Abstract
Biodirected epitaxial nanodeposition of polymers was achieved on a template with an oriented molecular surface. Acetobacter xylinum synthesized a ribbon of cellulose I microfibrils onto a fixed, nematic ordered substrate of glucan chains with unique surface characteristics. The substrate directed the orientation of the motion due to the inverse force of the secretion during biosynthesis, and the microfibrils were aligned along the orientation of the molecular template. Using real-time video analysis, the patterns and rates of deposition were elucidated. Field emission scanning electron microscopy revealed that a strong molecular interaction allowed for the deposition of nascent biosynthesized 3.5-nm cellulose microfibrils with inter-microfibrillar spacings of 7–8 nm on the surface of the template. The cellulose was deposited parallel to the molecular orientation of the template. Directed cellulose synthesis and ordered movement of cells were observed only by using a nematic ordered substrate made from cellulose, and not from ordered crystalline cellulose substrates or ordered cellulose-related synthetic polymers such as polyvinyl alcohol. This unique relationship between directed biosynthesis and the ordered fabrication from the nano to the micro scales could lead to new methodologies for the design of functional materials with desired nanostructures.
Interfacial surface structure and interaction of materials at the nanoscale have attracted much attention in the field of nanotechnology (1). Microbiological systems have been investigated as a microscale process (2); however, recent studies showing the unique interaction of biological systems with entirely synthetic molecular assemblies have prompted consideration of a new generation of approaches for controlled nanoassembly (3). We report on an interaction of a biological system with an artificially oriented molecular substrate. The preparation of such an ordered substrate recently was achieved by Kondo and coworkers (4, 5) by dissolving native cellulose and reprecipitating it in a unique manner to form a distinctive structure termed nematic ordered cellulose (NOC) (Fig. 1). This is a unique type of glucan chain association. The structure is highly ordered but not crystalline and therefore may have exclusive properties as a material product over the native form of crystalline cellulose. The unique surface of NOC can be used as a template for the construction of nanocomposites. The methods for producing NOC also have been applied to other biopolymers, e.g., chitin, xylan, their derivatives, and blends of these polymers, and these will be reported in future papers.
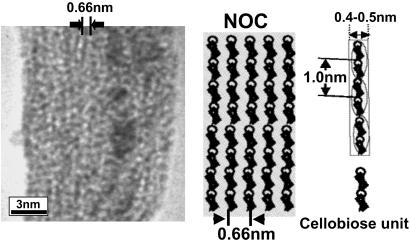
Fig 1.
(Left) High-resolution TEM of NOC, the molecular ordering template. The film was negatively stained by uranyl acetate and spans a region of the copper support grid. Note the individual glucan chains that are separated by an average distance of 0.66 nm (arrows). (Right) Schematic diagram of NOC showing the arrangement of glucan chains and the linear spacing of the cellobiose units (circled). The polymer chains lie on their narrow axes.
Native biopolymer assembly has been shown to be a complex process involving two separate, but integrated, steps of polymerization and crystallization (6). In particular, cellulose has been shown to be assembled by a macromolecular complex of enzymes located on the cell surface (7). Nature has designed an efficient system for regulating the molecular weight, crystallinity, size, and shape of the nanostructure of cellulose; however, this can be changed only by either genetic engineering (8) or conventional postharvesting methods of processing (9). Methods for direct intervention in the natural nanofabrication process are explored by the research reported herein, using the biosynthesis of cellulosic films by the Gram-negative bacterium Acetobacter xylinum. This organism secretes crystallized cellulose microfibrils directly into the culture medium from fixed enzyme complexes arranged in a row parallel to the long axis of the cell. The arrangement and position of the enzyme complexes determines the native hierarchical aggregation of the polymer chains into the final crystalline product, a ribbon of cellulose microfibrils. The average microfibril size is 3.5 nm but occasionally, highly ordered glucan chain aggregates more than 10 nm have been observed with lattice imaging (10).
Brown and coworkers (11) first described the movement of Acetobacter in relation to cellulose biosynthesis in 1976. They found a cell movement rate of 2.0 μm per min at 25°C to correspond with the actual synthesis and extrusion of cellulose microfibrils. This motion is due to an inverse force derived from secretion of crystalline cellulose microfibrils by the bacterium. When the bacterium moves in a certain direction, the secreted microfibrils are oriented in the same direction simultaneously. We have assumed that if the close interaction between the synthesized microfibrils and specific sites of the oriented molecules on the actual surface of NOC is very strong, the growth direction of the product would be controlled by the epitaxial deposition of the microfibrils. Therefore, in this study, we have synthesized cellulose microfibrils with Acetobacter cells positioned on a solid artificial and oriented substrate to demonstrate a close relationship between native synthesis of bacterial cellulose and its subsequent modification by an oriented substrate, including the real-time movements of the bacteria on the template. The goal of this study is to offer a novel concept to create altered nanostructures by using biological systems combined with templates having a specific molecular orientation.
Materials and Methods
Materials.
Bleached cotton linters with a degree of polymerization of 1,300 were used as the starting cellulose sample. The cellulose was first dried under vacuum at 40°C. N,N-dimethylacetamide purchased from Katayama Chemicals, Osaka (99+%) was dehydrated with molecular sieve 3A and used without further purification. Lithium chloride (LiCl) powder (Katayama Chemicals) was oven-dried at least for 3 days at 105°C. Methylcellulose with a degree of substitution of 1.6 and polyvinyl alcohol with a degree of polymerization of 2,000 were purchased from Shin-Etsu Chemicals (Tokyo) and Katayama Chemicals, respectively. Cellulose acetate with a degree of substitution of 2.45 (L-70) and purified chitin were provided by Daicel Chemicals (Tokyo) and Katakura Chikkarin (Tokyo), respectively.
Preparation of NOC Template in Schramm-Hestrin Medium.
NOC templates were prepared according to a previous method (5). Basically NOC was prepared by stretching a water-swollen cellulose film that had been coagulated from a dimethylacetamide/LiCl solution. Dissolution of cellulose and preparation of the template was accomplished by a previously described procedure (4) using a solvent exchange technique (12). Strips ≈30 mm long and 5 mm wide of the water-swollen gel-like films were stretched in a manual stretching device and elongated uniaxially to a draw ratio of 2.0 at room temperature. The entire drawing process was completed while the specimen was still in a wet state. The thickness of the dried films was ≈80 μm. Then the never-dried NOC template was put into Schramm-Hestrin medium (13) at pH 6.0, and the solvent was exchanged and finally maintained in a Petri dish until used for the study.
Other noncrystalline ordered substrates were prepared in the same manner from different polymers (Table 1). Oriented cellulose II and IV crystalline films with the same orientation as NOC were prepared by a mild mercerization at room temperature overnight or by heating in water, respectively. The dried NOC was mounted on the drawing device to be kept under a stressed state. The NOC in the stressed state was immersed in 7% aqueous NaOH solution and followed by water washing at room temperature. The treated NOC film was transformed into a cellulose II crystalline film. The cellulose IV film from NOC was crystallized at 140°C in an autoclave. The dried NOC under a stressed state in the same manner as for the cellulose II preparation was placed into a stainless steel tube (50 ml volume) with distilled water. Typically, the NOC film with ≈30 mm in length, 2 mm in width, and 0.15 mm in thickness fixed on the device was immersed into 30 ml of water in the tube. The tube was then heated in an oil bath at 140°C for 2 h. The treated NOC film was air-dried still in the stressed state overnight and subjected to a subsequent vacuum-drying process. Nematic ordered blend films of cellulose and its derivatives were prepared as follows: N,N-dimethylacetamide/LiCl was used as the common solvent for all samples to be mixed. Solution concentrations were 1 wt % for cellulose and 5 wt % for the cellulose derivatives. All solutions were filtered and stored in a closed container. Separately prepared solutions of the cellulose and its derivatives were mixed at room temperature in the desired proportions. The relative composition of the two polymers in the mixed solutions was 70:30, 50:50, and 30:70 by weight (cellulose/derivatives). After stirring for more than 3 days, the mixed solutions were used to prepare nematic ordered blend films by coagulation. The slow coagulation to prepare the gel-like film from an N,N-dimethylacetamide/LiCl solution was carried out according to a previous technique (4, 5). The solution was poured into a surface-cleaned glass Petri dish with a flat bottom and placed in a closed box containing saturated ethanol vapor at room temperature. In the blend solution, saturated ethanol vapor slowly diffusing into the solution was used to precipitate the cellulose/cellulose derivative blend gel, instead of water vapor for NOC. It was allowed to stand at room temperature for a few days until precipitation under a saturated ethanol vapor atmosphere was complete to obtain the gel-like film. The precipitated gel-like film (at this stage fixation of the film appeared to be complete) was washed thoroughly with ethanol to remove the solvent, and then the film was put in water to exchange the solvent to obtain a water-swollen transparent gel-like film. This water-swollen cellulose/derivative blended film was stretched in the same manner for NOC to be formed into nematic ordered blended films.
Table 1.
Qualitative evaluation of adhesion between substrates and biosynthesized cellulose microfibrils
| NOC | NOC precursor (gel) | Cellulose I crystal | Cellulose II crystal | Cellulose IV crystal | NOC acetate/cellulose blend | Nematic ordered methylcellulose/cellulose blend | Nematic ordered chitin | Nematic ordered chitin/cellulose blend | Oriented PVA |
|---|---|---|---|---|---|---|---|---|---|
| +++ | ++ | − | − | − | ++++ | ++++ | ++ | ++ | − |
−, No adhesive effect observed (normal spirals, no oriented synthesis); ++, mild adhesion effect (contact time reduced); +++, strong adhesion (maintains strong contact over long distances before jumping off track); ++++, very strong adhesion (exceptionally strong contact; no separations from the molecular track). PVA, polyvinyl alcohol.
Bacterial Cultures.
A. xylinum (NQ-5: ATCC 53582) was cultured in Schramm-Hestrin medium at pH 6.0 (13). After 3 days cultivation, the synthesized pellicle was removed, and the remaining medium solution was stirred by using a mixing instrument (Vortex) to obtain the active bacteria.
Transmission Electron Microscopy (TEM).
Specimens were prepared in the same manner as described (5). For sample specimens to be observed by high-resolution TEM, the cellulose solutions were also prepared in the same way as described above and diluted 100 times with water-free N,N-dimethylacetamide (1.0 × 10−2 wt %). After the solution covered a 3.0-mm TEM grid coated without any supporting film on a glass plate, the grid was left for more than 3 days at room temperature under a saturated water vapor. During this period, a cellulose gel-like film was precipitated and at the same time was naturally stretched across the 3.0-mm diameter of the TEM grid. It was then washed thoroughly with distilled water to exchange the solvent and finally air-dried. The sample on the grid was negatively stained with aqueous 2% uranyl acetate. Interaction with uranyl acetate appears to protect samples from electron beam damage. Specimens were observed with a Philips EM 420 transmission electron microscope operated at 100 kV with a beam current <5 μA. TEM images were acquired at instrumental magnifications of 210 and 450 K by using a Gatan (Pleasanton, CA) model 622 camera. The magnification factor for conversion to the digital image was ×23. The images were digitized, saved, and processed by IMAGE PRO PLUS 4.1 (Media Cybernetics, Silver Spring, MD).
Atomic Force Microscopy (AFM).
AFM images of the cellulose film specimens were acquired on a Nanoscope IIIa (Digital Instruments, Santa Barbara, CA) microscope as described (5). AFM was performed at room temperature, being controlled in contact (DC) mode with a scan rate from 1 to 3 Hz to observe 5 × 5 μm2 areas. The image was statistically analyzed with the cross-sectional images over 50 areas. In this observation, as height of the aggregates was smaller than radius of the tip, the following correction equation was used: E = 2 × (RH − H2)1/2, w = W − E, where E is the geometrical enhancement on the real width value (w), W is the apparent width observed in AFM, H is the height of the object observed in AFM, and R is the AFM tip radius (10 nm).
Real-Time Video Analysis.
Light microscopy images were acquired with ×20 or ×40 objective lenses, coupled with a ×1.6 optivar lens by using a Zeiss light microscope with an Optronix Laboratories (Orlando, FL) cooled charge-coupled device camera, attached with a ×0.55 camera lens (HR 055-CMT) from Diagnostic Instruments, Sterling Heights, MI. Frames were captured, digitized, saved, and processed with IMAGE PRO PLUS 4.1 (Media Cybernetics) at a rate that resulted in the acquisition of 1 frame/20 s (three frames per min). Samples observed were specimens of bacteria on substrates, covered with Schramm-Hestrin medium kept just above the surface at 24°C. The focus of the camera was set between the bacteria and the surface of the substrates, so that both bacteria and the synthesized cellulose ribbons could be observed. Incident light in the system was minimized to prevent drying of the surface by the heat produced. Using Adobe PREMIER 5.1, each captured frame was sequenced to produce a movie at the rate of 30 frames per sec (Movies 1 and 2, which are published as supporting information on the PNAS web site, www.pnas.org).
Field Emission Scanning Electron Microscopy (FE-SEM).
The sample specimens for FE-SEM were prepared according to the following: (i) the sample was rapidly frozen at a desired stage with liquid nitrogen and then transferred to glutaraldehyde/acetone (3% vol/vol) at −80°C. After 3 days at −80°C and a slow warming to room temperature, the specimen was exchanged to ethanol then dried by using a critical point drying device, mounted on a stab, and coated with Au for FE-SEM observation; or, (ii) the sample at a desired stage was placed into a container with several drops of a 4% OsO4/H2O solution, so that osmium vapor could fix the sample. The specimen was dehydrated with an ethanol series (30%, 50%, 70%, 90%, 100%), then dried and coated as described above.
The samples prepared as described above were observed with a Hitachi S-4500 FE-SEM at 5 or 8 kV. The SEM images were acquired as digitalized .tif files at 8-bit radiometric resolution. The images were processed (contrast enhancement, calibration of the scale, statistical size data collection, and cross section along the desired line) by IMAGE PRO PLUS 4.1 (Media Cybernetics).
Results and Discussion
The NOC Molecular Ordering Template.
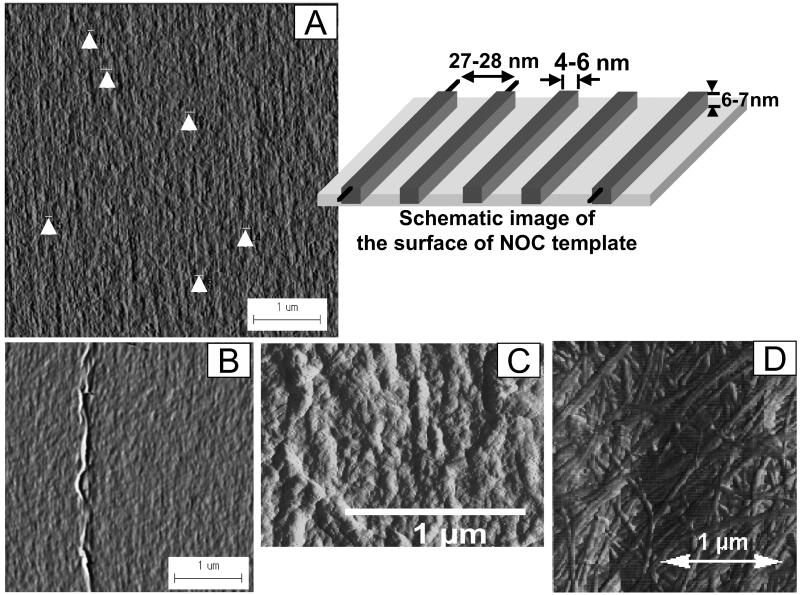
The template used in our studies was made from cellulose with molecular ordering as shown in Fig. 1. The high-resolution image obtained from the transmission electron microscope shows a preferentially oriented direction on the surface. These observations confirmed the mean width of a single glucan chain corresponding to its known dimensions as viewed from its narrow axis of the anhydroglucose ring. The average chain width was 0.462 nm (SD = ±0.0517 nm). Thus, the true width seems to be approximately 0.4–0.5 nm when taking into account the negative stain. The average distance between two parallel chains was 0.660 nm (SD = ±0.068 nm), which is wider than values for any crystalline cellulose. The AFM image analyses of the NOC surface without the negative stain demonstrate that well-aligned molecular aggregates with a width of 4–6 nm and a height of ≈6 nm in average are oriented uniaxially (see Fig. 6A). Without negative staining with uranyl acetate, it may be difficult to obtain a high-resolution AFM image of the NOC surface, particularly the glucan chain images shown in the TEM micrograph of Fig. 1.
Fig 6.
Atomic force micrographs showing the surface of the NOC template with a schematic representation (A), bacterial cellulose that continues to be oriented parallel to the molecular surface of the NOC template (B), the surface of the bacterial cellulose pellicle deposited epitaxially for 48 h (C), and the surface of normal bacterial cellulose pellicles (D). After 48 h of deposition shown in C, this ribbon is at least three layers removed from the template surface and still reflects the orientation of the underlying NOC template. The arrows indicate the areas examined statistically for the imaging analyses.
Bacterial Movements on Ordered and Nonordered Substrates.
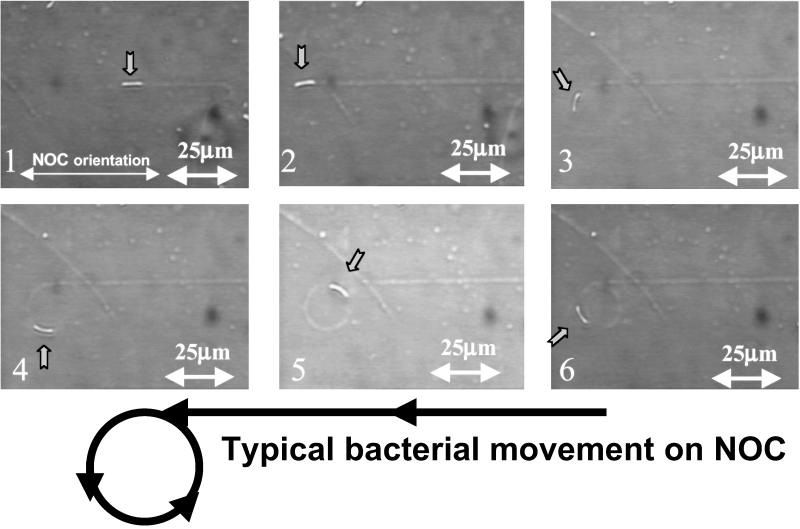
When active A. xylinum cells are transferred to the oriented surface, they synthesize cellulose ribbons that parallel the molecular orientation of the substrate. This is evidenced by direct video imaging of the motion of the bacteria as they synthesize the cellulose ribbon. The cell movement (at a constant rate of 4.5 μm/min at 24°C) is the result of an inverse force imposed by the directed polymerization and crystallization of the cellulose. This biodirected epitaxial nanodeposition can be studied over time and under varied conditions including different substrates. For example, in Fig. 2, a time-course analysis of an NOC template shows a bacterium moving in contact with a stretched NOC substrate. Midway through this series, the cell “jumps off” of the oriented substrate and continues to secrete its cellulose ribbon but now in the form of a spiral that is the normal pattern of formation when not in contact with any organized substrate (14). When the interaction between the bacterium and the surface of NOC is strong enough, the bacterium follows the track of the molecular template. As the polymer orientation of the NOC is not necessarily perfect, the bacterium may jump off the track at a structural defect (Fig. 2.3). After the bacterium leaves the track (Fig. 2.3–2.6), two forces, the inverse force of the secretion and the close interaction of adjacent microfibrils, affect the bacterium, possibly resulting in the synthesis of a spiral ribbon of microfibrils. In addition, the bacterium begins to rotate on its own axis. This rotation is the visible result of ribbon twisting that occurs to relieve strain induced by the absence of interaction with the substrate. When the ribbon is assembled in direct contact with the oriented molecular NOC substrate, ribbon twisting is prevented, thus suggesting a control of this oriented solid surface over the final physical interaction of polymer chains immediately after synthesis and during the early stages of crystallization.
Fig 2.
Successive images showing the motion of a bacterium as it secretes a cellulose ribbon with real-time video analysis. In 2.1, the bacterium is attached to and synthesizing its cellulose on the monomolecular rail track. In 2.2, the bacterium has jumped the track and is beginning to change its orientation. In 2.3–2.5, the bacterium is generating the first complete spiral. In 2.6, the bacterium is on the second rotation of a spiral.
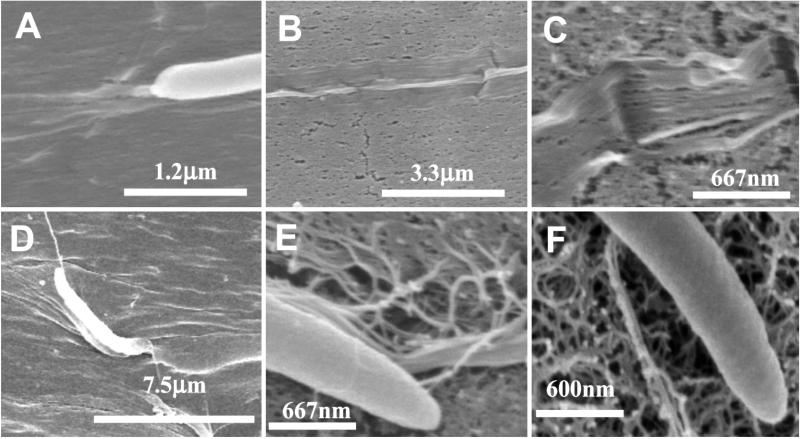
Ribbon interaction is more clearly shown by FE-SEM in Fig. 3. In this experiment, the motion of the same sample is observed by using light microscopy and then the sample is prepared for FE-SEM observations. In Fig. 2.1 and 2.2 the time-course sequence demonstrates an intense, directed nanodeposition that is exclusively linear and without cell rotation as the bacterium follows the track. This is confirmed by the ribbon structure shown by the FE-SEM images in Fig. 3 A–C that reveal no twisting and a more perfect alignment with the substrate. (See results from image analyses schematically shown in Fig. 5 Center.) The width of the NOC-altered cellulose ribbons is 500–700 nm with splayed microfibrils 7–8 nm apart. Likewise, when the bacterium “jumps off” the track, the cellulose ribbon is twisted. On the other hand, when Acetobacter is cultured on the surface of a nonstretched cellulose swollen gel substrate (precursor for NOC), it moves in a random manner including spirals (data not shown). An FE-SEM image on the nonoriented substrate is shown in Fig. 3E. The image suggests a random of microfibrils with the substrate surface. When Acetobacter cells are placed on an agar surface as a control, their interaction seems to be reduced, and twisted ribbons are always deposited, also in a random fashion (Fig. 3F). Table 1 summarizes the adhesion and orientation of Acetobacter cellulose with various substrates.
Fig 3.
FE-SEM images of the cellulose ribbon deposition process. (A–C) Examples of bacteria synthesizing cellulose ribbons on the oriented molecular track of NOC. (A) A bacterium shows a flat ribbon immediately behind its site of synthesis. (B and C) The tight association between the molecular track and the cellulose ribbon. (D) An example where the bacterium has just jumped off the track and follows a spiral path. (E) Random cellulose deposition on the surface of a nonstretched NOC precursor. (F) Random cellulose deposition on the surface of agar.
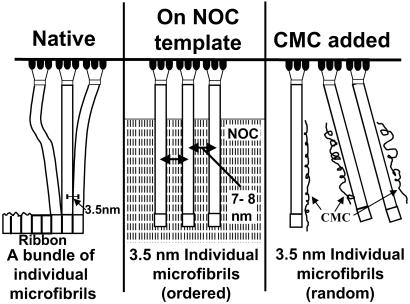
Fig 5.
Schematic diagram comparing three patterns of ribbon assembly. The enzyme sites from the bacterial surface are shown at the top, with aggregating glucan chains to form three microfibrils. Native condition (Left) occurs when the cell is freely suspended in the growth medium and produces a tight ribbon of microfibrils that are closely associated with each other. Ribbons produced in association with the NOC template track (Center) tightly adhere to the substrate and are not as tightly aggregated as in the native state. With the addition of CMC, individual microfibrils are immediately coated and prevented from aggregation (Right).
Concept of the “Molecular Track” and Its Function in the Control of Nanofabrication.
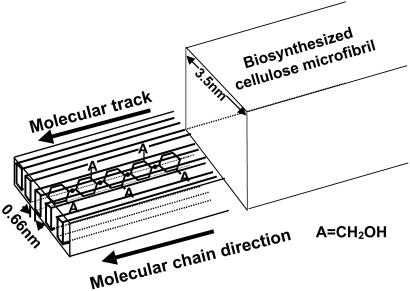
Depending on the O(6) rotational position with respect to the O(5) and C(4), the type of hydroxymethyl conformation at the C(6) position in NOC is assumed to be gauche-gauche (gg), which is typical for the C(6)positions in carbohydrates (5). As cellulose backbone is 21 screwed, the primary hydroxyl groups at the C(6) position and the secondary hydroxyl groups at the C(2) and C(3) positions appear alternately on the surface of NOC. The C(6) primary hydroxyl groups with gg conformation locate in the higher position of the NOC surface in the vertical axis when compared with the position of the C(2) and C(3) secondary hydroxyl groups of neighboring anhydroglucose units. Namely, in this gg conformation, the C(6) hydroxymethyl group is suggested to project from the oriented NOC surface with a lateral distance of 0.66 nm between chains for controlling movements of the bacterium as shown in Fig. 4. Thus, only the C(6) hydroxyl groups are hypothesized to be oriented as tracks on the top of the surface of NOC, whereas the ordered C(2) and C(3) hydroxyl groups do not appear there. Then the adhesion occurs between biosynthesized cellulose microfibrils and the C(6) primary hydroxyl groups. Thus, the important factor for controlling bacterial movements may be close contact and strong interaction between the C(6) hydroxyl groups of NOC and the newly synthesized cellulose microfibrils. A schematic figure explaining this concept is shown in Fig. 4. A cellulose microfibril would be synthesized along the molecular track. The width of the NOC-altered cellulose ribbons is 500 nm with splayed microfibrils 7–8 nm apart. When carboxymethylcellulose (CMC) is used as an additive to the bacterial culture medium, it prevents the aggregation of cellulose microfibrils from assembling into a normal ribbon and produces bundles 6–12 nm wide comprised of 3-nm fibrils (15). Thus, the same effect observed with CMC occurs for bacterial cellulose synthesis on the NOC surface, but in this case, it is a solid, oriented molecular surface.
Fig 4.
Schematic expression of molecular track concept. On the right is a cellulose microfibril aligned on a track of at least five oriented glucan chains of the NOC. Note that C6 projects above and below the plane of the substrate.
Epitaxial deposition.
The oriented C(6) hydroxyl groups allow a close contact with the glucan chains, as they are being secreted and crystallized. Thus, in theory, this interaction is predicted to alter the self-assembly of glucan chains into the crystalline microfibril (15–19). In fact, alteration of the aggregation of cellulose microfibrils on NOC has been confirmed in this work (Fig. 3 B and C). Just as Tinopal binds with cellulose and alters its crystallinity (16), CMC and other polysaccharides bind with the cellulose, and they alter its integration into the ribbon (15, 17–19). NOC has the same effect, but is a solid surface that interacts with the secreted cellulose microfibrils before they become associated to form the ribbons. This idea is summarized in Fig. 5. As described previously, such interactions with the substrate alter the production of the cellulose ribbon and also the motion of the bacterium and provides support for the epitaxial nature of cellulose deposition on NOC. In fact, both the FE-SEM image of Fig. 3B and the AFM image of Fig. 6B clearly demonstrate the epitaxial deposition of the cellulose ribbon on the NOC surface shown in Fig. 6A. Thus, nanoscale interactions induce microscale structural changes. Such interactions could be used on a larger scale to selectively modify biopolymers for many applications.
Hierarchical order.
The NOC imparts a molecular orientation and surface that perpetuates the hierarchical deposition process. This is demonstrated through the use of AFM 48 h after culture as shown in Fig. 6C when compared with the AFM image (Fig. 6D) of the surface of the dried pellicle formed by normal bacterial cellulose deposition. Here one can observe that additional layers secreted over time continue to mimic the original orientation initiated in the first deposited layer (e.g., the epitaxial deposition).
Critical factors.
The adhesion for epitaxial growth of microfibril deposition requires several different critical factors relating to molecular “order” of the substrate. This order is represented by three factors: (i) molecular chain orientation, (ii) specific polymer chain conformation, and (iii) O(6) rotational position with respect to the O(5) and C(4) in a β-glucan chain. The importance of molecular chain orientation is supported by the observation that unstretched NOC precursor does not cause epitaxial deposition. As already described, we consider that the C(6) hydroxymethyl conformation of NOC is also of importance. The role is like an anchor for molecular tracks to direct the epitaxial deposition of cellulose ribbons. The conformation of C(6) hydroxyl groups is believed to contribute to the crystalline forms depending on either gt (cellulose II) or tg (native cellulose). In this study, when the NOC with gg of hydroxymethyl conformation was crystallized, it was converted to ordered films containing cellulose II or cellulose IV crystalline forms with gt hydroxymethyl conformation. When Acetobacter is incubated on the crystallized NOC, there is no adhesion of bacterial cellulose ribbons. The crystallized films did not exhibit the molecular track effect to direct bacterial movements any longer. The same lack of adhesion was observed with substrates prepared from native crystalline cellulose I from Valonia (data not shown). In the crystalline samples, the specific polymer chain conformation is changed, and the rotational conformation at C(6) is gt in crystallized NOC and tg in Valonia cellulose (20–24). Considering the above, we conclude that the role of primary hydroxyl groups at the C(6) position is greater than those of hydroxyl groups at the C(2) and C(3) positions.
The Future.
In biological systems, skeletal materials such as cell walls, bones, and shells are made primarily of a nanoscale building block of polysaccharides, proteins, and inorganic salts. The assembly of these building blocks facilitates the production of a hierarchical framework structure. The formation dynamics observed in this study could be applicable to the design of nanoscale-controlled, hierarchically structured materials with specific properties. We have used a biological system combined with a polymer platform to directly fabricate hierarchically ordered materials from the nano level up to the μm level. Our proposed method is predicted to provide a unique type of nanotechnology by using biological systems with molecular nanotemplates to design molecular structures.
Supplementary Material
Acknowledgments
We thank Mr. Richard Santos at the University of Texas at Austin and Ms. Wakako Kasai at the Forestry and Forest Products Research Institute for their assistance with the experiments. This research was supported in part by a Joint Research Program between Japan and the United States through the Science and Technology Agency of Japan, the Welch Foundation (F-1217), and Johnson & Johnson Centennial Chair Funds.
Abbreviations
NOC, nematic ordered cellulose
TEM, transmission electron microscopy
AFM, atomic force microscopy
FE-SEM, field emission scanning electron microscopy
CMC, carboxymethylcellulose
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Drexler K. E., (1992) Nanosystems: Molecular Machinery, Manufacturing, and Computation (Wiley Interscience, New York).
- 2.Zhao X.-M., Whitesides, G. M., Qin, D., Xia, Y., Rogers, J. A. & Jackman, R. J. (1998) in Microsystem Technology in Chemistry and Life Sciences, eds. Manz, A. & Becker, H. (Springer, Berlin), Vol. 194, pp. 1–20. [Google Scholar]
- 3.Whaley S. R., English, D. S., Hu, E. L., Barbara, P. F. & Belcher, A. M. (2000) Nature 405, 665-668. [DOI] [PubMed] [Google Scholar]
- 4.Togawa E. & Kondo, T. (1999) J. Polym. Sci. 37, 451-459. [Google Scholar]
- 5.Kondo T., Togawa, E. & Brown, R. M., Jr. (2001) Biomacromolecules 2, 1324-1330. [DOI] [PubMed] [Google Scholar]
- 6.Cousins S. K. & Brown, R. M., Jr. (1997) Polymer 38, 903-912. [Google Scholar]
- 7.Brown R. M. (1996) Pure Appl. Chem. 10, 1345-1373. [Google Scholar]
- 8.Hu W.-J., Lung, J., Harding, S. A., Popko, J. L., Ralph, J., Stokke, D. D., Tsai, C.-J. & Chiang, V. L. (1999) Nat. Biotechnol. 17, 808-812. [DOI] [PubMed] [Google Scholar]
- 9.Kennedy J. F., Phillips, G. O. & Williams, P. A., (1989) Wood Processing and Utilization (Ellis Horwood, London).
- 10.Kuga S. & Brown, R. M., Jr. (1987) J. Electron Microsc. Technique 6, 349-356. [Google Scholar]
- 11.Brown R. M., Willison, J. H. M. & Richardson, C. L. (1976) Proc. Natl. Acad. Sci. USA 73, 4565-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chanzy H., Peguy, A., Chaunis, S. & Monzie, P. (1980) J. Polym. Sci. Polym. Phys. Ed. 18, 1137-1141. [Google Scholar]
- 13.Hestrin S. & Schramm, M. (1954) Biochem. J. 58, 345-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson N. S., Kaustinen, H. M., Carlson, J. A. & Uhlin, K. I. (1988) Int. J. Biol. Macromol. 10, 126-127. [Google Scholar]
- 15.Haigler C. H., White, A. R., Brown, R. M., Jr. & Cooper, K. M. (1982) J. Cell Biol. 94, 64-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benziman M., Haigler, C. H., Brown, R. M., Jr., White, A. R. & Cooper, K. M. (1980) Proc. Natl. Acad. Sci. USA 77, 6678-6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto H., Horii, F. & Hirai, A. (1996) Cellulose 3, 229-242. [Google Scholar]
- 18.Tokoh C., Takabe, K., Fujita, M. & Saiki, H. (1998) Cellulose 5, 249-261. [Google Scholar]
- 19.Haigler C. H., Brown, R. M., Jr. & Benziman, M. (1980) Science 210, 903-906. [DOI] [PubMed] [Google Scholar]
- 20.Dudley R. L., Fyfe, C. A., Stephenson, P. J., Deslandes, Y., Hamer, G. K. & Marchessault, R. H. (1983) J. Am. Chem. Soc. 105, 2469-2472. [Google Scholar]
- 21.Fyfe C. A., Stephenson, P. J., Veregin, R. P., Hamer, G. K. & Marchessault, R. H. (1984) J. Carbohydr. Chem. 3, 663-673. [Google Scholar]
- 22.Isogai A., Usuda, M., Kato, T., Uryu, T. & Atalla, R. H. (1989) Macromolecules 22, 3168-3172. [Google Scholar]
- 23.Horii F., Hirai, A., Kitamaru, R. & Sakurada, I. (1985) Cellulose Chem. Technol. 19, 513-523. [Google Scholar]
- 24.Langan P., Nishiyama, Y. & Chanzy, H. (1999) J. Am. Chem. Soc. 121, 9940-9946. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.