Abstract
Ndt80 is a transcriptional activator required for meiosis in the yeast Saccharomyces cerevisiae. Here, we report the crystal structure at 2.3 Å resolution of the DNA-binding domain of Ndt80 experimentally phased by using the anomalous and isomorphous signal from a single ordered Se atom per molecule of 272-aa residues. The structure reveals a single ≈32-kDa domain with a distinct fold comprising a β-sandwich core elaborated with seven additional β-sheets and three short α-helices. Inspired by the structure, we have performed a mutational analysis and defined a DNA-binding motif in this domain. The DNA-binding domain of Ndt80 is homologous to a number of proteins from higher eukaryotes, and the residues that we have shown are required for DNA binding by Ndt80 are highly conserved among this group of proteins. These results suggest that Ndt80 is the defining member of a previously uncharacterized family of transcription factors, including the human protein (C11orf9), which has been shown to be highly expressed in invasive or metastatic tumor cells.
Keywords: transcription factor, sporulation, meiotic, β-sandwich
Meiosis is a conserved process in eukaryotes in which haploid cells are generated from diploid cells. This process requires complex regulation, ensuring that the necessary genes are expressed at the correct times and levels. In Saccharomyces cerevisiae, diploid cells undergo meiosis and sporulation in response to nitrogen starvation and in the presence of a poor carbon source to form a tetrad of haploid cells packaged into spores (reviewed in ref. 1). The beginning of the sporulation pathway involves the exit of cells from the mitotic cell cycle. Premeiotic DNA replication then occurs followed by prophase in which the homologous chromosomes undergo meiotic recombination, pair, and form a synaptonemal complex. A key step in this process is the progression out of pachytene, the penultimate stage of prophase, and the initiation of meiotic divisions. Before the meiotic divisions, cells are not committed to meiosis and can return to mitotic growth under favorable nutritional conditions. However once the cells have initiated the meiotic divisions, they must complete the sporulation process (2).
Ndt80 is a transcriptional regulatory protein that plays a key role in activating expression of genes that are required for the meiotic divisions during the middle stages of sporulation (3). The Ndt80 protein binds specifically to an element called the middle sporulation element (MSE) present in the promoters of these genes (4–8). The pachytene checkpoint controls the activity of Ndt80 by preventing its accumulation and phosphorylation until meiotic recombination and formation of the synaptonemal complex have been completed, thus ensuring the production of viable meiotic products (8, 9).
The Ndt80 protein does not share sequence homology with any known DNA-binding proteins. We have determined the crystal structure and performed a mutational analysis of the DNA-binding domain of Ndt80 as a first step toward understanding how this molecule recognizes the MSE. Our studies reveal that the DNA-binding domain of Ndt80 has a distinct structure and DNA-binding motif. We have identified a number of homologous proteins from higher eukaryotes, including a human protein associated with cancer, which have significant sequence similarity to the DNA-binding domain of Ndt80. The conservation of residues involved in DNA binding by Ndt80 suggests that these homologous proteins also bind DNA and therefore defines a previously uncharacterized family of transcription factors.
Materials and Methods
Crystal Structure Determinations.
Cloning, expression, purification, and crystallization of the DNA-binding domain of Ndt80 have been reported elsewhere (10). Two data sets for the form I selenomethionyl substituted Ndt80 crystals were collected at the Se peak wavelength at the Advanced Photon Source Industrial Macromolecular Crystallography Association beamline 17-ID. An isomorphous native data set was collected at the National Synchrotron Light Source beamline X25. The Se sites (two sites, one Se per molecule) were identified by inspection of anomalous difference Patterson methods as well as by direct methods (11). Phases were calculated from the isomorphous and anomalous Se signal by using sharp (12) and then subjected to density modification by using solomon (13). A chain trace and initial model were built by using o (14). A second Se site in each molecule was identified in an anomalous difference Fourier map phased with the partial protein model and was useful for confirming the chain trace but was not useful for improving the experimental phasing. The crystallographic refinement was done by using all data to 2.8 Å resolution in cns (15).
The form II crystal structure was determined by molecular replacement implemented in amore (16). One molecule from the form I structure was used as the search model. This structure determination has been completed at 2.3 Å resolution following multiple rounds of manual rebuilding in o (14) and refinement by using cns (15). Difference Fourier maps (2Fo-Fc, Fo-Fc) and composite-omit maps have been used to evaluate the final model. The A molecule of the form II structure is the most complete and has been used for the analysis and figures reported here. This model includes 265 of the 272 residues in the molecule starting with residue 63 and ending with residue 327. Because of lack of side chain density, 12 residues have been modeled as Ala in this model. The remaining residues have well-ordered side chain density. The A and B molecules of form I and B molecule of form II include 249, 240, and 250 of 272 residues, respectively, in the current models.
Figures and Model.
Fig. 1C was generated by using molscript (17) and RASTER 3D (18). Fig. 2 C and D were made by using grasp (19). Positioning of Ndt80 interacting with idealized B-form DNA was done by using o (14) based on the constraints provided by the mutational analysis.
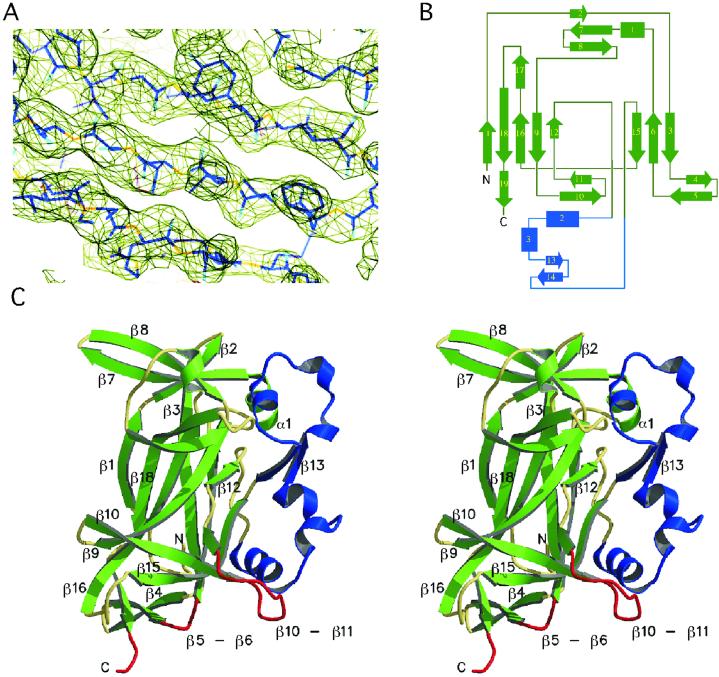
Fig 1.
The DNA-binding domain of Ndt80 has a previously uncharacterized structure. (A) A representative region of the experimental electron density map is shown contoured at 1.8 σ with the model in the form I lattice. (B) A topological diagram of the DNA-binding domain of Ndt80 reveals a distinct fold comprising 19 β-strands, three short α-helices, and two 310 helices. The central core is a β-sandwich comprising eight antiparallel β-strands. (C) The structure is shown here as a stereo ribbon rendering with the conserved β core in green. An additional structural feature (shown in blue), which we refer to as “florets,” contains two helix-loop regions held in place by two antiparallel β-strands. The proposed DNA-binding regions of the structure are highlighted in red.
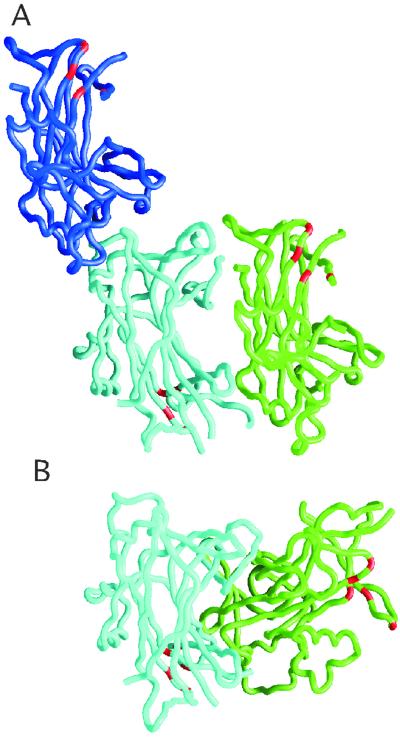
Fig 2.
Analysis of interacting molecules in the two different crystalline lattices. (A) Possible pairs of interacting molecules are shown for the form I lattice, one in a head-to-tail arrangement shown as backbone traces in green and cyan and the other in a head-to-head arrangement in blue and cyan. (B) The two molecules in the asymmetric unit of the form II lattice are packed in a head-to-tail arrangement shown in cyan and green. The cyan molecule is shown in a similar orientation to that of the cyan molecule in A. Although there are head-to-tail arrangements in both the form I and II lattices, the details of the interactions between the two molecules differ significantly as seen readily by comparing the relative positions of the green molecules in the two lattices. The residues involved in DNA binding are highlighted in red for the trace renderings of each molecule in both A and B.
DNA Binding and Sporulation Assays.
Point mutations were introduced into the NDT80-coding region of the bacterial and yeast expression vectors by using the QuikChange mutagenesis kit (Stratagene). Substituted proteins were expressed in BL21 Codon Plus cells (Invitrogen), lysed by sonication, and purified by Ni-nitrilotriacetic acid affinity chromatography by using the manufacturer's recommended procedures (Qiagen). Electrophoretic mobility-shift assays (EMSAs) were used to measure the binding affinity of the purified mutant proteins to the SMK1 MSE as described (20). In brief, the binding reactions included 10 mM Tris⋅HCl (pH 7.5)/40 mM NaCl/4 mM MgCl2/6% glycerol/10 mg/ml BSA/10 μg/ml sonicated salmon sperm DNA, 32P-labeled 29-mer duplex oligonucleotides ≈9 fmol/reaction, and Ndt80 samples (final concentrations 0.001–0.2 mg/ml) in 24-μl volumes at room temperature for 30 min. Samples were analyzed on 6% polyacrylamide gels. Complementation by Ndt80 mutants in a Δndt80/Δndt80 diploid strain was assayed by the ability of the transformed cells to undergo meiotic divisions as determined by the number of 4′,6-diamidino-2-phenylindole-stained foci in each cell (4). The formation of spore walls was monitored by the presence of ditryrosine as described (3).
Results
The DNA-binding domain of Ndt80 (residues 59–330) was defined by deletion analysis and limited proteolysis and subsequently shown to retain specific DNA-binding activity in EMSAs (10). Crystals of the DNA-binding domain were obtained in two distinct lattices, each with two molecules in the asymmetric unit (see Table 1). The structure was determined for the form I crystals by using the anomalous and isomorphous phasing from a single ordered Se atom per molecule of 272 residues (see Materials and Methods). The resulting experimental electron density map calculated at 2.8 Å resolution was of sufficient quality to build an initial chain trace and assign ≈70% of the amino acid residues (Fig. 1A). The structural model has been completed and refined to 2.8 Å resolution for the form I crystal. The form II crystal structure was determined by molecular replacement and refined to 2.3 Å resolution.
Table 1.
Data collection, phasing, and refinement statistics for Ndt80 (residues 59–330) crystal forms I and II
| Form II native | Form I native | Form I Se-Met 1 | Form I Se-Met 2 | |
|---|---|---|---|---|
| Data collection statistics | ||||
| Wavelength, Å | 1.54 | 1.10 | 0.979 | 0.979 |
| Resolution limit, Å | 2.3 | 2.8 | 2.8 | 2.8 |
| Space group | C2 | P3121 | P3121 | P3121 |
| Cell dimensions | ||||
| a, Å | 101.28 | 63.96 | 64.14 | 64.16 |
| b, Å | 64.90 | 63.96 | 64.14 | 64.16 |
| c, Å | 92.42 | 285.16 | 285.97 | 286.04 |
| β, ° | 107.52 | — | — | — |
| Rsym | 0.084 (0.241) | 0.067 (0.209) | 0.077 (0.250) | 0.075 (0.153) |
| I/σ | 15.8 (3.40) | 18.6 (6.80) | 26.3 (7.90) | 27.4 (10.6) |
| Completeness, % | 95.7 (84.9) | 98.7 (99.9) | 96.1 (92.3) | 92.3 (92.7) |
| SIRAS phasing statistics | ||||
| Phasing power iso | — | — | 0.505 | 0.666 |
| Phasing power ano | — | — | 1.77 | 1.72 |
| Refinement statistics | ||||
| Rwork, % | 22.6 | 23.2 | ||
| Rfree, % | 27.1 | 29.0 | ||
| rms deviation | ||||
| Bond length, Å | 0.006 | 0.008 | ||
| Bond angles, ° | 1.37 | 1.43 | ||
| B-factorave | 35.3 (A) | 36.7 (A) | ||
| 53.3 (B) | 42.5 (B) |
Rsym = {∑|(I − 〈I〉)|}/{∑(I)}, where 〈I〉 refers to the average intensity of multiple measurements of the same reflection. Phasing power = {[|Fh(calc)|/phase-integrated lack of closure]}. Rwork and Rfree = (∑|Fobs − Fcalc|)/(∑|Fobs|) where Rfree was calculated over 5% of the amplitudes not used in refinement.
Values for the highest resolution shell are enclosed in parentheses, 2.90–2.80 Å for form II and 2.38–2.30 Å for form I.
Values given are based on acentric reflections. (A) and (B) refer to the A or B molecule within the asymmetric unit of each lattice. In the final Ramachandran plot, 81.7% of residues are in the most favored region with an additional 17.2% in the additionally favored region for the 2.3 Å resolution form I structure. For the form II structure, the values are 80% and 19.1%, respectively. SIRAS, single isomorphous replacement with anomalous scattering.
The DNA-binding domain (Fig. 1 B and C) comprises a single structural domain. The core of the domain is a β-sandwich with two antiparallel β sheets (β1β18β16β9β12 and β15β6β3) that is similar in topology to the N-terminal domain of NFκB (21, 22), which was found to be the best match in a dali search (23) with an rms deviation for 103 Cα atoms of 3.1 Å. The second best match in the dali search was with β-galactosidase resulting in an rms deviation of 3.0 Å for 100 Cα atoms. Surprisingly, our structure includes only three short α-helical segments. Beyond the β-sandwich core, the DNA-binding domain of Ndt80 has no structural similarity to any previously reported protein based on a dali search. It is significantly larger than β-sandwich domains found in other proteins (which are ≈100 residues) and has a distinct structure shown in Fig. 1C. The structure includes nine β-sheets in total, eight antiparallel and one parallel (β5β19) with several strands participating in two different sheets. The antiparallel sheets in addition to those comprising the core include β7β2β3, β2β7β8, β10β11β15, β13β14, β17β18, and β4β5. There are two, three-stranded antiparallel sheets positioned on either end of the β-sandwich core (β10β11β15 and β2β7β8). The strands in these two sheets are approximately perpendicular to the direction of those found in the β-sandwich. The N-terminal part of β5 forms an antiparallel β-sheet with β4 and the C-terminal part forms a parallel β-sheet with β19. A unique structural feature that we refer to as the “florets” comprises a continuous polypeptide chain with two short α-helices, helical turns, and loop regions held in place by an antiparallel β-sheet β13β14 (Fig. 1).
There are two molecules in the asymmetric unit of each crystal lattice for which α-carbon atoms of the secondary structural elements superimpose with an rms deviation of 1.2 Å in the form I lattice and 1.0 Å in the form II lattice. The structural model is complete for the A molecule of the form II lattice and has been used for the structural analysis reported here. Several of the loops are partially disordered in the other molecules found in the form I and II structures (see Materials and Methods). The molecule has approximate dimensions of 55 Å × 40 Å × 30 Å.
We suggest that a monomer of the DNA-binding domain of Ndt80 binds to the MSE based on the following results. Despite the fact that there are two molecules in the asymmetric unit in each of the two lattices as shown in Fig. 2, the protein–protein interactions are distinct in the two crystal lattices and are not consistent with a biological dimer. In the form I lattice, there are two pairs of plausible interacting molecules, one in a head-to-head arrangement and the other in a head-to-tail arrangement. In the form II lattice, the two most closely associated molecules are packed in a head-to-tail arrangement. In the head-to-tail pairs of molecules in both crystalline lattices, the DNA-binding regions are located on opposite faces. Although there are head-to-tail arrangements of molecules in both the form I and form II lattices, the details of these interactions are substantially different as readily seen in Fig. 2. Neither of the head-to-tail packing arrangements suggests a mechanism for interactions of a dimer with DNA. In addition, none of the pairs of interacting molecules makes a particularly compelling dimer interface that would be predicted to be stable in solution.
The DNA-binding domain of Ndt80 and its complex with an MSE-containing oligonucleotide have been characterized by size exclusion gel chromatography. Based on calibration of the column, the DNA-binding domain elutes as a monomer with a molecular mass of 35 kDa (the predicted molecular mass is 32 kDa). The complex of Ndt80 and the MSE DNA elutes as a single peak with a molecular mass of 55 kDa, consistent with a 1:1 complex of the protein with the 22-mer DNA duplex (data not shown). In addition, Ndt80 binds to a 9-bp consensus MSE, which lacks dyad symmetry (4, 10). EMSA analysis of a mixture of two Ndt80 fragments, which differ in length and result in Ndt80-DNA complexes that migrate with different mobilities after electrophoresis, does not result in a heterodimer complex with intermediate mobility (M.P., unpublished results). These results are consistent with a monomer of this domain of Ndt80 binding DNA.
The structure of the DNA-binding domain has provided the basis for mutational analysis designed to define the DNA-interacting surface of the molecule. We have used in vitro DNA-binding assays as well as in vivo meiosis and sporulation assays to identify functionally important residues in Ndt80 (Fig. 3). We mutated several conserved surface residues and found that R111A and R177A substitutions in Ndt80 decreased DNA-binding activity 19-fold and 27-fold, respectively, as compared to the WT protein. Substitution of proximal residues K214A/K215A, R202A, and R208A did not affect DNA-binding activity significantly nor did substitution of residues R73, H82, H226, and K264/K265 found on distant surfaces of the protein (Fig. 3). We have also tested the ability of the Ndt80 mutants to complement a Δndt80/Δndt80 diploid strain (Fig. 3C; ref. 10). The R111A and R177A mutants, which had significantly reduced DNA-binding activity in vitro, failed to undergo meiotic divisions or sporulate in vivo. We conclude that these residues are functionally important.
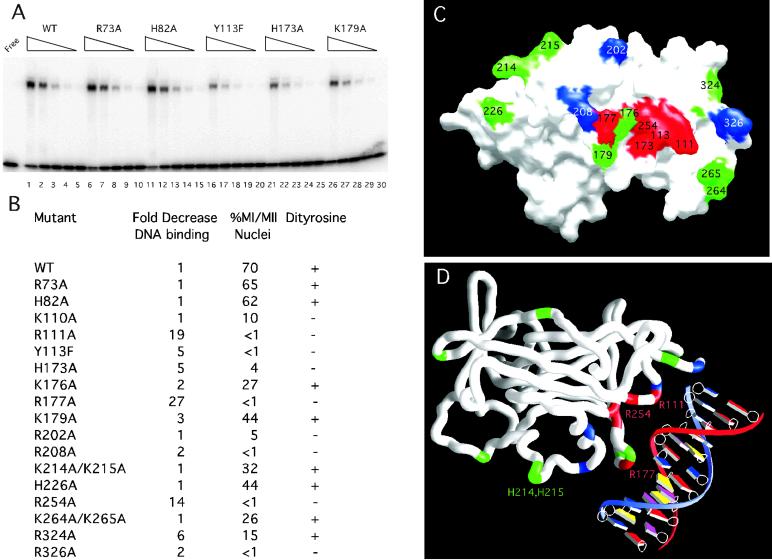
Fig 3.
Mutational analysis and modeling of Ndt80–DNA interactions. (A) The DNA-binding activity is shown for the WT and mutant Ndt80 proteins. WT (lanes 1–5), R73A (lanes 6–10), H82A (lanes 11–15), Y113F (lanes 16–20), H173A (lanes 21–25), and K179A (lanes 26–30) Ndt80 proteins were purified and assayed by EMSA. Lanes 1, 6, 11, 16, 21, and 26 contain the same amount of purified protein. The wedges indicate decreasing concentrations of each protein corresponding to fivefold serial dilutions in successive lanes. (B) The relative decrease in DNA-binding activity analyzed by EMSA, progression through meiosis I and II (MI/MII), and spore-wall formation (Dityrosine) was compared for 18 mutant Ndt80 proteins and the WT protein. (C) A molecular surface rendering of the DNA-binding domain of Ndt80 is shown looking directly at the proposed DNA-interacting surface of the molecule. Residues shown in red result in at least a fivefold decrease in relative DNA-binding affinity and loss of the ability to sporulate in vivo when substituted with Ala or Phe in the case of Y113. Those in green have near WT DNA-binding activity and retain the ability to sporulate, and those shown in blue retain near WT DNA-binding activity but fail to sporulate in vivo. Note: 12 residues in the A molecule for the form II structure lack clear side chain density (including 176, 177, and 179) and have been modeled as Ala. (D) A model for Ndt80 bound to the MSE reveals a previously uncharacterized DNA-binding motif. A worm rendering of the protein is shown with the DNA. Residues R111, Y113, H173, R177, and R254 (red) are predicted to interact with the DNA based on the model and the mutational analysis. Residues in green are not predicted to interact significantly with the DNA. Those in blue may have a role distinct from DNA binding that is required for activation in vivo, i.e., they may interact with other proteins.
The mutational analysis suggests that residues in the β5–β6 loop and the β10–β11 loop are involved in DNA-binding and allowed us to model the interaction of Nd80 with DNA (Fig. 3D). Circular permutation DNA-bending experiments suggest that Ndt80 bends the DNA 40° on binding (I.F., unpublished results). As the exact nature of the DNA-bending is not defined by this assay (i.e., the position and direction of the bend), we have not attempted to model the bend. However, the shape complementarity of the model with the β10–β11 loop including R177 placed in the major groove of idealized B-form DNA and the β5–β6 loop including R111 in the minor groove is quite striking. The proposed interactions of Ndt80 in the major and minor grooves may serve to induce the observed bend in the DNA.
Based on the model of Ndt80 bound to DNA, we have introduced substitutions for a number of residues that are predicted to be within hydrogen-bonding distance of the DNA. Substituted Ndt80 proteins were purified from Escherichia coli for EMSA analysis resulting in yields and purity similar to the WT protein. All substituted Ndt80 proteins were compared directly by EMSA to the WT protein, which has a Kd of 1.9 × 10−7 M for the SMK1 MSE. Consistent with the model, the Y113F, H173A, and R254A substitutions found in β6, β10, and β15, respectively, (in addition to R111A and R177A) decreased DNA-binding affinity by at least fivefold (Fig. 3 B and C). These same mutants fail to sporulate in vivo. The positions of these residues relative to the DNA clearly define a DNA-interacting surface on the protein (Fig. 3D). Proposed interactions of Y113, H173, and R254 with the DNA involve the sugar-phosphate backbone that lies between the major and minor grooves. Our data also suggest that R324 and R326 may contribute to the DNA-binding activity of the protein. As defined by our structural and mutational analysis, the DNA-binding motif found in Ndt80 is distinct from those previously reported.
Residues that are within the DNA-binding region of the protein but that do not appear to have a direct role in interactions with the DNA are K110, K176, and K179. Substitution of Ala for K176 or K179 results in proteins that retain near WT affinity for the DNA and the ability to sporulate. K110A, R202A, and R208A mutants retain near WT DNA binding but do not sporulate. Of these, R202 and R208 are found in the floret region [present only in yeast and fungal members of this family of transcription factors (Fig. 4)] and may be involved in interactions with other proteins or domains required for transcriptional activation in vivo.
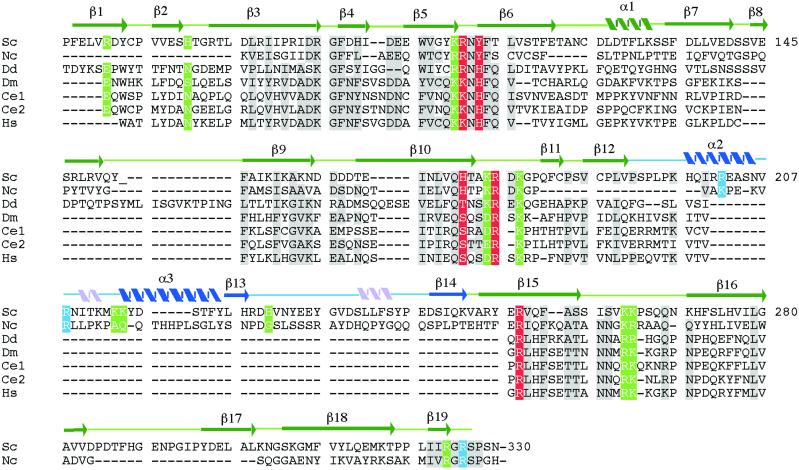
Fig 4.
Sequence alignment of the DNA-binding region of Ndt80 and homologs with assigned secondary structure. Sequence homologs were identified by using psi-blast (27) and aligned by using the program clustal w (28). The sequences are from Sc, S. cerevisiae (U35122); Nc, N. crassa (AL355929); Dd, D. discoideum (U66369.1); Dm, D. melanogaster (AE003463.1); Ce1, C. elegans (F59B10.1); Ce2, C. elegans (F21A10.2), and Hs, humans (KIAA0954/C11orf9). Secondary structural elements in the conserved core of the molecule are indicated in green. Those in blue are in the “florets.” Two 310 helices are indicated in pink. Residues shaded in gray are identical or have strong sequence similarity among the seven proteins. Residues shaded in red, green, and blue are the same as in Fig. 3.
Ndt80 is not homologous to any reported DNA-binding proteins, and sequence homology even within yeast species is limited to the DNA-binding domain. However, we have identified several sequence homologs from higher eukaryotes including proteins from Neurospora crassa, Dictyostelium discoideum, Caenorhabditis elegans, Drosophila melanogaster, and humans (Fig. 4). The sequence conservation (sequence identities of 17–23%, 31–42% conserved, for individual pairings with Ndt80) is sufficiently high to suggest that the proteins are in fact structurally and functionally related. Notably, of the residues that we have identified as interacting with the DNA, R177 and R254 are absolutely conserved and R111 is highly conserved (R or K) in all of the proteins. Y113 and H173 are conserved in the family members as H or S/T, respectively, maintaining the ability to form hydrogen bonds with the DNA. The sequence similarity of the residues in Ndt80 that we have shown are important for DNA binding suggests that these proteins may be members of a distinct family of transcription factors. Although these proteins are largely uncharacterized, available data suggest that the C. elegans homologs are developmentally regulated (24). In addition, expression of the human homolog (C11orf9) is strongly induced in an array analysis of several invasive or metastatic tumor cell lines (25), indicating that this protein may be of importance in a diagnostic capacity and potentially as a target molecule. Based on our analysis, we suggest that the human protein is a transcription factor and not a transmembrane protein as previously suggested (26).
Ndt80 plays a critical role in meiosis in yeast by activating expression of middle sporulation genes. The crystal structure of the DNA-binding domain of Ndt80 has provided a starting point for understanding the mechanism of DNA recognition and suggests that Ndt80 and its homologs contain a previously uncharacterized DNA-binding motif. In addition, the structure provides the basis for defining a new family of transcription factors, which are expected to have a conserved structure similar to that of the DNA-binding domain of Ndt80.
Acknowledgments
We thank Mike Becker, Lonnie Berman, and Bill Nolan from the National Synchrotron Light Source (NSLS) X25; Andy Howard and Jorge Rios from the Industrial Macromolecular Crystallography Association Collaborative Access Team (IMCA CAT); the members of the Georgiadis and Vershon groups for helpful discussions; Richard Leidich and Tom Weaver for maintenance of home source x-ray equipment; and Attilio Defalco and Greg Listner for systems support. Data were collected at beamline X25 at the NSLS, Brookhaven National Laboratory, which is supported by the U.S. Department of Energy, Division of Materials Sciences and Division of Chemical Sciences, and at beamline 17-ID in the facilities of the IMCA CAT at the Advanced Photon Source. The IMCA CAT facilities are supported by the companies of the Industrial Macromolecular Crystallography Center for Synchrotron Radiation Research and Instrumentation. This work was funded by grants from the National Institutes of Health (to M.M.G. and A.K.V.).
Abbreviations
MSE, middle sporulation element
EMSA, electrophoretic mobility-shift assay
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes and ).
References
- 1.Kupiec M., Byers, B., Esposito, R. & Mitchell, A. (1997) in The Molecular and Cellular Biology of the Yeast Saccharomyces, eds. Pringle, J., Broach, J. & Jones, E. (Cold Spring Harbor Lab. Press, Plainview, NY), Vol. 3, pp. 889–1036. [Google Scholar]
- 2.Honigberg S. M. & Esposito, R. E. (1994) Proc. Natl. Acad. Sci. USA 91, 6559-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu L., Ajimura, M., Padmore, R., Klein, C. & Kleckner, N. (1995) Mol. Cell. Biol. 15, 6572-6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu S. & Herskowitz, I. (1998) Mol. Cell 1, 685-696. [DOI] [PubMed] [Google Scholar]
- 5.Chu S., DeRisi, J., Eisen, M., Mulholland, J., Botstein, D., Brown, P. O. & Herskowitz, I. (1998) Science 282, 699-705. [DOI] [PubMed] [Google Scholar]
- 6.Hepworth S. R., Ebisuzaki, L. K. & Segall, J. (1995) Mol. Cell. Biol. 15, 3934-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozsarac N., Straffon, M. J., Dalton, H. E. & Dawes, I. W. (1997) Mol. Cell. Biol. 17, 1152-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hepworth S. R., Friesen, H. & Segall, J. (1998) Mol. Cell. Biol. 18, 5750-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tung K. S., Hong, E. J. & Roeder, G. S. (2000) Proc. Natl. Acad. Sci. USA 97, 12187-12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montano, S. P., Pierce, M., Coté, M. L., Vershon, A. K. & Georgiadis, M. M. (2002) Acta Crystallogr. D, in press. [DOI] [PubMed]
- 11.Sheldrick G. M. (1990) Acta Crystallogr. A 46, 467-473. [Google Scholar]
- 12.de La Fortelle E. & Bricogne, G. (1997) Methods Enzymol. 276, 472-494. [DOI] [PubMed] [Google Scholar]
- 13.Abrahams J. P. & Leslie, A. G. W. (1996) Acta Crystallogr. D 52, 30-42. [DOI] [PubMed] [Google Scholar]
- 14.Jones T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 15.Brunger A. T., Adams, P. A., Clore, G. M., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, N., Pannu, N. S., Read, R. J., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 16.Navaza J. (1994) Acta Crystallogr. A 50, 157-163. [Google Scholar]
- 17.Kraulis P. J. (1991) J. Appl. Cryst. 24, 946-950. [Google Scholar]
- 18.Merritt E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505-524. [DOI] [PubMed] [Google Scholar]
- 19.Nicholls A., Sharp, K. & Honig, B. (1991) Proteins Struct. Funct. Genet. 11, 281-296. [DOI] [PubMed] [Google Scholar]
- 20.Xie J., Pierce, M., Gailus-Durner, V., Wagner, M., Winter, E. & Vershon, A. K. (1999) EMBO J. 18, 6448-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller C. W., Rey, F. A., Sodeoka, M., Verdine, G. L. & Harrison, S. C. (1995) Nature 373, 311-317. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh G., Van Duyne, G., Ghosh, S. & Sigler, P. B. (1995) Nature 373, 303-310. [DOI] [PubMed] [Google Scholar]
- 23.Holm L. & Sander, C. (1993) J. Mol. Biol. 233, 123-138. [DOI] [PubMed] [Google Scholar]
- 24.Hill A. A., Hunter, C. P., Tsung, B. T., Tucker-Kellogg, G. & Brown, E. L. (2000) Science 290, 809-812. [DOI] [PubMed] [Google Scholar]
- 25.Kiemer A. K., Takeuchi, K. & Quinlan, M. P. (2001) Oncogene 20, 6679-6688. [DOI] [PubMed] [Google Scholar]
- 26.Stohr H., Marquardt, A., White, K. & Weber, B. H. F. (2000) Cytogenet. Cell Genet. 88, 211-216. [DOI] [PubMed] [Google Scholar]
- 27.Altschul S. F., Madden, T. L., Achaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]