mobilities as the two ABA-activated MAP kinases present in seedlings. However, it is premature to conclude either that ABI5 is the direct MAPK substrate or that MAPK signaling is sufficient for ABI5-mediated growth arrest.
Taken together, the results of the present experiments suggest that the ability of ABA to inhibit postgermination growth arrest depends on the relative concentrations or activities of several factors, including ABA, activated MAPKs, and relevant transcription factors such as ABI5. Although the dominant negative ABI1 and ABI2 mutations in homologous 2C protein phosphatase (PP2C) genes decrease ABA sensitivity, recently isolated null mutations in these PP2C genes are ABA hypersensitive (28). There is evidence that PP2Cs are involved in the negative regulation of ABA response (29, 30) and that at least one PP2C gene is expressed at an unusually low level in each of two recently described ABA-hypersensitive mutants designated abh1 and sad1 (31, 32). This finding implies that a reduction in protein phosphatase activity can increase the sensitivity of seedlings to ABA-triggered growth arrest, an observation that is consistent with our evidence that increased MAPK activity enhances ABA sensitivity. Much remains to be discovered about both the ABA signaling pathway and the mechanism of postgermination growth arrest, including identification of the proteins that detect the ABA signal and and transmit it to the ABA-activated kinase cascade, as well as those that cause growth arrest.
Acknowledgments
We thank S. Assmann, L. Lopez-Molina, and R. Mahalingam for critical comments on the manuscript. We also thank Luis Lopez-Molina for performing the Western blot experiment shown in Fig. 1A. This work was supported by National Science Foundation Grant IBN-0091650. A.G.-G. was partially supported by Consejo Nacional de Ciencia y Technología (Mexico) Postdoctoral Fellowship 141238.
Abbreviations
ABA, abscisic acid
MAPK, mitogen-activated protein kinase
MAPKK, MAPK kinase
MBP, myelin basic protein
MS, Murashige and Skoog
ABI5, ABA insensitive5 mutant
HA, hemagglutinin
References
- 1.MacRobbie E. A. (1998) Philos. Trans. R. Soc. London B 353 1475-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assmann S. M. & Wang, X.-Q. (2001) Curr. Opin. Plant Biol. 4 421-428. [DOI] [PubMed] [Google Scholar]
- 3.Hetherington A. M. (2001) Cell 107 711-714. [DOI] [PubMed] [Google Scholar]
- 4.Bewley J. D. & Black, M., (1994) Seeds: Physiology of Development and Germination (Plenum, New York).
- 5.Rock C. D. & Quatrano, R. S. (1995) in Plant Hormones: Physiology, Biochemistry and Molecular Biology, ed. Davies, P. J. (Kluwer Academic, London), pp. 671–697.
- 6.Finkelstein R. R., Gampala, S. S. L. & Rock, C. D. (2002) Plant Cell 14 S15-S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingram J. & Bartels, D. (1996) Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 377-403. [DOI] [PubMed] [Google Scholar]
- 8.Koornneef M., Bentsink, L. & Hilhorst, H. (2002) Curr. Opin. Plant Biol. 5 33-36. [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Molina L., Mongrand, S. & Chua, N. H. (2001) Proc. Natl. Acad. Sci. USA 98 4782-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fedoroff N. V., (2002) Sci. STKE, http://stke.sciencemag.org/cgi/content/full/OC_sigtrans;2002/140/re10. [DOI] [PubMed]
- 11.Hirt H. (2000) Results Probl. Cell Differ. 27 1-9. [DOI] [PubMed] [Google Scholar]
- 12.Tena G., Asai, T., Chiu, W.-L. & Sheen, J. (2001) Curr. Opin. Plant Biol. 4 392-400. [DOI] [PubMed] [Google Scholar]
- 13.Lu C. & Fedoroff, N. (2000) Plant Cell 12 2351-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bechtold N. & Pelletier, G. (1998) Methods Mol. Biol. 82 259-266. [DOI] [PubMed] [Google Scholar]
- 15.Clough S. J. & Bent, A. F. (1998) Plant J. 16 735-743. [DOI] [PubMed] [Google Scholar]
- 16.McBride K. E. & Summerfelt, K. R. (1990) Plant Mol. Biol. 14 269-276. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y., Li, R. & Qi, M. (2000) Plant J. 22 543-551. [DOI] [PubMed] [Google Scholar]
- 18.Tsugeki R., Kochieva, E. Z. & Fedoroff, N. V. (1996) Plant J. 10 479-489. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S. & Klessig, D. F. (1997) Plant Cell 9 809-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knetsch M. L., Wang, M., Snaar-Jagalska, B. E. & Heimovaara-Dijkstra, S. (1996) Plant Cell 8 1061-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirt H. (2000) Proc. Natl. Acad. Sci. USA 97 2405-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ligterink W. & Hirt, H. (2001) Int. Rev. Cytol. 201 209-275. [DOI] [PubMed] [Google Scholar]
- 23.Ichimura K., Mizoguchi, T., Yoshida, R., Yuasa, T. & Shinozaki, K. (2000) Plant J. 24 655-665. [DOI] [PubMed] [Google Scholar]
- 24.Kovtun Y., Chiu, W. L., Tena, G. & Sheen, J. (2000) Proc. Natl. Acad. Sci. USA 97 2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuasa T., Ichimura, K., Mizoguchi, T. & Shinozaki, K. (2001) Plant Cell Physiol. 42 1012-1016. [DOI] [PubMed] [Google Scholar]
- 26.Asai T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W. L., Gomez-Gomez, L., Boller, T., Ausubel, F. M. & Sheen, J. (2002) Nature 415 977-983. [DOI] [PubMed] [Google Scholar]
- 27.Grant J. J., Yun, B. W. & Loake, G. J. (2000) Plant J. 24 569-582. [DOI] [PubMed] [Google Scholar]
- 28.Merlot S., Gosti, F., Guerrier, D., Vavasseur, A. & Giraudat, J. (2001) Plant J. 25 295-303. [DOI] [PubMed] [Google Scholar]
- 29.Sheen J. (1998) Proc. Natl. Acad. Sci. USA 95 975-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gosti F., Beaudoin, N., Serizet, C., Webb, A. A. R., Vartanian, N. & Giraudat, J. (1999) Plant Cell 11 1897-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hugouvieux V., Kwak, J. M. & Schroeder, J. I. (2001) Cell 106 477-487. [DOI] [PubMed] [Google Scholar]
- 32.Xiong L., Gong, Z., Rock, C. D., Subramanian, S., Guo, Y., Xu, W., Galbraith, D. & Zhu, J. K. (2001) Dev. Cell 1 771-781. [DOI] [PubMed] [Google Scholar]